Abstract
Theranostics was coined originally as a term used to describe a system that combines diagnosis and therapy, aiming to provide the tools for personalized medicine. This review reasserts the grounds for regarding non-coding RNAs as theranostics in human cancers. MiRNAs are the most well studied non-coding RNAs in recent years; their pivotal role in orchestrating tumor initiation and progression has been confirmed in all types of cancers. Hence, these small non-coding RNAs have emerged as attractive therapeutic targets and diagnostic tool. Various approaches to use their therapeutic potential have been taken, here we summarize the most important ones. In the near future, the focus of theranostics will be shifted towards longer and mechanistically more versatile ncRNAs, and we included some recent advances supporting this view.
The discovery that around 98% of all transcriptional output in humans is actually non-coding RNA, questioned the traditional opinion that RNA is a simple intermediate between DNA and protein1. The biological complexity of higher organisms renders in these RNA species that orchestrate all fundamental cell processes, rather than in the number of protein-coding genes.
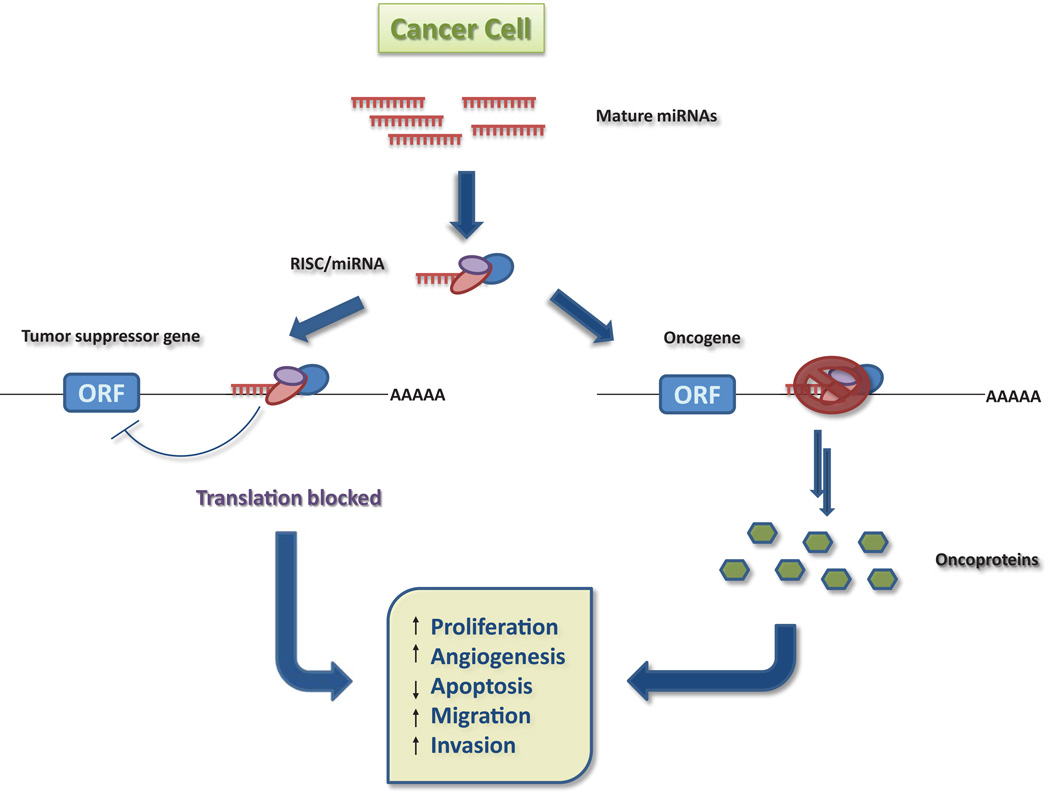
Non-coding RNAs can be devided into two major classes based on transcript size: small ncRNAs (e.g. microRNAs, siRNAs or piRNAs), and long ncRNAs (e.g. long intergenic or intronic ncRNAs, pseudogens or trascribed ultraconserved regions). Of this class of non-coding RNAs, microRNAs have captured the spotlight in the past decade. These microRNAs (miRNA) are phylogenetically conserved, single stranded RNAs of 19–25 nucleotides, mostly transcribed from intragenic or intergenic regions by RNA polymerase II into primary transcripts, termed primary miRNAs2. The pri-miRNAs are then processed to a smaller, hairpin intermediates, called pre-miRNAs (precursor miRNA), by Drosha RNase III endonuclease and exported to the cytoplasm by Exportin 5. In the cytoplasm, the pre-miRNAs are further cleaved by Dicer, also an RNase III endonuclease, resulting in mature double-stranded miRNAs. After strand separation, the mature miRNA is incorporated in the RNA-induced silencing complex (RISC), whereas the other strand commonly undergoes degradation. The RISC complex contains the proteins necessary for the degradation and/or silencing of mRNA targets, such as argonautes, helicases, deadenylases and methyltransferases3. For target recognition and incorporation into the RISC, the mature miRNAs are essential. As perfect complementarity is required only between the positions 2 to 8 from the 5’ miRNA (seed sequence) with the 3’ untranslated region (UTR) of their target mRNA for efficient silencing, each miRNA can potentially target a large number of mRNAs, and each mRNA can be targeted by more then one miRNA2. Thus, miRNAs can function in cancer cells as tumor suppressor or as oncogenes, or in some cases, both, rendering them the capability of reprogramming molecular pathways and networks in cancer (Figure 1).
Figure 1.
miRNAs as oncogenes and tumor suppressors.
It is then not surprising that these small non-coding RNAs have emerged as appealing therapeutic targets and diagnosis and prognosis tools.
MiRNAs and cancer
A plethora of studies linked by now the abnormal expression of these non-coding RNAs to the pathogenesis of several human diseases, including solid and hematopoietic tumors. MiRNA frequent location at amplified, deleted or translocated chromosomal regions (fragile sites), further supports their role in cancer development4. It was the discovery by Calin et. al (2002) that miR15a/16-1 are located in 13q14, a region frequently either deleted or dowregulated in CLL (chronic lymphocytic leukaemia) patients, that provided the first link of miRNAs to cancer5. Expression of miR15a/16-1 was inversely correlated to the levels of the anti-apoptotic protein, BCL-2 in CLL, supporting the previous findings6. Furthermore, Klein et. al (2010) have recently reported that miR-15a/16-1 knockout mice develop CLL-like diseases and lymphomas7. MiR-29 and miR-181 were also reported to be downregulated in CLL and to target TCL1, a gene overexpressed in 25–35% of CLL cases8. Whereas, in HCC (hepatocellular carcinoma) these microRNAs exhibited opposite expression levels. While miR-29 is downregulated and regulating apoptosis through a mitochondrial pathway that involves MCL-1 and BCL-2 9, miR-181 upregulation by TGFbeta promotes carcinogenesis by targeting TIMP3 and enhanced resistance to anticancer drug Doxorubicin10. Moreover, Ji J et al. (2009) found high expression of miR-181 in EpCAM-positive hepatic cancer stem cells, and determined that inhibition results in cell differentiation and suppression of tumorigenicity11.
MiR-17/92a cluster, also know as oncomir-1, is among the most potent oncogenic miRNAs, carrying out pleiotropic functions during malignant transformation. O’Donnell et al. (2005) reported that transcription of this cluster is directly transactivated by MYC, a transcription factor frequently hyperactive in cancer cells12. MYC transgenic mice developed lymphomas more rapidly when infected with murine haematopoietic stem cells with a retrovirus carrying miR-17/92a cluster13. Ventura et al (2008) showed that miR-17/92a knockout mice die shortly after birth of lung hypoplasia and ventricular septal defect14. Moreover, it was recently demonstrated that miR-19 is the key oncogenic component of the cluster, promoting cell survival by repressing PTEN and activating the AKT-mTOR pathway15.
Similar, miR-21 has an integral role in tumor pathogenesis, and extensive studies indicate its involvement in all know processes of cancer. It is overexpressed in most solid tumors with a wide range of targets. In lung cancer, it was demonstrated that overexpression of miR-21 increased K-RAS tumorigenesis in vivo16, while in glioblastoma, Chan JA et al (2005) identified miR-21 as an anti-apoptotic factor17. Recently, Liu LZ et al (2011) presented in their study on a prostate cancer cell line, that miR-21 induces carcinogenesis by targeting PTEN, which leads to AKT and ERK1/2 signaling pathways activation, and thereby enhancing HIF-1α and VEGF expression18.
Frank Slack’s group confirmed the large body of in vitro evidence nominating miR-21 as a powerfull oncogene, in a NesCre8, mir-21LSL-Tetoff mouse model. They showed that overexpression of miR-21 can lead to a pre-B malignant lymphoid-like phenotype, while inactivation of the oncomiR resulted in regression of the tumors19. Similar, Hatley ME et al (2010) used a miR-21 knock-in and a knock-out KRASLA2 mouse model of non-small-cell lung cancer (NSCLC) to evaluate the role the miRNA has in tumor development. Their findings not only confirmed miR-21 as a tumor promoter, but identified it as an important regulator of the Ras/MEK/ERK pathway and of apoptosis, by targeting APAF1, PDCD4, RhoB and FASLG20.
In contrast, members of the miR-34 family function as tumor suppressor downstream of the p53 pathway, and dysregulation of their expression occurs in various types of cancer. Of the three members of the family, miR-34a, which is expressed at higher levels then miR-34b/c, resides in 1p36 which is commonly deleted in neuroblastomas and its’ epigenetic inactivation was identified in cell lines derived from some of the most common tumors (breast, lung, colon, kidney, bladder, pancreatic cancer and melanoma)21 In human colon cancer cells, Tazawa H et al. (2007) reported suppression of cell proliferation and senescence-like growth arrest through modulation of E2F pathway, when miR-34a was induced, and reduction of tumor growth in vivo22. The effect miR-34 on apoptosis in lung cancer was recently determined by Duan W et al (2010); the data revealed the implication of miR-34a in the apoptotic network generated by PRIMA-1 (p53-dependent reactivation and induction of massive apoptosis)23.
The let-7 family, with 13 members located on 9 different chromosomes, is widely viewed as the longest family of tumor suppressor microRNA. Consistent with this activity, the expression of let-7 family members is downregulated in many cancer types when compared to normal tissue and during tumor progression and indicates a poor survival. This direct targeting of RAS by let-7 was confirmed in non-small-cell lung cancer (NSCLC), where it was demonstrated in a mouse model that let-7g inhibited tumor growth via suppression of RAS24. While in ovarian cancer, Ratner et al (2010) reported a variant in the KRAS 3’UTR that interferes with let-7 binding, increasing the risk of developing the disease25. In breast cancer, let-7g was identified as a posible prognostic marker, its’ diminished expression was associated with lymph node metastasis and poor survival in breast cancer patiens. The same study showed that abrogation of let-7g expression in otherwise non-metastatic mammary carcinoma cells elicits rapid metastasis from the orthotopic location, through preferential targets, GAB2 and FN1, and consequent activation of p44/42 MAPK and specific matrix metalloproteinases26.
MicroRNAs involvement in chemoresistance
Chemotherapy is the main strategy for cancer treatment; however it can fail in eliminating all malignant cells due to drug resistance. Resistance to therapy can be classified in 2 categories: intrinsic – the factors that would make the therapy ineffective exist prior to administration, and acquired – the tumors are not initially resistant to a particular drug, but develop resistance during the course of the treatment.
There are a number of mechanisms known to be involved in anticancer drug resistance, such as increased expression of target proteins, alteration of drug target, increased repair of DNA damage, reduced apoptosis, failure of the drug to reach or enter the target cell, ejection of the drug from the cell, drug induced karyotypic changes or altered metabolism of the drug27. Genes often involved in these processes were shown to be affected by miRNA pathways28. Several recent findings strongly support the miRNA involvement in chemoresistance.
Zhao JJ et al. (2008) reported that miR-221 and miR-222 modulate the ERα status in breast cancer cell lines. Overexpression of the microRNAs in ERα-positive cell lines resulted in decreased mRNA levels of the receptor and induced resistance to Tamoxifen, whereas downregulation of the miRs in a ERα-negative cell line had the opposite effects29.
The increased expression of the already mentioned, miR-21, has been shown to generate chemoresistance through 2 pathways: downregulating PDCD4 (programmed cell death 4), which leads to increased expression of IAP (inhibitors of apoptosis proteins) and MDR1/P-glycoprotein (multi drug resistance 1)30 observed in breast cancer, and repression of tumor suppressor PTEN in non-small cell lung cancer31.
In a different study, in a Doxorubicin resistant cancer cell line, expression of miR-451 was inversely correlated to expression of MDR1 and, more importantly, increasing levels of miR-451 led to higher cell sensitivity to Doxorubicin32. Fujita Y et al (2008) found miR-34a to be downregulated in drug-resistant prostate cancer cells, and ectopic expression of the microRNA resulted in increased sensitivity to Camptothecin33. Recently, Port M et al (2011) reported miR-371/373 cluster as a promising target for explaining the Cisplatin resistance in germ cell tumor cell lines. By upregulation, the cluster prevents p53-driven cellular senescence through several target genes (NEO1, LATS2), leading to cell proliferation34.
Several more examples of miRNAs involved in chemoresistance are summarized in Table 1.
Table 1.
MicroRNAs involved in chemoresistance.
| MiRNA | Target or pathway |
Tumor type | Mechanism of resistance to therapy |
Ref |
|---|---|---|---|---|
| miR-146a | BRCA1 | Breast cancer | Increased proliferation and resistance to Cisplatin | [71] |
| miR-200c | ZEB1 | NSCLC1 | Suppresion of EMT program and restoring sensitivity to Cisplatin and Cetuximab | [72] |
| miR-221/222 | EGFR/ErbB2 pathway Wnt/β-catenin pathway |
Breast cancer | Activation of the pathways suports estrogen-independent cell growth and Fulvestrant resistance | [73] |
| miR-9* | SOX2 | Glioblastoma | ID42 renders GSC3 resistance to Doxorubicin through the ID4-miR-9*-SOX2-ABCC3/ABCC64 regulatory pathway | [74] |
| miR-21 | PTEN and PI3K/Akt pathway | Leukaemia | Activation of the PI3K/Akt pathway by decrease in the PTEN protein level induces resistance to Daunorubicin | [75] |
| miR-181a | Bim | Non-Hodgkin lymphoma | Downregulation of the Bim-apoptosis pathway | [76] |
| miR-34a | SIRT1, BCL-2 | Prostate cancer | SIRT1 and BCL-2 reduction of expression levels induces sensitivity to Paclitaxel | [77] |
| miR-143 | ERK5/NF-kβ pathway | Colon cancer | Increases sensitivity to 5-Fluorouracil by downregulation the ERK5/NF-kβ pathway | [78] |
NSCLC= non-small-cell lung cancer.
ID4=Inhibitor of differentiation 4
GSC= Glioma stem cells
ABC= ATP-binding cassette transporters
Strategies for targeting miRNAs
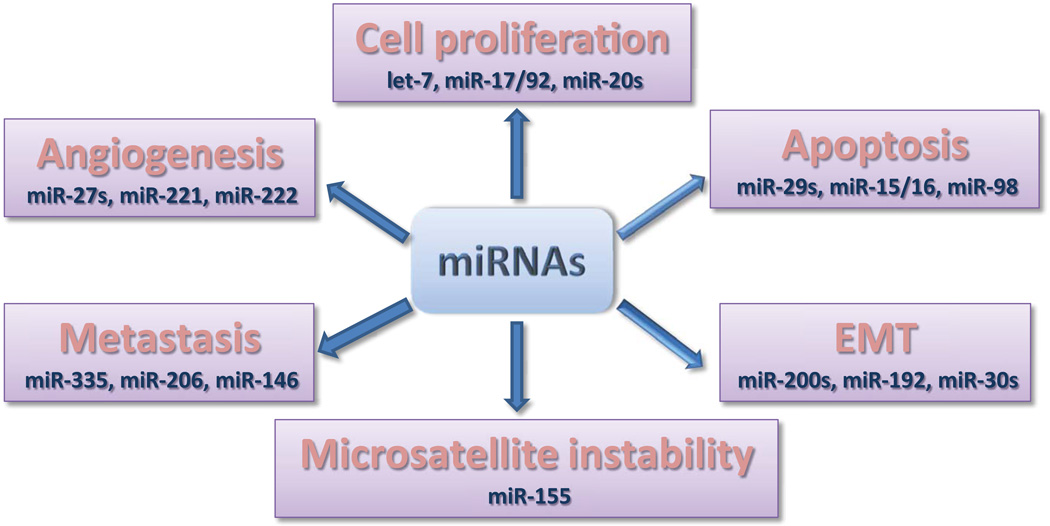
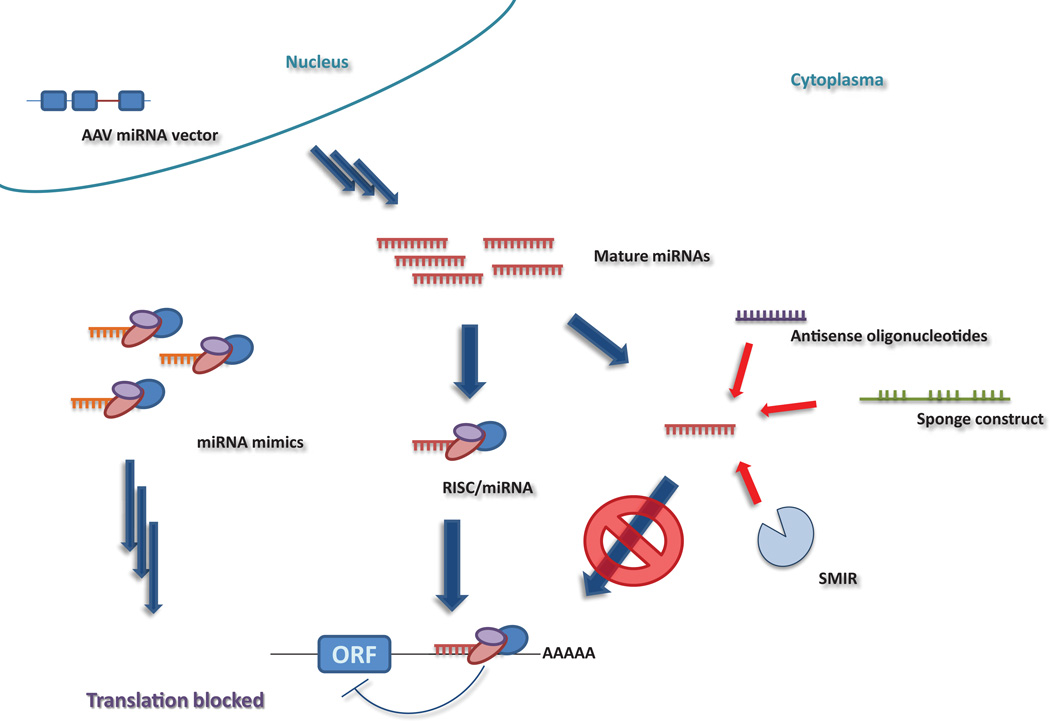
In light of the potential that lays in microRNAs (Figure 2), it is not surprising that several approaches to employ microRNAs as therapeutic targets have been developed. Mainly there are 2 strategies to target miRNA expression: by blocking the expression of an oncomiR or re-expression of a tumor suppressor miRNA, or by targeting the genes involved in their transcription and processing (Figure 3).
Figure 2.
miRNAs as key regulators of tumor initiation and progression
Figure 3.
Stretegies of targeting miRNAs.
Anti-miRNA oligonucleotides (AMOs)
Artificially reducing the expression level of microRNAs by using synthetic nucleotides represents a new application of antisense technology. AMOs are designed to bind specifically to the miRNA ‘seed region’ and sterically block their mechanism of action. Chemical modifications can be used to alter the properties of these synthetic oligonucleotides by conferring increasing binding affinity, nuclease resistance, aiding in cellular uptake and altering the ability to trigger an immune response35.
In 2004, Meister et al. employed 2’-O-methyl (2’OMe) RNA oligonucleotides to regulate the expression of miR-21 in HeLa cells36. The advantages that the 2’OMe RNA chemistry offers regard a higher binding affinity for the duplex formation with RNA targets, and higher nuclease resistance compared to DNA oligonucleotides. Endo- and exonuclease degradation proved to be problematic and to overcome this impediment several phosphorothioate (PS) bonds were used to link nucleotides at each end of the molecule. Krutzfeldt et al. (2005) were the first to introduce this modification; a cholesterol group was attached to the 3’-end to assist in vivo delivery and the new molecule was named ‘antagomir’. MiR-16 and miR-122 antagomirs were administrated to mice and showed good efficiency in lowering the expression levels of the miRNAs and consequentially of their gene targets37,38. However, PS linkages reduce binding affinity and could compromise the potential of 2’OMe. The 2’-O-methyoxyethyl (2’MOE) modification improved the nuclease resistance and increased binding affinity, as shown in an in vivo model targeting miR-12239.
Besides the AMOs with chemical modifications of the ribose nucleic acid backbone, other non-chemical backbones have been described, such as peptide nucleic acids (PNAs) or phosphorodiamidate morpholino oligonucleitides (PMOs), which rendered good results in both in vitro40 and in vivo41 studies.
Locked nucleic acids (LNA)
LNAs are generally considered to be RNA mimics in which the ribose sugar moiety is locked by an oxymethylene bridge connecting the 2’-C- and 4’-C-atoms which conformationally restricts LNA monomers into an N-type conformation42. This modification provides stabilization against nucleases, higher binding affinity and low toxicity in biological systems, making LNAs a versatile tool not only in anti-cancer therapy. The first to report miRNA inhibition by LNA-antimiR was Orom UA et al (2006), who knocked down bantam miRNA in Drosophila cells43. Following, Fabani MM et al. (2008) described a miR-122 LNA/2’OMe mixer AMO, containing 10 LNA bases and 13 2’OMe bases with PO backbone, which displayed increased potency 40. Furthermore, Elmen J et al. (2008) used in their study a similar AMO but fully PS modified to suppress miR-122 in mice44.
In a preclinical trial on non-human primates (African green monkeys), Santarias Pharma (Horsholm, Denmark) employed an optimized version of the anti-miR-122 LNA/DNA-PS compound, and was able to denote a 40% decrease in total plasma cholesterol. The effect could be detected for 3 months after the last dose45.
As inhibition of entire miRNA families may be of interest in some cases, short LNA, which bind solely to the seed sequence, have been developed recently46. These tiny compounds efficaciously downregulated the expression levels of miR-221/222 and let-7 families in cell cultures.
Small-molecule inhibitors (SMIRs)
The use of heterocyclic derivatives to modulate the expression of microRNAs may be closer to reallity as anticipated. The complex three-dimensional structure of the microRNAs allows formation of defined pockets suitable for binding of these small-molecules, leading to disrupture of their biological function47.
However, the identification of such molecules can be challenging. One approach is efficient screening of chemical libraries. Gumireddy K et al. (2008) discovered diazobenzene and its’ derivatives as inhibitors of pri-miR-21 formation, by applying this method48. Complementary sequences to miR-21 were cloned into a luciferase reporter gene, and the construct was transfected into HeLa cells, resulting in low luciferase activity ascribable to the high levels of miR-21. When cells were treated with diazobenzene, a 250% increase in the intensity was observed.
Another approach suggested by Zhang S et al. (2010) is to use an integrated drug discovery platform that can provide the 3D structure of the miRNA and perform molecular docking-based virtual high-throughput screening (vHTS), identifing potential hits based on RNA-compatible scoring functions49.
The advantages of small-molecule inhibitors, such as cost-efficiency and their pharmacokinetic and pharmacodymanic properties, will push these molecules to the top of anti-cancer drug research, if specific hits will be identified and confirmed.
miRNA sponges or decoys
MicroRNA sponges or decoys represent transcripts that contain multiple tandem binding sites for microRNAs and are transcribed from mammalian expression vectors, such as adenovirus, lentivirus or retrovirus. The binding site of a sponge is complementary to the seed sequence of the microRNA, therefore making it possible to target entire families50. Sponges can inhibit microRNA function at least as effectively as chemically modified AMOs, but have the advantage of being stably integrated into the host’s genome.
Valastyan S et al. (2009) identified miR-31 as strongly down-regulated in aggressive metastatic cancer and using a retroviral eGFP sponge demonstrated in an in vivo model, that loss of miR-31 leads to lung metastasis and 10 times more lessions51. A similar approach was taken by Ma L et al (2010) to show the miR-10b promotes metastasis in breast cancer52.
With the recent development of the technology, trangenic vertebrates expressing sponges are a work in progress.
Nanoparticles
The nanotechnology platforms have been primarily used for siRNA-therapeutics and it has only recently expended to the delivery of miRNA into target cells. Nanocarriers (ranging from 1 to 1000 nm) are usually made from biodegradable nanomaterials, such as natural or synthetic lipids (e.g. liposomes, micelles or solid lipid nanoparticles – SLN) and polymers (e.g. poly lactic co-glycolic acid, polyethilenimin or atellocollagen) or iron oxide magnetic nanoparticles. The safety, biodistribution and uptake of these particles by cells and tissues vary according to size, surface charge and hydrophobicity53.
Chen Y et al (2010) developed a liposome-polycation-hyaluronic acid (LPH) nanoparticle modified with a tumor-targeting single-chain antibody fragment (scFv) for the delivery of miR-34a into a murine model of metastatic melanoma. The authors observed a significant downregulation of survivin expresson in the metastatic tumor and reduction of tumor load in the lung54.
Aside from delivering the actual miRNAs, nanoparticles can be suitable vehicles for carring oligonucelotides, as they can protect the oligonucleotides against endo- and exonucleases. Two different groups have recently exploited this benefit in their studies. One group produced an AMO-CLO (cationic lipid binded oligonucelotide loaded SLN) which efficiently targeted miR-21 in an in vitro model of lung cancer, subsequently decresing proliferation, migration and invasion of tumor cells55.
The other group reported antiangiogenesis activity and suppressed cell migration in HUVEC (human umbilical vein endothelial cells) by employing a PEGylated LPH (liposome-polycation-hyaluronic acid) nanoparticle funcionalized with cyclic RGD peptide for delivery of anti-miR-296 AMO into αvβ3 integrin-positive endothelial cells. The same results were presented in vivo using Matrigel plug assay56.
miRNA mimics and adenovirus-assosiated (AAV) vectors
A competent way of restoring expression of tumor-suppressor miRNAs is by introducing miRNA mimics. A miRNA mimic has the same sequence as the depleted, naturally occuring miRNA and is therefore expected to have the same mRNA targets, making nonspecific, off-target effects unlikely57.
MiRNA mimics have been used in various in vitro studies resulting in induced cell death or blocked proliferation. In vivo data using miRNA mimics was provided by Takeshita F et al (2010) in a prostate cancer metastasis model. MiR-16 chemically modified precursor and complexed with atelocollagen, was administrated into the tail vein of mice bearing bone metastasis from prostate cancer cells. Significant inhibition of tumor growth by restoration of the microRNA expression was reported at the end of the experiment58.
An alternative to miRNA mimics are the adenovirus-assosiated vectors (AAV). They present the advantage of efficiently transducting the target cells, without integrating into the genome. The advances made with self-complementary AAV vectors and the availability of AAV serotypes for improved transduction of specific target tissues has nominated them for being ideally suited for therapeutic gene delivery59. With he use of AAV vectors, Kota J et al (2009) were the first to provide the evidence that restoring the expression of a tumor-suppressor miRNA can stop cancer progression in vivo. MiR-26a was packed into a AAV vector system and injected into the tail vein of tet-o-MYC/LAP-tTA mice, leading to suppression of tumorigenicity by repressing proliferation and inducing apoptosis60.
These studies conclude that restoring expression of tumor-suppressor miRNAs seems to be a promising perspective in cancer therapy.
Future perspectives – other non-coding RNAs as therapeutic targets
Originally thought to be just, transcriptional noise’, these lnRNAs are emerging as new, essential players in the cancer paradigm, with roles in both oncogenic and tumor-suppressive pathways. Various studies have shed light into their potential functions, and revealed their involvement in high-order chromosomal dynamics, telomere biology and subcellular structural organization61. One of their major roles seems to be the regulation of neighbouring protein-coding genes and recent findings suggest that lncRNA can act as natural, miRNA sponges’ to reduce miRNA levels62.
Amoung the better characterized lncRNAs that have been associated with cancer biology, is HOTAIR (HOX antisense intergenic RNA), involved in metastasis. The lncRNA, located in the mammalian HOXC locus on chr 12q13.13, was found to be highly upregulated in primary, as well as metastatic breast tumors and it’s elevated levels were correlated with both metastasis and poor survival rate63.
Also associated with metastasis and poor prognosis for patients with non-small cell lung cancer64 is MALAT1 (Metastasis-associated lung adenocarcinoma transcript 1), residing at 11q13.1, which has been found to harbor chromosomal translocation breakpoints linked to cancer65. Other in vitro studies, have implicated MALAT1 in the regulation of the invasive potential of cancer cells, in cervical66 and lung67 cancer.
As mentioned before, lncRNAs can act as decoys for miRNAs and HULC (highly upregulated in liver cancer) is one of them. Transcribed from chr 6p24.3, with extremlly high expression levels in liver cancer68, seems to function as a miRNA sponge, for miR-372, of which one function is the translational repression of PRKACB, a kinase targeting cAMP response element binding protein (CREB). A feedback loop was also found, the activated CREB protein is able to promote HULC transcription by maintaining and open chromatin structure at the HULC promoter69.
Nevertheless, a more comprehensive understanding of their mechanism of action will provide novel approaches of regulating genes, including mimetics to compete with binding sites for miRNAs, chromatin remodelers or DNA. Furthermore, their cancer type-specific expression can be used to reduce the risk of affecting normal tissue during transgene-mediated therapy or it may potentially correlate with patient response to therapy.
As disscusted in this review, there are well-founded arguments for exploiting non-coding RNAs as therapeutic targets and the studies conducted so far show promising results. However, it must be admitted that our yet limited understanding of the biology and function of these ncRNAs burdens the clinical translation of these new strategies and further studies are necessary to improve our ability to utilize their full potential.
Acknowledgements
G.A.C. is supported as a Fellow at The University of Texas MD AndersonResearch Trust, as a University of Texas System Regents Research Scholar and by the CLL Global Research Foundation. Work in Dr. Calin’s laboratory is supported in part by the NIH/NCI (CA135444), a Department of Defense Breast Cancer Idea Award, Developmental Research Awards in Breast Cancer, Ovarian Cancer, Brain Cancer, Multiple Myeloma and Leukemia SPOREs, a 2009 Seena Magowitz–Pancreatic Cancer Action Network AACR Pilot Grant, and the Laura and John Arnold Foundation and RGK Foundation.
References
- 1.Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism and Function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Di Leva G, Calin GA, Croce CM. MicroRNAs: fundamental facts and involvement in human diseases. Birth Defects Res C Embryo Today. 2006;78:180–189. doi: 10.1002/bdrc.20073. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancer. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of microRNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL-2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein U, Lia M, Crespo M, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukaemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Pekarsky Y, Santanam U, Cimmino A, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 9.Xiong Y, Fang JH, Yun JP, et al. Effects of microRNA-29 on apoptosis, tumorigenicity and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 10.Wang B, Hsu SH, Majumder S, et al. TGFbeta-mediated upregulation of hepatic miR-181 promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29:1787–1797. doi: 10.1038/onc.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji J, Yamashita T, Budhu A, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positice hepatic cancer stem cells. Hepatology. 2009;50:472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Donnell KA, Wentzel EA, Zeller KI, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 13.He L, Thomas JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olive V, Bennett MJ, Walker JC, et al. miR-19 is a key oncogenic component of miR-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatley ME, Patrick DM, Garcia MR, et al. Modulation of K-RAS dependent lung tumorigenesis by microRNA-21. Cancer Cell. 2010:1-282–1-293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan JA, Krichevsky AM, Kosik KS, et al. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 18.Liu LZ, Li C, Chen Q, et al. MicroRNA-21 induced angiogenesis through AKT and ERK activation and HIF1A expression. Plos One. 2011;6:e19139. doi: 10.1371/journal.pone.0019139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell-lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 20.Hatley ME, Patrick DM, Garcia MR, et al. Modulation of K-Ras-Dependent Lung Tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lodygin D, Tarasov V, Epanchintsev A, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 22.Tazawa H, Tsuchiya N, Izumiya M, et al. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. PNAS. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan W, Gao L, Wu X, et al. MicroRNA-34a is an important component of PRIMA-1-induced apoptotic network. Int J Cancer. 127:313–320. doi: 10.1002/ijc.25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar MS, Erkeland SJ, Pester RE, et al. Suppresion of non-small cell lung tumor development by the let-7 microRNA family. PNAS. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratner E, Lu L, Boeke M, et al. A KRAS-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010;70:6509–6515. doi: 10.1158/0008-5472.CAN-10-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian PX, Zuo Z, Wu ZS, et al. Pivotal role of reduced let-7g expression in breast cancer invasion and metastasis. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-11-1322. (epub) [DOI] [PubMed] [Google Scholar]
- 27.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 28.Wu X, Xiao H. miRNAs modulate drug responce in tumor cells. Sci China C Life Sci. 2009;9:797–801. doi: 10.1007/s11427-009-0114-4. [DOI] [PubMed] [Google Scholar]
- 29.Zhao JJ, Lin J, Yang H, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with Tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Bourguignon LY, Spevak CC, Wong CC, et al. Hyaluronan-CD44 interaction with protein kinase C (epsilon) promotes oncogenic signaling by the stem cell marker Nanog and the production of microRNA-21, leading to downregulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J Biol Chem. 2009;284:26533–26546. doi: 10.1074/jbc.M109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang JG, Wang JJ, Zhao F, et al. microRNA-21 represses tumor supressor PTEN and promotes growth and invasion in non-small cell lung cancer. Clin Chim Acta. 2010;411:11–12. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 32.Kovalchuk O, Filkowski J, Meservy J, et al. Involvement of microRNA-451 in resistance of the MCF7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7:2152–2159. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 33.Fujita Y, Kojima K, Hamada N, et al. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cell. Biochem Biophys REs Commun. 2008;377:114–119. doi: 10.1016/j.bbrc.2008.09.086. [DOI] [PubMed] [Google Scholar]
- 34.Port M, Glaesener S, Ruf C, et al. Micro-RNA expression in cispaltin resistant germ cell tumor cell lines. Mol Cancer. 2011;10:52–60. doi: 10.1186/1476-4598-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lennox KA, Behlke MA. Chemical modification and design of anti-miRNA oligonucleotides. Gene Therapy. 2011 doi: 10.1038/gt.2011.100. [DOI] [PubMed] [Google Scholar]
- 36.Meister G, Landthaler M, Dorsett Y, et al. Sequence-specific inhibition of microRNA and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krutzfeldt J, Rajewsky N, Braich R, et al. Silincing of microRNAs in vivo with antagomirs. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 38.Krutzfeldt J, Kuwajima S, Braich R, et al. Specificity, duplex degradation and subcellular localization of anatagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Fabani MM, Gait MJ. miR-122 targeting with LNA/2’-O-methyl oligonucleotide mixers, peptide nucleotides (PNA) and PNA-peptide conjugates. RNA. 2008;14:336–346. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabani MM, Abreu-Goodger C, Williams D, et al. Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acid Res. 2010;38:4466–4475. doi: 10.1093/nar/gkq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koshkin AA, Rajwanshi VK, Wengel J, et al. Novel convenient syntheses of LNA [2.2.1]bicycle nucleotides. Tetrahedron Lett. 1998;39:4381–4384. [Google Scholar]
- 43.Orom UA, Kaupinnen S, Lund AH, et al. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 44.Elmen J, Lindow M, Silahtaroglu A, et al. Antagonism of miR-122 in mice by systemically administered LNA-antimiR leads to upregulation of a large set of predcted target mRNAs in liver. Nuclei Acid Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elmen J, Lindow M, Schutz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 46.Obad S, dos Santos CO, Petri A, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 48.Gumireddy K, Young DD, Xiong X, et al. Small-molecule inhibitors of microRNA miR-21 function. Angew Chem Int Ed. 2008;47:7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang S, Chen L, Jung EJ, et al. Targeting microRNAs with small molecules: from dream to reality. Clin Pharmacol Ther. 2010;87:754–758. doi: 10.1038/clpt.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valastyan S, Reinhardt F, Benaich N, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Ma L, Reinhardt F, Pan E, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozpolat B, Sood AK, Lopez-Berestein G. Nanomedicine based approaches for delivery of siRNA in cancer. J Intern Med. 2010;267:44–53. doi: 10.1111/j.1365-2796.2009.02191.x. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y, Zhu X, Zhang X, et al. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Ther. 2010;18:1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi SJ, Zhong ZR, Liu J, et al. Solid Lipid Naonparticles loaded with anti-microRNA oligonulotides (AMOs) for suppression of microRNA-21 functions in human lung cancer cells. Pharm Res. 2011 doi: 10.1007/s11095-011-0514-6. Epub: [DOI] [PubMed] [Google Scholar]
- 56.Liu XQ, Song WJ, Sun TM, et al. Targeted delivery of antisense inhibitor of miRNA for antiangiogenesis therapy using cRGD-functionalized nanoparticles. Mol Phar. 2010;8:250–259. doi: 10.1021/mp100315q. [DOI] [PubMed] [Google Scholar]
- 57.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer REs. 2010;70:7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takeshita F, Patrawala L, Osaki M, et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastic prostate tumors via downregulation of multiple cell-cycle genes. Mol Ther. 2010;18:181–187. doi: 10.1038/mt.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu Z, Asokan A, Samulski RJ. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol Ther. 2006;14:316–327. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 60.Kota J, Chivukula RR, O’Donnell KA. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 62.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gupta RA, Shah N, Wang KC, et al. Long-non coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 65.Rajaram V, Knezevich S, Bove KE, et al. DNA sequence of the translocation breakpoints in undifferentiated embryonal sarcoma arising in mesenchymal hamartoma of the liver harboring the t(11;19)(q11;q13.4) translocation. Genes Chromosome Cancer. 2007;46:508–513. doi: 10.1002/gcc.20437. [DOI] [PubMed] [Google Scholar]
- 66.Guo F, Li Y, Liu Y, et al. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochem Biophys Sin (Shangai) 2010;42:224–229. doi: 10.1093/abbs/gmq008. [DOI] [PubMed] [Google Scholar]
- 67.Tano K, Mizuno R, Okada T, et al. MALAT-1 enhances cell motolity of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 68.Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking upregulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 69.Wang J, Liu X, Wu H, et al. CREB upregulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pogribny IP, Filkowski JN, Tryndyak VP, et al. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer. 2010;127:1785–1794. doi: 10.1002/ijc.25191. [DOI] [PubMed] [Google Scholar]
- 71.Ceppi P, Mudduluru G, Kumarswamy R, et al. Loss of miR-200c expression induces an aggressive, invasive and chemoresistant phenotype in non-small cell lung cancer. Mol Cancer Res. 2010;8:1207–1216. doi: 10.1158/1541-7786.MCR-10-0052. [DOI] [PubMed] [Google Scholar]
- 72.Ao X, Leva GD, Li M, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011;30:1082–1097. doi: 10.1038/onc.2010.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jeon HM, Sohn YW, Oh SY, et al. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 2001;71:3410–3421. doi: 10.1158/0008-5472.CAN-10-3340. [DOI] [PubMed] [Google Scholar]
- 74.Bai H, Xu R, Cao Z, et al. Involvement of miR-21 in resistance to daunorubicin by regulating PTEN expression in the leukaemia K562 cell line. FEBS Lett. 2011;585:402–408. doi: 10.1016/j.febslet.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 75.Lwin T, Lin J, Choi YS, et al. Follicular dendritic cell-dependent drug resistance of non-Hodgkin lymphoma involves cell adhesion-mediated Bim down-regulation through induction of microRNA-181a. Blood. 2010;116:5228–5236. doi: 10.1182/blood-2010-03-275925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kojima K, Fujita Y, Nozawa Y, et al. MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanism. Prostate. 2010;70:1501–1512. doi: 10.1002/pros.21185. [DOI] [PubMed] [Google Scholar]
- 77.Borralho PM, Kren BT, Castro RE, et al. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS. 2009;276:6689–6700. doi: 10.1111/j.1742-4658.2009.07383.x. [DOI] [PubMed] [Google Scholar]