Summary
Nuclear pore complexes (NPCs) are large macromolecular structures embedded in the nuclear envelope (NE), where they facilitate exchange of molecules between the cytoplasm and the nucleoplasm. In most cell types, NPCs are evenly distributed around the NE. However, the mechanisms dictating NPC distribution are largely unknown. Here, we used the model organism C. elegans to identify genes that affect NPC distribution during early embryonic divisions. We found that down-regulation of the Sm proteins, which are core components of the spliceosome, but not down-regulation of other splicing factors, led to clustering of NPCs. Down-regulation of Sm proteins also led to incomplete disassembly of NPCs during mitosis, but had no effect on lamina disassembly, suggesting that the defect in NPC disassembly was not due to a general defect in nuclear envelope breakdown. We further found that these mitotic NPC remnants persisted on an ER membrane that juxtaposes the mitotic spindle. At the end of mitosis, the remnant NPCs moved toward the chromatin and the reforming NE, where they ultimately clustered by forming membrane stacks perforated by NPCs. Our results suggest a novel, splicing-independent, role for Sm proteins in NPC disassembly, and point to a possible link between NPC disassembly in mitosis and NPC distribution in the subsequent interphase.
Keywords: Nuclear pore complex, Nuclear envelope breakdown, Sm proteins, early embryonic divisions
Introduction
The nuclear envelope (NE), which separates the nucleoplasm from the cytoplasm, consists of two distinct lipid bilayers, the inner and outer nuclear membranes. The inner nuclear membrane is associated with a meshwork of intermediate filaments and associated proteins that make up the nuclear lamina. The outer nuclear membrane is continuous with the endoplasmic reticulum (ER). Embedded in the NE, at sites where the outer and inner nuclear membranes fuse, are nuclear pore complexes (NPCs) that mediate all macromolecule transport between the nucleus and the cytoplasm (reviewed in (Hetzer, 2010)). NPCs are large protein complexes and their structure is conserved from yeast to mammals. The NPC has an eight-fold rotational symmetry; it is composed of a central core in the plane of the NE, a nuclear basket and cytoplasmic filaments (reviewed in (D’Angelo and Hetzer, 2008)). Although very large in size, the NPCs are composed of only 30 different proteins, referred to as nucleoporins (Nups), each present in multiple copies in the complex. The NPCs are modular, and most nucleoporins associate in biochemically stable subcomplexes (reviewed in (Brohawn et al., 2009; D’Angelo and Hetzer, 2008)).
In higher eukaryotes that undergo open mitosis, the nuclear envelope breaks down between prophase and metaphase, allowing the mitotic spindle to access the chromosomes. During NE breakdown (NEBD), the NPCs disassemble into their subcomplexes and the nuclear lamina de-polymerizes. The mechanisms that drive and coordinate NEBD are not fully understood, but this process involves the phosphorylation of multiple targets by the mitotic cyclin-Cdk complex (reviewed by (Burke and Ellenberg, 2002)). For example, the disassembly of NPCs, which coincides with an increase in NE permeability and is probably required to initiate NEBD (Lenart et al., 2003), is triggered by mitotic cyclin-Cdk dependent phosphorylation (reviewed in (Rabut et al., 2004)). The phosphorylation of at least one nucleoporin, Nup98, significantly increases the efficiency of NPC disassembly (Laurell et al., 2011). NPC disassembly is an ordered process: it starts with Nup98 dissociation, followed by the dissociation of Nup153, Nup214 and the Nup107-160 complex. The membrane nucleoporin Pom121 and the Nup62 complex dissociates only after the membrane is completely permeable (Dultz et al., 2008; Lenart et al., 2003). While the order of NPC disassembly has been described, the mechanisms that regulate this ordered process are largely unknown.
At the end of mitosis, the NE reassembles around the segregated chromosomes to form the two daughter nuclei. NPC assembly occurs during two cell cycle phases: at the end of anaphase, concomitantly with NE reassembly, and during interphase, when NPCs are inserted into an intact NE (Antonin et al., 2008). The mechanisms for NPC assembly at these two cell cycle stages are distinct. At the end of mitosis, NPC assembly starts with the recruitment of the Nup107-160 complex to chromatin, followed by the recruitment of ER membrane and the association of the membrane Nups Pom121 and Ndc1 with the forming pore. Next, Nup93 and Nup62 complexes associate concomitant with the formation of an import competent NPC. The last step in NPC assembly is the addition of peripheral Nups and the membrane-associated nucleoporin gp210 in early G1 (reviewed by (Antonin et al., 2008)). During interphase, early steps in NPC assembly require the integral membrane nucleoporin Pom121, the ER membrane bending proteins reticulons, the inner NE membrane protein Sun1, and the insertion of the Nup107-160 complex from both the cytoplasmic and nuclear sides of the NE ((Doucet et al., 2010; Dultz and Ellenberg, 2010; Funakoshi et al., 2011; Talamas and Hetzer, 2011) reviewed by (Doucet and Hetzer, 2010)). It should be noted, however, that a recent study by Lu et al suggested that even at the end of anaphase, NPCs are inserted into an intact NE rather than form on the chromatin (Lu et al., 2011). Nonetheless, what controls the sites of NPC assembly, the number of NPCs assembled in either mitosis or interphase, and the distribution of NPCs remains to be discovered.
In interphase nuclei, NPCs are uniformly distributed throughout the NE, but how this uniform distribution is achieved is not known. Defects in NPCs distribution were mainly studied in the yeast Saccharomyces cerevisiae, where it was found that defects in NPC assembly or stability, as well as defects in membrane fluidity and alteration of NE structure and dynamics, lead to abnormal localization of NPCs (Chadrin et al., 2010; Dawson et al., 2009; Doye and Hurt, 1997). However, there are several significant differences between NPC dynamics in yeast and higher eukaryotes: unlike in mammalian cells, in which the NPCs are largely immobile in the NE, the NPCs in yeast cells are mobile (Daigle et al., 2001; Winey et al., 1997). In addition, since yeast cells undergo a closed mitosis (e.g. without NEBD), NPCs are always assembled into an intact NE, in contrast to the post-mitotic assembly of NPCs in higher eukaryotes. NPC clustering was also observed in C. elegans embryos and animal cells following depletion of certain nucleoporins (for example (Cohen et al., 2003; Funakoshi et al., 2007; Galy et al., 2003)). Abnormal localization of NPCs was also described in apoptotic cells (Fahrenkrog, 2006; Reipert et al., 1996), but the processes that affect NPC distribution in proliferating higher eukaryotic cells have yet to be investigated. Here, we used the model organism C. elegans to identify genes that affect NPC distribution. Our observations uncover a novel role for Sm proteins in NPC dynamics, and lead to a possible link between NPC disassembly in mitosis and NPC distribution in the subsequent interphase.
Material and Methods
Worms Strains
All strains were derived from the C. elegans Bristol strain N2. OCF3: unc-119(ed3); jjIs1092[pNUT1:npp-1::gfp + unc-119(+)]; ltIs37 [pAA64: pie-1p::mCHERRY::his-58 + unc-119 (+)] (Golden et al., 2009); OCF17: unc-119(ed3); ocfIs3[pie-1 prom::GFP::npp-5::pie-1 3′UTR +unc119 (+)]; JH2184: unc-119(ed3); axIs1595 [pie-1 prom::GFP:npp-9::npp-9 3′UTR] (Voronina and Seydoux, 2010); AZ212: unc-119(ed3); [pie-1 prom::GFP::hisH2B::pie-1 3′UTR +unc119 (+)]; OCF22: unc-119(ed3); ocfIs5[pie-1 prom::mCherry::npp-1::pie-1 3′UTR +unc119 (+)]; OCF15: unc-119(ed3); ocfIs2[pie-1 prom::mCherry::sp12::pie-1 3′UTR +unc119 (+)]; OCF25(expressing mCherry::NPP-1,GFP::NPP-5): generated by crossing OCF22 with OCF17; OCF26(expressing mCherry::NPP-1,YFP::LMN-1): generated by crossing OCF4 (Golden et al., 2009) with OCF22 and backcrossing with N2; OCF28(expressing mCherry::NPP-1,GFP::TBA-2): generated by crossing OCF22 with OD57: unc-119(ed3); ltIs37 [pAA64: pie-1p::mCHERRY::his-58 + unc-119 (+)]; ltIs25 [pAZ132; pie-1p::GFP::tba-2 + unc-119 (+)] (McNally et al., 2006); OCF16(expressing GFP::NPP-1, mCherry::SP12): generated by crossing OCF15 with LW1094: unc-119(ed3); jjIs1094[pNUT1:npp-1::gfp + unc-119(+)]; MSN146(expressing CR::SP12, GFP::TBA-2): unc-119(ed3) III; ltIs76 [pAA178: pie-1p::mCHERRY::sp12 + unc-119 (+)]; ltIs25 [pAZ132; pie-1p::GFP::tba-2 + unc-119 (+)]. The nucleoporin-GFP and mCherry fusion proteins used in this study were not tested for rescuing the functionality of mutant alleles and therefore it is not known whether they are functional.
Generation of transgenic worms
Transgenes were constructed by Gateway cloning (Invitrogen, CA). Coding sequences of npp-1 and npp-5 were amplified from N2 genomic DNA by PCR using the primers attB1_npp1_L, attB2_npp1_L and attB1_npp5, attB2_npp5 (Table S1) and sp12 sequence was amplified by PCR using the primers attB1_SP12 and attB2_SP12 (Table S1). The PCR products were cloned into the entry vector pDONR221 and then into destination vectors pGateway mCherry (for npp-1 and sp12) and pID3.01 (for npp-5) to create N-terminal mCherry and GFP fusion respectively. Both pGateway mCherry and pID3.01 use pie-1 promoter and 3′UTR sequences. Transgenic lines were generated by microparticle bombardment method (Praitis et al., 2001).
RNAi experiments
RNAi was performed by feeding worms with bacteria expressing a control dsRNA against smd-1 (Golden et al., 2009) or dsRNA from the RNAi feeding library (Open Biosystems, Huntsville, AL). The identity of each clone was verified by sequencing. L4-staged larvae were grown on RNAi plates at 20°C (all strains except OCF26 and OCF28) or 24°C (OCF26 and OCF28) and embryos were examined after 30-48 hours of RNAi treatment. To determine embryonic lethality, 6 worms were transferred individually following RNAi treatment for 24 or 32 hours to 6 new RNAi plates and removed after additional 8 hours. Hatching was scored 24 hours later.
Immunofluorescence
Worms from RNAi plates were dissected on poly-L-lysine-treated slides and embryos’ eggshell was opened by freeze cracking on dry ice. Samples were fixed in methanol (−20°C) for 20 minutes, blocked for 30 minutes in PBS with 0.5% Tween 20 and 0.5% BSA (PBST/BSA) and incubated overnight at 4°C with primary antibodies diluted in PBST/BSA. mAb414 (Covance) was diluted at 1:400 and anti-Ce-gp210 Ab (Galy et al., 2008) at 1:500. Samples were washed in PBST/BSA, incubated for 2 hours at room temperature with the secondary antibodies Alexa Fluor 568 and Alexa Fluor 488 (Invitrogen) each at 1:1000 dilutions, washed as described above and mounted in Vectashield with DAPI (Vector Laboratories).
Microscopy
Confocal images were captured by spinning-disk confocal microscopy, using a Nikon Eclipse TE2000U microscope equipped with a 60× 1.4 NA Plan Apo objective. This system is outfitted with a Spectral Applied Research LMM5 laser merge module to control the output of four diode lasers (excitation at 405, 491, 561 and 655 nm), a Yokogawa CSU10 spinning-disk unit, and a Hamamatsu C9100-13 EM-CCD camera. Images were acquired using IPLab 4.0.8 software (BD Biosciences). Images were also captured using a Nikon E800 microscope, equipped with a 60× 1.4 NA Plan Apo objective, using a charge-coupled device camera (CCD; C4742-95; Hamamatsu Photonics) and operated by IPLab 3.9.5 software (BD Biosciences). Images were processed with Imaris 7.0.0 (Bitplane) and Adobe Photoshop CS.
Transmission Electron Microscopy (TEM)
GFP-H2B expressing control and snr-1 RNAi worms were dissected in M9 buffer and embryos were pulled into capillary tubes and imaged with Leica TCS SP2 confocal microscope. At the appropriate cell cycle stage, the capillary tubes were placed in 100 μm membrane carriers filled with 20% BSA in M9 buffer and frozen with the EMPACT2 high pressure freezing machine (Leica Microsystems, Vienna). Embryos were then freeze substituted in 0.1% uranyl acetate in acetone and embedded in Lowicryl, HM20 according to (Cohen et al., 2008). Serial sections were cut at 60 nm and placed on formvar coated grids. Grids were stained with 2% uranyl acetate and lead citrate and imaged on a CM120 Biotwin microscope.
RESULTS
Down-regulation of SNR-1, a core spliceosome component, leads to abnormal NPC distribution
We performed an RNAi screen in C. elegans to identify genes that affect nuclear morphology. We reasoned that alteration in nuclear morphology will likely cause defects in embryonic development, and therefore we focused our screen on 1870 genes that were identified as causing embryonic lethality according to Wormbase (http://www.wormbase.org/). Changes in nuclear morphology were monitored in embryos of a strain expressing the C. elegans homolog of the nuclear pore protein Nup54 (NPP-1, referred to here as CeNup54) fused to GFP (CeNup54::GFP) and histone H2B fused to the monomeric red fluorescent protein mCherry (H2B::CR) (Golden et al., 2009). In addition to the identification of genes that affected nuclear morphology (e.g. abnormal nuclear shape, clustered nuclei), the tagged CeNup54 allowed us to identify genes that affected NPC distribution in the NE. Overall, down-regulation of 350 genes by RNAi led to abnormal nuclear phenotypes. This included the down-regulation of 67 genes that were scored as causing abnormal distribution of NPCs and are the focus of this study (the remaining hits are currently being verified and will be described in a future study).
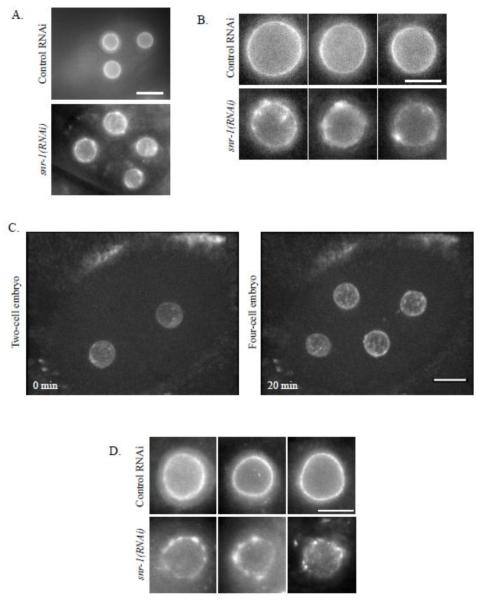
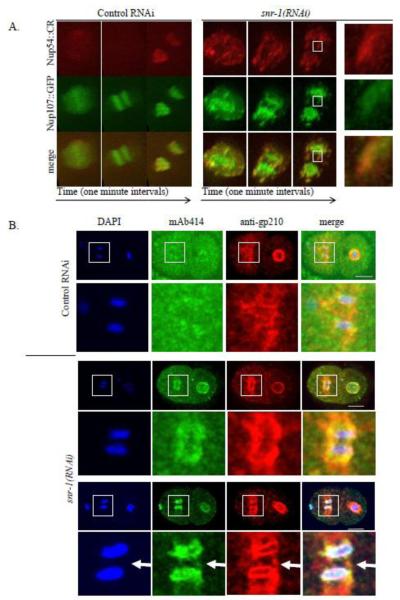
Normally, NPCs are distributed evenly around the nucleus ((Galy et al., 2003) and Figures 1A and 1B, top rows). In contrast, down-regulation of several genes from our RNAi screen led to uneven distribution and clusters of CeNup54::GFP (for example, see Figures 1A and 1B, bottom rows). One of the genes that had a prominent effect on CeNup54::GFP distribution was snr-1, an ortholog of human small ribonucleoprotein Sm D3 (Barbee et al., 2002) (Figure 1). The abnormal distribution phenotype of CeNup54 following down-regulation of SNR-1 was either absent or weak in the one cell embryo, clearly apparent in two-cell embryos, and even more pronounced in four-cell embryos and in later embryonic stages (Figure 1C, Supplemental Figure S1 and see Discussion). To confirm that the abnormal distribution of CeNup54 was not a result of the ectopic expression of the GFP fusion construct (which may or may not be fully functional), embryos from N2 worms treated with control RNAi or snr-1(RNAi) were immunostained with mAb414, an antibody against NPC proteins. The staining with mAb414, which in C. elegans was shown to recognize CeNup98/96 (NPP-10) (Galy et al., 2003), also showed NPC clustering following snr-1(RNAi) treatment, but not after control RNAi (Figure 1D). Therefore, this result also suggested that CeNup54 was not the only NPC protein whose localization is disrupted following SNR-1 down-regulation (see below).
Figure 1. snr-1(RNAi) leads to abnormal distribution of CeNup54.
A. Four-cell embryos from worms expressing CeNup54::GFP, treated with either control RNAi (top) or snr-1(RNAi) (bottom). Bar= 10 μm B. Individual nuclei from different embryos expressing CeNup54::GFP, treated as described in panel A. Bar= 3 μm C. Images from time-lapse experiment of an embryo expressing CeNup54::GFP, after treatment with snr-1(RNAi). Shown is the maximum intensity projection of confocal sections (acquired at 0.8 μm intervals). Bar=10μm. D. Nuclei from different embryos from N2 worms that were treated as described in panel A and stained using mAb414 antibodies. Bar= 3 μm.
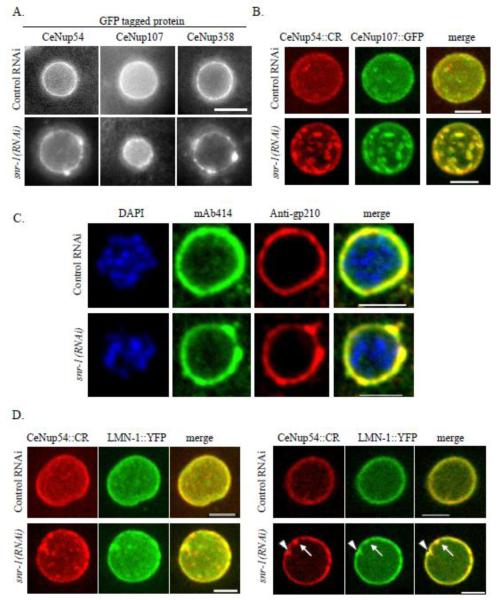
To examine the effect of SNR-1 down-regulation on the distribution of additional components of the NPC, worms expressing CeNup358 (NPP-9) fused to GFP (CeNup358::GFP) or CeNup107 (NPP-5) fused to GFP (CeNup107::GFP) were treated with control RNAi or snr-1(RNAi) and the distribution of these nucleoporins was examined in 2-4 cell stage embryos. In addition, N2 worms were treated with control RNAi or snr-1(RNAi) and immunostained with antibodies against gp210 (Galy et al., 2008). Nup54 is a member of the nuclear pore complex Nup62, which is part of NPC central channel, Nup358 is a component of NPC cytoplasmic filaments, Nup107 is a member of the Nup107-160 subcomplex, which is part of the NPC scaffold, and gp210 is one of the two known C. elegans transmembrane nucleoporins (reviewed in (D’Angelo and Hetzer, 2008)). snr-1(RNAi) led to abnormal distribution and clustering of all three nucleoporins (Figures 2A and 2C). Furthermore, in a strain expressing both CeNup54::CR and CeNup107::GFP, the two nucleoporins colocalized in snr-1(RNAi)-induced clusters (Figure 2B), and immunostaining with both anti-gp210 and mAb414 antibodies showed that gp210 also colocalized with clusters (Figure 2C). Interestingly, in embryos from a strain expressing the single C. elegans lamin fused to YFP (LMN-1::YFP) and CeNup54::CR, there appeared to be an enrichment of LMN-1::YFP at some but not all NPC clusters (Figure 2D).
Figure 2. snr-1(RNAi) leads to abnormal NPC distribution.
A. Nuclei of embryos from worms expressing CeNup54::GFP (left column), CeNup107::GFP (middle column), or CeNup358::GFP (right column) following treatment with control RNAi (top row) or snr-1(RNAi) (bottom row). Bar= 3 μm. B. Maximum intensity projection of confocal sections (acquired at 0.6 μm intervals) of typical nuclei from worms expressing CeNup54::CR (red) and CeNup107::GFP (green). C. Nuclei of embryos from N2 worms were treated with control RNAi (top row) or snr-1(RNAi) (bottom row) and subjected to immunofluorescence using mAb414 antibodies (green) and antibodies against gp210 (red). DNA is stained with DAPI (blue). D. Nuclei of embryos from worms expressing CeNup54::CR (red) and LMN-1::YFP (green) following treatment with control RNAi (top row) or snr-1(RNAi) (bottom row). Shown are the maximum intensity projection of confocal sections acquired at 0.5 μm intervals (left panel) and a single confocal section (right panel). Some (arrowhead) but not all (arrow) NPC clusters are enriched with LMN-1::YFP. Bars= 5μm.
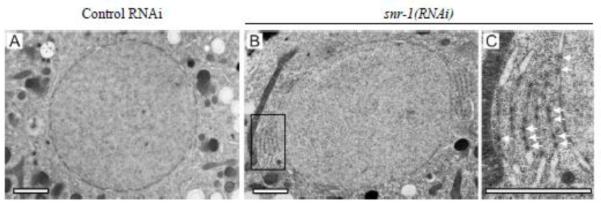
To further characterize these NPC clusters, two-cell stage embryos from worms that were treated with control RNAi or snr-1(RNAi) were analyzed by Transmission Electron Microscopy (TEM). As expected, in embryos from both control and snr-1(RNAi) treated worms, NPCs were properly embedded in the NE. However in embryos from snr-1(RNAi) worms, in addition to NPCs that localize to the NE, NPC-like structures were also found in stacks of double membranes, akin to annulate lamellae, that were adjacent to the NE (Figure 3B and 3C). The presence of clusters of NPCs on membranes outside the NE is also consistent with the lack of LMN-1::YFP enrichment at some NPC clusters (Figure 2D). Although it is possible that the structure of the clustered NPCs is altered in some way, the fact that these NPCs are embedded in a double membrane, and that every nup that we have tested is present in the clusters, suggest that the clusters in snr-1(RNAi) embryos are composed of intact, or nearly intact, NPCs rather than of nuclear pore subcomplexes or protein aggregates.
Figure 3. NPCs in snr-1(RNAi)-induced clusters localize to stacked membranes adjacent to the NE.
Two-cell stage control(RNAi) (A) and snr-1(RNAi) (B,C) embryos were imaged and fixed during interphase by high-pressure freezing. 60 nm thin sections were analyzed by TEM. snr-1(RNAi) embryos contained densely packed NPCs in their NE and in annulate lamellae, the large membrane stacks fenestrated by NPCs, that were closely associated with the NE. C is enlargement of the boxed area in B. The arrows point to some representative NPCs. Bars= 2 μm.
Down-regulation of some, but not all, splicing factors results in NPC clustering
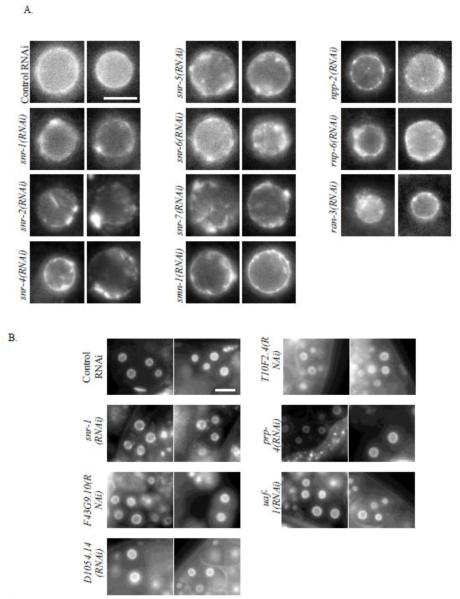
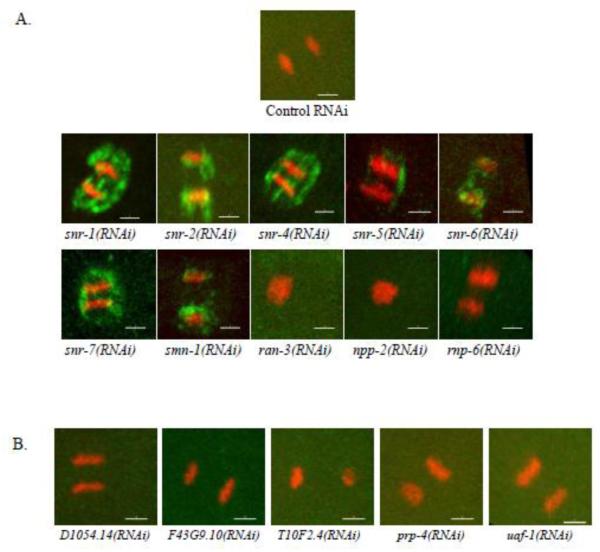
Unlike the NPC clustering phenotype seen following snr-1(RNAi), where nuclear morphology appeared otherwise normal, in the majority of genes whose down-regulation led to NPC clustering, the NPC clustering phenotype was accompanied by severe alterations to nuclear morphology (e.g. nuclear shape deformations). Since, in these cases, NPC clustering could have been a secondary consequence, for example, due to cell death, we decided to focus on those genes whose RNAi led to NPC clustering that was apparent in early (2 or 4 cells) embryos without any additional major nuclear morphology aberrations (Figure 4A). 10 such genes were identified in our screen. These included genes coding for 6 out of the 7 proteins that make up the Sm protein complex: snr-1 (Y116A8C.42), snr-2 (W08E3.1), snr-4 (C52E4.3), snr-5 (ZK652.1), snr-6 (Y49E10.15), and snr-7 (Y71F9B.4), which are orthologs of the mammalian SMD3, SMB, SMD2, SMF, SME and SMG, respectively. Interestingly, down-regulation of the seventh Sm protein, SNR-3, did not lead to NPC clustering, but did result in 100% embryonic lethality (data not shown). The screen also identified smn-1 (C41G7.1), npp-2 (T01G9.4), rnp-6 (Y47G6A.20), and ran-3 (C26D10.1) as genes whose down-regulation resulted in NPC clustering (Figure 4A). The Sm proteins are core components of the spliceosome (Barbee et al., 2002; Will and Luhrmann, 2001). Together with snRNAs, they form a core component of uridine-rich small nuclear ribo-nucleoproteins (snRNPs) that are essential for splicing. SMN-1 has been demonstrated to facilitate the assembly of Sm proteins into snRNPs (Kolb et al., 2007). Since the Sm proteins are core components of the spliceosome, their effect on NPC distribution could be either through their role in mRNA splicing, or through a splicing-independent function. If splicing defects were the cause of the NPC clustering phenotype, then down-regulation of other essential components of the spliceosome would also affect NPC distribution. However, in our initial screen, down-regulation of 19 essential splicing genes by RNAi resulted in embryonic lethality but had no effect on NPC distribution (see below and data not shown). To confirm this result, worms expressing CeNup54::GFP were treated with RNAi targeting each of five essential splicing factors, and the distribution of NPCs was reexamined in embryos after 48 hours of RNAi treatment (Figure 4B). For all five genes, 100% of embryos that were laid between 32 and 40 hours of RNAi treatment failed to hatch (Supplemental Figure S2), indicating a strong inhibition of the target genes by the respective RNAi. NPC distribution, however, was normal (Figure 4B). These results are consistent with the possibility that the Sm proteins affect NPC distribution through a novel mechanism, not related to their role in splicing.
Figure 4. Down regulation of some, but not all, splicing genes results in NPC clustering.
A. Nuclei from two different embryos expressing CeNup54::GFP, following treatment with the indicated RNAi. Bar= 3 μm. B. Two- and four-cell embryos from worms expressing CeNup54::GFP treated with RNAi against essential splicing factors as indicated. Bar= 10 μm.
Down-regulation of Sm proteins leads to defects in NPC disassembly during mitosis
To further characterize the NPC clusters formed following the down-regulation of Sm proteins by RNAi, we performed time-lapse microscopy of embryos from worms treated with control or snr-1(RNAi). In C. elegans early embryos, NPCs completely dissociate from the NE only after metaphase (Lee et al., 2000). In control embryos, CeNup54::GFP was completely dispersed during anaphase, whereas in snr-1(RNAi) treated worms, CeNup54::GFP remnants were visible between and around the anaphase chromosomes (Figure 5A). To quantify the occurrence of the phenotype, we monitored CeNup54::GFP through the first three embryonic divisions. Following treatment with control RNAi, CeNup54::GFP completely dispersed during mitosis in all 21 divisions that were imaged. In contrast, in embryos from snr-1(RNAi) treated worms, remnants of CeNup54::GFP were apparent in 16 out of 23 mitotic divisions (Figure 5B).
Figure 5. Down regulation of SNR-1 leads to incomplete dispersal of CeNup54 during mitosis.
A. Time-lapse microscopy of a two-cell embryo expressing CeNup54::GFP (green) and histone H2B::CR (red), after treatment with control RNAi (top row) or snr-1(RNAi) (bottom row). Time points are from the first embryonic metaphase. Shown is the maximum intensity projection of confocal sections acquired at 0.6 μm intervals. On the far right is an enlarged image of areas marked in the corresponding second time points. Bar= 10 μm. B. One-cell embryos, expressing CeNup45::GFP (green) and histone H2B::CR (red), from worms treated with control or snr-1(RNAi) were followed through up to three nuclear divisions, and the number of anaphases with remnants of CeNup54::GFP was scored. Shown are the first three divisions of an embryo from a control-treated worm. The number of mitoses with remnant NPCs, during each cell division, following control or snr-1(RNAi) is indicated below.
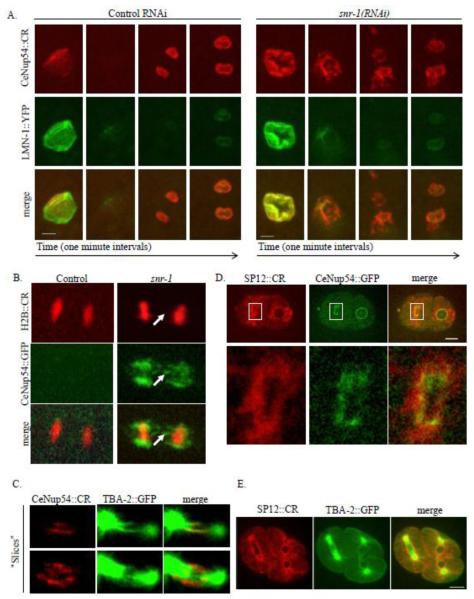
The failure to completely disperse following SNR-1 down-regulation could have been a property of CeNup54 alone, or it could have reflected a general defect in NPC disassembly during mitosis. To examine the behavior of other NPC components following snr-1(RNAi), we performed live cell imaging of worms expressing CeNup54::CR and CeNup107::GFP. It was previously shown that while Nup54 is completely dispersed into the cytoplasm during mitosis, a fraction of Nup107 relocalizes to the kinetochores (Belgareh et al., 2001). As expected, in both control and snr-1(RNAi) embryos, a fraction of CeNup107::GFP localized to the kinetochores (Figure 6A). However, in snr-1(RNAi) embryos, in addition to its kinetochore localization, CeNup107::GFP was still present around the anaphase chromosomes, and these abnormal remnants colocalized with CeNup54::CR remnants (Figure 6A).
Figure 6. Down regulation of SNR-1 leads to defects in NPC disassembly during mitosis.
A. Time-lapse images in one-minute intervals (from left to right) of embryos expressing CeNup54::CR (red) and CeNup107::GFP (green), from control worms (left panels) or worms treated with snr-1(RNAi)(right panels), as they undergo anaphase. Shown is the maximum intensity projection of confocal sections acquired at 0.4 μm intervals. Images on the far right are enlargements of the boxed areas in the last snr-1(RNAi) time point. B. Representative images of an indirect immunofluorescence experiment of embryos from N2 worms treated with control or snr-1(RNAi). The antibodies used were anti-gp210 (red) and mAb414 (green). DNA was stained with DAPI (blue). For each embryo, the bottom row is an enlarged image of the boxed area in the top row. The arrow points to remnant NPCs not associated with DNA. Shown is the maximum intensity projection of confocal sections acquired at 0.4 μm intervals. Bars= 10 μm.
To further investigate the nature of the nucleoporin remnants observed in snr-1(RNAi) embryos, we examined the localization pattern of gp210, a transmembrane nucleoporin. gp210 remains attached to the ER membrane throughout mitosis, after other nucleoporins have dispersed and can no longer be detected near the mitotic chromosomes (Galy et al., 2008; Yang et al., 1997). Embryos from N2 worms that were treated with control RNAi or snr-1(RNAi) were immunostained with both anti-gp210 antibodies and mAb414 antibodies. As expected, following control RNAi treatment, gp210, but not the mAb414 epitope, remained in the general vicinity of the segregating chromosomes (Figure 6B, top). In contrast, following snr-1(RNAi) treatment, gp210 colocalized with the mAb414 epitope in the mitotic remnants (Figure 6B, bottom), suggesting not only that the mitotic remnants are comprised of intact NPCs, but also that they are attached to a membrane. Thus, our results indicate that in snr-1(RNAi) embryos, a fraction of the NPCs does not disassemble during mitosis and remains attached to gp210, and thus to a membranous structure (see below).
Our results show that down-regulation of SNR-1 affects both NPCs distribution during interphase and NPC disassembly during mitosis. To determine whether other genes that affected NPC distribution during interphase (Figure 4A) also affect NPC disassembly during mitosis, CeNup54::GFP distribution was examined following treatment with RNAi to each of the ten genes that caused uneven NPC distribution during interphase (Figure 7A). Incomplete dispersal of CeNup54::GFP during anaphase was observed following treatment with RNAi to the six Sm genes, as well as to smn-1. In contrast, mitotic CeNup54::GFP dispersal appeared to be normal in embryos from ran-3, npp-2 or rnp-6(RNAi) treated worms. Depletion of five essential splicing factors also did not affect NPC disassembly during mitosis (Figure 7B). Taken together, these results suggest the involvement of the Sm core complex in a unique pathway that affects NPCs during both interphase and mitosis.
Figure 7. Down regulation of Sm proteins, but not other splicing genes, leads to defects in NPC disassembly during mitosis.
A. Embryos expressing CeNup54::GFP (green) and histone H2B::CR (red) from worms treated with the indicated RNAi’s (which caused NPC clustering in interphase, see Figure 4A). Images are from cells in anaphase. Shown is the maximum intensity projection of confocal sections acquired at 0.6 μm intervals. Note that there are chromosome segregation defects following RNAi of ran-3 and npp-2 (bottom row). Nonetheless, mitotic chromosomes from these embryos are not decorated with remnant CeNup54::GFP. B. Embryos were treated as in A, using RNAi to different essential splicing genes (see Figure 4B and Supplemental Figure S2). Bars= 3 μm.
Down-regulation of Sm proteins does not cause a general NEBD defect
The mechanisms underlying NEBD are poorly understood, but it is known that the disassembly of both NPCs and the nuclear lamina is triggered by mitotic cyclin/Cdk activity (Anderson and Hetzer, 2008; Laurell et al., 2011). Thus, the incomplete disassembly of NPCs observed in snr-1(RNAi) embryos could have reflected a more general defect in NEBD. We therefore examined the effect of snr-1(RNAi) on nuclear lamina disassembly in mitosis, by following the single C. elegans lamin protein, LMN-1. As reported previously (Lee et al., 2000), shortly after entering mitosis, (as evidenced by the “wrinkled” NE, Figure 8A, control, left column), both the nuclear lamina and the NPCs disperse and are no longer visible, with nucleoporins, such as CeNup54, dispersing slightly earlier than LMN-1 (Figure 8A, control, second column from the left). snr-1(RNAi) had no effect on LMN-1 disassembly during mitosis: LMN-1::YFP in both control and snr-1(RNAi) treated embryos disassemble by the time cells reached late anaphase (Figure 8A, third column from the left). This was in contrast to the distribution of CeNup54::GFP, which dispersed in the control embryos, but was present throughout mitosis in embryos from snr-1(RNAi) treated worms. These results suggest that SNR-1 does not have a general effect on NEBD, but is required specifically for NPC disassembly.
Figure 8. snr-1(RNAi)-induced NPC remnants localize to a membranous structure near the mitotic spindle.
A. Time-lapse images at one-minute intervals (from left to right) of embryos expressing CeNup54::CR (red) and LMN-1::YFP (green), from worms treated with control RNAi (left panels) or worms treated with snr-1(RNAi)(right panels), as they undergo mitosis. Shown is the maximum intensity projection of confocal sections acquired at 0.5 μm intervals. Bars= 4 μm. B. Anaphase in embryos expressing CeNup54::GFP (green) and histone H2B::CR, from worms treated with control or snr-1(RNAi). The arrow points to remnant CeNup54::GFP not associated with DNA. C. snr-1(RNAi)- induced NPC remnants (as detected by CeNup54::CR, in red) and the mitotic spindle (as detected by TBA-2::GFP, in green). Shown are two “slices” from images acquired using confocal sections at 0.5 μm intervals and obtained using oblique slicer in Imaris Software. D. snr-1(RNAi)- induced NPC remnants (as detected by CeNup54::GFP, in green) and the ER membrane (as detected by SP12::CR, in red). The bottom row is an enlarged image of the boxed area in the top row. Bar= 10μm. E. Presence of ER membranes (detected by SP12::CR in red) adjacent to the spindle (detected by GFP::TBA-2 in green). Bar=10 μm.
The NPC remnants caused by Sm protein down-regulation accumulate on ER membranes adjacent to the mitotic spindle
The presence of NPCs during anaphase, induced by the down-regulation of Sm proteins, led us to search for the structure to which the remnant NPCs could be attached. Subcomplexes of the NPC bind to kinetochores during mitosis (Belgareh et al., 2001) and the first step of post-mitotic NPC assembly is the binding of nucleoporins to the chromatin (Antonin et al., 2008). We therefore examined whether the mitotic NPC remnants co-localize with DNA. However, no DAPI staining or fluorescently tagged histone H2B (H2B::CR) could be detected with CeNup54::GFP remnants during mitosis (Figures 6B and 8B), suggesting that the remnant NPCs are not attached to chromatin.
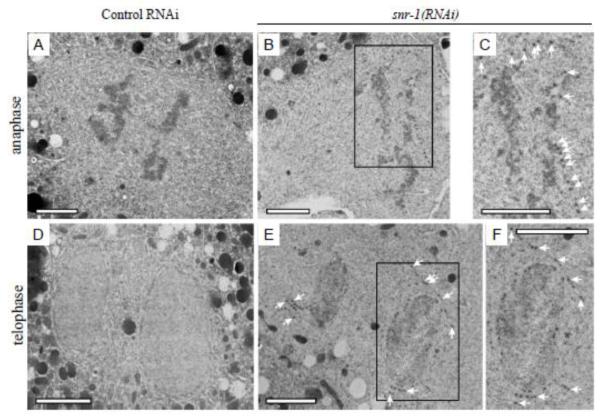
CeNup54::GFP remnants in early anaphase form a pattern of lines that are roughly perpendicular to the anaphase chromosomes (for example, see Figures 5A and 7A). This pattern is somewhat similar to that observed by tubulin, and suggested that a fraction of the NPC remnants could be associated with, or in the vicinity of, the mitotic spindle. We also found that the remnant NPCs contained the membrane nucleoporin gp210 (Figure 6B) suggesting that the remnants could be attached to a membranous structure. We therefore examined the localization of mitotic NPC remnants relative to spindle microtubules in a strain expressing GFP labeled alpha tubulin (TBA-2::GFP) and CeNup54::CR, and to the ER membrane in a strain expressing the ER marker SP12 (Rolls et al., 2002) fused to mCherry (SP12::CR) and CeNup54::GFP. Snr-1(RNAi) induced NPC remnants coincided with a fraction of spindle microtubules (Figure 8C) and ER membrane that is adjacent to the spindle (Figure 8D). Indeed, the ER membrane in C. elegans embryos is present at the periphery of the mitotic spindle (Figure 8E), as was previously shown for Drosophila embryos (Bobinnec et al., 2003). TEM analysis confirmed the abnormal presence of NPC-like structures embedded in the mitotic ER in embryos from snr-1(RNAi)-treated worms but not in the control (Figure 9). Taken together, these results suggest that down-regulating Sm proteins results in the persistence of NPCs during mitosis in a membranous structure that juxtaposes the spindle. These membrane structures are likely to be the remnants of the previous NE containing the non-disassembled NPCs (see below).
Figure 9.
Two-cell stage control(RNAi) (A,D) and snr-1(RNAi) (B,C,E,F) embryos were imaged and fixed during third zygotic division at anaphase (upper row) or telophase (lower row) by high-pressure freezing. 60 nm thin sections were analyzed by TEM. In snr-1(RNAi) embryos during anaphase (B,C) and telophase (E,F) some NPCs do not disassemble and they persist in the NE membrane remnants as indicated by the arrows (panels C and F are enlargements of the boxed areas in B and E, respectively). Bars= 2 μm.
Failure to disassemble NPCs in anaphase leads to NPC clusters during interphase
In later stages of mitosis, during late anaphase and telophase, the NPC remnants seemed to move alongside microtubules toward the chromosomes, until they reached the NEs forming around the daughter nuclei (Supplemental Figure S3). This led us to investigate the relationship between NPC clusters during interphase and the incomplete disassembly of NPCs during mitosis. To this end, we monitored NPCs throughout the cell cycle in embryos expressing CeNup107::GFP following treatment with snr-1(RNAi) (Figure 10). At the end of mitosis, the abnormal remnants of CeNup107::GFP (see arrows and arrowheads) moved toward the newly assembled NE, until they appeared as clusters of NPCs in the NEs of the daughter cells. Similar results were obtained when snr-1(RNAi)- induced NPC remnants were followed by CeNup54::GFP (data not shown). Taken together, our data suggest that the Sm proteins are required for NPC disassembly at the onset of mitosis, and that NPCs that failed to disassemble form NPC clusters in the nuclei of the daughter cells.
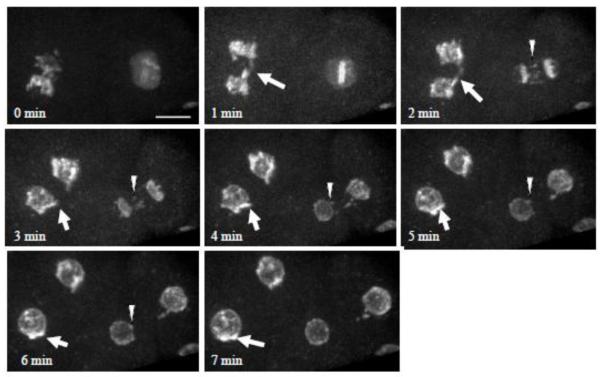
Figure 10. Failure to disassemble NPC in mitosis leads to NPC clustering in the subsequent interphase.
Time-lapse images of a 2-cell embryo expressing CeNup107::GFP from an snr-1(RNAi)-treated worm. Shown is the maximum intensity projection of confocal sections acquired at 0.4 μm intervals. The arrow and arrowhead point to NPC remnants that move towards the daughter chromosomes and eventually form an NPC cluster in the daughter NE. Bar= 8 μm.
Discussion
In this study we showed that down-regulation of Sm proteins, but not other splicing factors, led to NPC clustering and defects in NPC disassembly. We further found that NPC clusters were in membrane stacks, and our data suggest that these formed, at least in part, from NPCs that did not disassemble in the previous mitosis. These observations point to a novel function for the Sm proteins, and suggest that disruption of NPC disassembly during mitosis leads to abnormal NPC distribution in the subsequent interphase.
The Sm proteins are core components of the spliceosome. In recent years, a growing body of literature has suggested that splicing proteins have diverse functions, independent of their role in splicing (Bayne et al., 2008) (Seto et al., 1999) (Barbee and Evans, 2006; Barbee et al., 2002) and large scale screens identified components of the splicing machinery as playing a role in mitosis ((Kittler et al., 2004; Neumann et al., 2010), reviewed in (Hofmann et al., 2010)). In our screen, we found that the down-regulation of six out of the seven Sm proteins results in an NPC disassembly defect and abnormal NPC distribution. We also observed similar phenotypes following down-regulation of SMN-1, which is involved in Sm protein complex assembly into snRNPs (Kolb et al., 2007). In contrast, efficient down-regulation of other core components of the spliceosome did not affect NPC disassembly or distribution.
NPC disassembly and distribution defects following Sm protein down-regulation were observed in embryos as early as the 2- and 4-cell stage, before gene expression resumes at the 4-cell stage of embryonic development (Seydoux and Dunn, 1997; Walker et al., 2007). Therefore, Sm proteins are required for NPC-related process before the need for zygotic splicing, and well before the need for zygotic-expressed splicing factors (inhibition of zygotic splicing and gene expression results in an embryonic arrest at the 50- to 100-cell stage (Barbee et al., 2002; Powell-Coffman et al., 1996)). Thus, for the Sm proteins to affect NPC behavior through splicing, one would have to assume that a specific set of maternally spliced RNAs are more sensitive to Sm protein down-regulation than to the down-regulation of other splicing component. While possible, a more parsimonious explanation is that the Sm proteins and SMN-1 affect NPCs through a splicing-independent process. Moreover, since splicing is needed for gametogenesis (Green et al., 2011), and because NPC clustering and disassembly defects were apparent under conditions that did not cause complete sterility (Supplemental Figure S2), the defects in NPC distribution and disassembly we observed occurred while the splicing machinery in the germ line was still active. This observation is also consistent with a splicing-independent role for Sm-proteins in NPC disassembly and distribution.
We found that the extent of NPC clustering following SNR-1 down-regulation increases in the course of early embryonic divisions (Figure 1, Supplemental Figure S1). We also found that mitotic remnant of NPCs, formed after the down-regulation of Sm proteins, are attached to membrane structure adjacent to the spindle (Figure 8), and time-lapse imaging showed that these remnant NPCs move toward the reforming NEs of the daughter nuclei (Supplemental Figure S3). Consistent with this, our TEM analysis revealed the abnormal presence of NPCs in the mitotic ER (Figure 9) adjacent to the reforming NE. Finally, we found that NPCs that failed to disassemble during mitosis formed clusters in the subsequent interphase (Figure 10), likely in the form of membrane stacks that are perforated with NPCs (Figure 3). Thus, our data suggest that NPC disassembly may be an important requirement for normal NPC distribution during the subsequent interphase, and that NPCs from the previous cell cycle accumulate, along with their surrounding membrane, in membrane stacks adjacent to the newly formed nuclei.
The precise step at which Sm proteins and SMN-1 affect NPCs remains to be established. It is unlikely that the defect in NPC disassembly was due to a general defect in NEBD because conditions that inhibited NPC disassembly did not prevent nuclear lamina disassembly (Figure 8). It is also unlikely that the Sm proteins affect NPC distribution indirectly through their known role in germ granule localization and germ cell specification (Barbee and Evans, 2006; Barbee et al., 2002), because under conditions of Sm protein down-regulation only, a subset of cells shows abnormal germ granule localization (Barbee et al., 2002), whereas NPC distribution is affected in all cells in the embryo. Thus, it is more likely that the Sm proteins affect a process that is specific to the NPC.
The involvement of the Sm proteins and SMN-1 in splicing makes it difficult to assess the consequences of NPC clustering. During early embryonic divisions, the NEs of nuclei with clustered NPCs appear functional, as they expand during interphase, support nuclear lamina formation and exclude tubulin from the nucleus at the end of mitosis. In addition, mitotic cell divisions continue, albeit with a short delay (data not shown). Thus, at least some of the NPCs in the affected nuclei are functional. However, it is possible that the consequences of NPC clustering manifest themselves later in development. Unfortunately, embryos defective in splicing cannot progress beyond the 50- to 100-cell stage (Barbee et al., 2002; Powell-Coffman et al., 1996), precluding us for determining the long-term consequences of NPC clustering.
Proper cell cycle progression depends on the timely execution of, and the precise coordination between, various cell cycle processes. There is evidence accumulating in support of crosstalk between components of the NPC and the cell cycle machinery (reviewed in (Antonin et al., 2008)). For example, a fraction of the Nup107/160 NPC subcomplex relocates to kinetochores during mitosis and contributes to kinetochore function (Loiodice et al., 2004; Zuccolo et al., 2007). It is possible that this couples certain kinetochore functions to the prior execution of NEBD. NPC disassembly and NEBD are early events in open mitosis that allow mixing of nuclear and cytoplasmic compartments, and are correlated with disassembly of nuclear structures and the arrest of basic nuclear functions (Shin and Manley, 2002), reviewed in (Hofmann et al., 2010). This process involves phosphorylation by the mitotic cyclin-Cdk complex, but the mechanisms coordinating NEBD with other mitotic events are poorly understood (Burke and Ellenberg, 2002). It is possible that as cells enter mitosis, certain splicing factors become available to serve in other capacities. Although the precise stage of Sm protein involvement in NPC disassmebly still has to be determined, we speculate that the requirement for Sm proteins during mitosis serves as a temporal link between the mitotic repression of splicing and the disassembly of NPCs.
Supplementary Material
Highlights.
Sm proteins affect nuclear pore complex (NPC) dynamics in C. elegans embryos
The effect of Sm proteins on NPCs is independent of Sm proteins’ role in splicing
Sm protein down-regulation causes NPC clustering and incomplete NPC disassembly
NPCs that fail to disassemble persist on an ER structure juxtaposed to the spindle
NPCs that fail to disassemble form clusters in nuclei of the daughter cells.
Acknowledgements
We thank Kevin O’Connell, Iain Mattaj and Geraldine Seydoux for advice and comments on the manuscript. We thank Andy Golden and members of the Golden lab for reagents and invaluable assistance. D. J. S. and O. C. F. are supported by an intramural NIDDK grant. A. A. is supported by an NIH grant GM088151. E. V. was supported by an NIH fellowship 5F32GM080923.
Abbreviations
- NPC
Nuclear pore complex
- NE
Nuclear envelope
- NEBD
Nuclear envelope breakdown
- GFP
Green fluorescent protein
- CR
mCherry fluorescent protein
- snRNP
small nuclear ribonucleoporotein
- ER
endoplasmic reticulum
- TEM
Transmission Electron Microscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson DJ, Hetzer MW. The life cycle of the metazoan nuclear envelope. Curr Opin Cell Biol. 2008;20:386–392. doi: 10.1016/j.ceb.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W, Ellenberg J, Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 2008;582:2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- Barbee SA, Evans TC. The Sm proteins regulate germ cell specification during early C. elegans embryogenesis. Dev Biol. 2006;291:132–143. doi: 10.1016/j.ydbio.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Barbee SA, Lublin AL, Evans TC. A novel function for the Sm proteins in germ granule localization during C. elegans embryogenesis. Curr Biol. 2002;12:1502–1506. doi: 10.1016/s0960-9822(02)01111-9. [DOI] [PubMed] [Google Scholar]
- Bayne EH, Portoso M, Kagansky A, Kos-Braun IC, Urano T, Ekwall K, Alves F, Rappsilber J, Allshire RC. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science. 2008;322:602–606. doi: 10.1126/science.1164029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, Ellenberg J, Doye V. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y, Marcaillou C, Morin X, Debec A. Dynamics of the endoplasmic reticulum during early development of Drosophila melanogaster. Cell Motil Cytoskeleton. 2003;54:217–225. doi: 10.1002/cm.10094. [DOI] [PubMed] [Google Scholar]
- Brohawn SG, Partridge JR, Whittle JR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B, Ellenberg J. Remodelling the walls of the nucleus. Nat Rev Mol Cell Biol. 2002;3:487–497. doi: 10.1038/nrm860. [DOI] [PubMed] [Google Scholar]
- Chadrin A, Hess B, San Roman M, Gatti X, Lombard B, Loew D, Barral Y, Palancade B, Doye V. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J Cell Biol. 2010;189:795–811. doi: 10.1083/jcb.200910043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Feinstein N, Wilson KL, Gruenbaum Y. Nuclear pore protein gp210 is essential for viability in HeLa cells and Caenorhabditis elegans. Mol Biol Cell. 2003;14:4230–4237. doi: 10.1091/mbc.E03-04-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Santarella R, Wiesel N, Mattaj I, Gruenbaum Y. Electron microscopy of lamin and the nuclear lamina in Caenorhabditis elegans. Methods Cell Biol. 2008;88:411–429. doi: 10.1016/S0091-679X(08)00421-4. [DOI] [PubMed] [Google Scholar]
- D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N, Beaudouin J, Hartnell L, Imreh G, Hallberg E, Lippincott-Schwartz J, Ellenberg J. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol. 2001;154:71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TR, Lazarus MD, Hetzer MW, Wente SR. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J Cell Biol. 2009;184:659–675. doi: 10.1083/jcb.200806174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet CM, Hetzer MW. Nuclear pore biogenesis into an intact nuclear envelope. Chromosoma. 2010;119:469–477. doi: 10.1007/s00412-010-0289-2. [DOI] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 2010;141:1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V, Hurt E. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- Dultz E, Ellenberg J. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol. 2010;191:15–22. doi: 10.1083/jcb.201007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol. 2008;180:857–865. doi: 10.1083/jcb.200707026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B. The nuclear pore complex, nuclear transport, and apoptosis. Can J Physiol Pharmacol. 2006;84:279–286. doi: 10.1139/y05-100. [DOI] [PubMed] [Google Scholar]
- Funakoshi T, Clever M, Watanabe A, Imamoto N. Localization of Pom121 to the inner nuclear membrane is required for an early step of interphase nuclear pore complex assembly. Mol Biol Cell. 2011;22:1058–1069. doi: 10.1091/mbc.E10-07-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi T, Maeshima K, Yahata K, Sugano S, Imamoto F, Imamoto N. Two distinct human POM121 genes: requirement for the formation of nuclear pore complexes. FEBS Lett. 2007;581:4910–4916. doi: 10.1016/j.febslet.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Galy V, Antonin W, Jaedicke A, Sachse M, Santarella R, Haselmann U, Mattaj I. A role for gp210 in mitotic nuclear-envelope breakdown. J Cell Sci. 2008;121:317–328. doi: 10.1242/jcs.022525. [DOI] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, Askjaer P. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell. 2003;14:5104–5115. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden A, Liu J, Cohen-Fix O. Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J Cell Sci. 2009;122:1970–1978. doi: 10.1242/jcs.044743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Kao HL, Audhya A, Arur S, Mayers JR, Fridolfsson HN, Schulman M, Schloissnig S, Niessen S, Laband K, Wang S, Starr DA, Hyman AA, Schedl T, Desai A, Piano F, Gunsalus KC, Oegema K. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell. 2011;145:470–482. doi: 10.1016/j.cell.2011.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW. The nuclear envelope. Cold Spring Harb Perspect Biol. 2010;2:a000539. doi: 10.1101/cshperspect.a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann JC, Husedzinovic A, Gruss OJ. The function of spliceosome components in open mitosis. Nucleus. 2010;1:447, 459. doi: 10.4161/nucl.1.6.13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, Fischer S, Konstantinova I, Habermann B, Grabner H, Yaspo ML, Himmelbauer H, Korn B, Neugebauer K, Pisabarro MT, Buchholz F. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- Kolb SJ, Battle DJ, Dreyfuss G. Molecular functions of the SMN complex. J Child Neurol. 2007;22:990–994. doi: 10.1177/0883073807305666. [DOI] [PubMed] [Google Scholar]
- Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, Kutay U. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 2011;144:539–550. doi: 10.1016/j.cell.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Lee KK, Gruenbaum Y, Spann P, Liu J, Wilson KL. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol Biol Cell. 2000;11:3089–3099. doi: 10.1091/mbc.11.9.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart P, Rabut G, Daigle N, Hand AR, Terasaki M, Ellenberg J. Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes. J Cell Biol. 2003;160:1055–1068. doi: 10.1083/jcb.200211076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Ladinsky MS, Kirchhausen T. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J Cell Biol. 2011;194:425–440. doi: 10.1083/jcb.201012063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K, Audhya A, Oegema K, McNally FJ. Katanin controls mitotic and meiotic spindle length. J Cell Biol. 2006;175:881–891. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, Cetin C, Sieckmann F, Pau G, Kabbe R, Wunsche A, Satagopam V, Schmitz MH, Chapuis C, Gerlich DW, Schneider R, Eils R, Huber W, Peters JM, Hyman AA, Durbin R, Pepperkok R, Ellenberg J. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell-Coffman JA, Knight J, Wood WB. Onset of C. elegans gastrulation is blocked by inhibition of embryonic transcription with an RNA polymerase antisense RNA. Dev Biol. 1996;178:472–483. doi: 10.1006/dbio.1996.0232. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Lenart P, Ellenberg J. Dynamics of nuclear pore complex organization through the cell cycle. Curr Opin Cell Biol. 2004;16:314–321. doi: 10.1016/j.ceb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Reipert S, Reipert BM, Hickman JA, Allen TD. Nuclear pore clustering is a consistent feature of apoptosis in vitro. Cell Death Differ. 1996;3:131–139. [PubMed] [Google Scholar]
- Rolls MM, Hall DH, Victor M, Stelzer EH, Rapoport TA. Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol Biol Cell. 2002;13:1778–1791. doi: 10.1091/mbc.01-10-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- Shin C, Manley JL. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111:407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- Talamas JA, Hetzer MW. POM121 and Sun1 play a role in early steps of interphase NPC assembly. J Cell Biol. 2011;194:27–37. doi: 10.1083/jcb.201012154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Seydoux G. The C. elegans homolog of nucleoporin Nup98 is required for the integrity and function of germline P granules. Development. 2010;137:1441–1450. doi: 10.1242/dev.047654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Winey M, Yarar D, Giddings TH, Jr., Mastronarde DN. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol Biol Cell. 1997;8:2119–2132. doi: 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Guan T, Gerace L. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J Cell Biol. 1997;137:1199–1210. doi: 10.1083/jcb.137.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, Sibarita JB, Fukagawa T, Shiekhattar R, Yen T, Doye V. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853, 1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.