Abstract
Liquid Chromatography Mass Spectrometry (LC-MS) is a powerful and widely applied method for the study of biological systems, biomarker discovery and pharmacological interventions. LC-MS measurements are, however, significantly complicated by several technical challenges, including: (1) ionisation suppression/enhancement, disturbing the correct quantification of analytes, and (2) the detection of large amounts of separate derivative ions, increasing the complexity of the spectra, but not their information content. Here we introduce an experimental and analytical strategy that leads to robust metabolome profiles in the face of these challenges. Our method is based on rigorous filtering of the measured signals based on a series of sample dilutions. Such data sets have the additional characteristic that they allow a more robust assessment of detection signal quality for each metabolite. Using our method, almost 80% of the recorded signals can be discarded as uninformative, while important information is retained. As a consequence, we obtain a broader understanding of the information content of our analyses and a better assessment of the metabolites detected in the analyzed data sets. We illustrate the applicability of this method using standard mixtures, as well as cell extracts from bacterial samples. It is evident that this method can be applied in many types of LC-MS analyses and more specifically in untargeted metabolomics.
Keywords: LC-MS, Metabolomics, Orbitrap, Metabolite identification
Introduction
Untargeted metabolomics aims to describe living systems by the set of metabolites present in a cell at certain moment of time and under specific environmental constraints (Fiehn 2002; Dettmer et al. 2007; Oldiges et al. 2007). Since metabolites are the final link between the gene expression and the phenotype exhibited by the cell, metabolomics represents a valuable tool to achieve a better understanding of an organism’s phenotype (Fiehn 2002; Oldiges et al. 2007). The study of the metabolome is complementary to the other “omics” sciences (genomics, transcriptomics, proteomics, fluxomics…) and fits well with the general approach of systems biology (Arita 2009).
Important advances have been realized in the past years for untargeted metabolite profiling in different research fields, from human health to nutrition (Scalbert et al. 2009; Kamleh et al. 2008). However, metabolomics is still an emerging field in the post-genomic arena. For example, due to the chemical diversity of cellular metabolites and the complexity of the cell extracts, there is no single method which can separate, detect and identify all small molecules present in a cell extract. Furthermore the Achilles’ heel of metabolomics remains the identification and structure elucidation of metabolites (Kind and Fiehn 2010). Sometimes, fragmentation patterns of the molecules can be used for identification. For metabolomics data the detected fragment patterns can, e.g., be matched to online databases, like Metlin (Smith et al. 2005), and assigned to a quality score. But in our experiments we have however observed that the scan time of the LTQ-Orbitrap is considerably affected by the inclusion of fragmentation steps, making the normal LC-MS data stream fragmentary and difficult to analyze automatically. As more convenient alternative, the Orbitrap Exactive platform (without the linear iontrap but with faster scan speeds) can be used to capture more data points using the positive–negative polarity switch mode (Lu et al. 2010). Thus, currently matching on mass alone to databases is the most commonly used method. Unfortunately, this approach to metabolite identification is very seriously hampered by the fact that the vast majority of the signals in the data set can be caused by contaminants in the sample or LC-MS system (Keller et al. 2008), technical artefacts and so-called “derivative peaks” (Scheltema et al. 2009). In many cases, several peaks or signals share the same identifications, even if signals are detected with an accuracy of better than 2 ppm, as is routinely possible using, e.g., modern Fourier Transform mass spectrometers, like the Orbitrap (Scheltema et al. 2008). Such spurious peaks need to be checked manually and assigned to their real identification or discarded if the signal shows typical artefacts.
Our goal was to develop an analytical method that would be able to eliminate a substantial part of the spurious signals from the data set. This required the development of new approaches and the collection of an unusual type of data on biological samples and mixtures of analytical standards, to distinguish real effects from spurious fluctuations in LC-MS analyses and peak detection algorithms. The strategies developed here will be generally useful for metabolomics.
Materials and methods
Amino acid standard mixture samples
A mixture of 38 physiological amino acid standards (Product No. A9906, Sigma) was used. In the stock solution, amino acids and related compounds are contained at a final concentration of 0.5 μmol/ml ± 4% in 0.2 N lithium citrate buffer, pH 2.20, containing thiodiglycol (2% w/v) and phenol (0.1% w/v) as antioxidant and preservative, respectively. The concentration in the injected diluted samples is described in Table 1.
Table 1.
Dilution factor and concentrations of the analysed samples
| Dilution factor | 1/8 | 1/16 | 1/32 | 1/64 | 1/128 | 1/256 | 1/512 | 1/1024 |
|---|---|---|---|---|---|---|---|---|
| Concentration (μmol/ml) | 0.0625 | 0.0313 | 0.0156 | 0.0078 | 0.0039 | 0.0020 | 0.0010 | 0.0005 |
| Injected on column (pmol) | 312.5000 | 156.2500 | 78.1250 | 39.0625 | 19.5313 | 9.7656 | 4.8828 | 2.4414 |
Biological samples
Analytical samples were obtained from Streptomyces coelicolor wild-type M145 strain (Bentley et al. 2002). Bacteria were grown in 50 ml liquid minimum medium (Nieselt et al. 2010) as described (Takano et al. 2001).
Cells from 25 ml of culture were collected on a 0.45 μm filter by vacuum filtration and washed twice with 25 ml of 2.63% NaCl solution. For cell quenching, the filter with the collected cells was quickly moved into 60% methanol solution (HPLC-grade, Boom, The Netherlands) pre-chilled at −20°C and frozen in liquid nitrogen. Samples were stored at –80°C until metabolite extraction was performed.
Metabolites were extracted by three freeze–thaw cycles. Cells were thawed in an ethanol bath at −20°C (~15 min), vortexed vigorously for 1 min and, right afterwards, frozen in liquid nitrogen for 5 min. The cycle was repeated three times. After the third cycle, the samples were centrifuged at 4500 rpm for 10 min at −9°C. The supernatant (cell extract) was collected and stored at −80°C until LC-MS analysis. Before analysis, obtained samples were diluted with the same dilution factor as for the analytical standards mixture, resulting in eight samples with different metabolite concentrations.
LC-Orbitrap MS analysis
The analytical mixtures and cell extracts were analyzed by liquid chromatography coupled to a high-accuracy LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Germany).
Two chromatographic columns were used: a reversed-phase Shim-pack XR-ODS C18 column (Achrom, Belgium) (3.0 × 75 mm, 2.2 μm, Shimadzu Corp.) and a ZIC-HILIC column (Achrom, Belgium) (150 × 2.1 mm, 3.5 μm, Merck Sequant AB) fitted with a ZIC-HILIC PEEK guard column (Achrom, Belgium) (15 × 1.0 mm; 5 μm, Merck Sequant AB).
For the C18 column, the flow rate was set to 0.6 ml/min; the mobile phase consisted of (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile. A gradient of 18 min was used. The elution of solvent B started at 2% for the first 2 min and was increased to 95% within 8 min. This composition was maintained for 2 min, after which the elution of B was decreased to 2% within 1 min. To re-equilibrate the system, the elution of B was held at 2% for 5 min.
For the ZIC-HILIC column, the flow rate was set to 0.1 ml/min; as buffers, (A) 0.1% formic acid in acetonitrile and (B) 0.1% formic acid in water were used. A gradient of 40 min was applied. Solvent A was set to 80% as starting condition. The elution fraction of solvent B was increased to 40% within 6 min and maintained at 40% for 12 min, after which solvent B was increased to 90% in a 4 min-interval. This composition was held for 2 min after which B was decreased to 20% in 2.5 min. The gradient was held at 20% B for 13.5 min to re-equilibrate the system.
The sample volume injected was 5 μl for both columns, and two technical replicates were recorded for the C18 analysis, and three replicates on the HILIC column.
The system was operated with the electrospray ionization source in positive mode. Full-scan spectra were obtained over an m/z range of 50–1000 Da.
ULC grade acetonitrile, formic acid and water were purchased at Biosolve (Netherlands).
Data processing
Raw data files from the mass spectrometer were converted into the mzXML format by the ReAdW.exe utility (a tool of the Trans-Proteomic Pipeline software collection, downloaded from http://tools.proteomecenter.org/wiki/index.php?title=Software:ReAdW).
The CentWave (Tautenhahn et al. 2008) feature detection algorithm from the XCMS (Smith et al. 2006) package was used on each individual data file. Further processing was handled by the flexible data processing pipeline mzMatch (Scheltema et al. 2011), performing noise removal (Windig 2004) and several steps of signal filtering and peak matching. The first matching step involved aligning of the chromatographic features between technical replicates of a single sample. Peaks that were not detected in all technical replicates were discarded from further analysis. In the second matching step, the chromatographic peaks, which were combined in single files containing technical replicates in the previous matching step, were aligned to each other for all eight dilutions. After combining the eight measurements in a single file, there were still peak sets that did not include peaks from every sample. Such gaps were filled by extracting ion chromatograms within the retention time and mass window of the given peak set directly from the raw data files.
Derivative signals (isotopes, adducts, dimers and fragments) were automatically annotated by correlation analysis on both signal shape and intensity pattern, as described (Scheltema et al. 2009). These peaks were not discarded and their assigned annotations were taken into account in the subsequent analysis.
Putative identifications were made by matching the detected masses to a database of Streptomyces coelicolor (ScoCyc) metabolites, a contaminants database (Keller et al. 2008), and the list of analytical standards in the standard mixture. The metabolite database was obtained from a genome annotation file created by Jonathan Moore as part of the SysMO STREAM project (https://www.wsbc.warwick.ac.uk/groups/sysmopublic/), which is also available for download from the BioCyc project page (Karp et al. 2009) as a flat-file in Pathway Tools format (Karp et al. 2002).
Pearson’s correlation of binary logarithm of the peak intensities was applied to evaluate dilution trends in the obtained data set. Samples for the 8 dilution points were ordered from highest to lowest concentration, so that metabolites matching the sample dilution trend would show high negative correlation values between intensity and sample number. Correlation values smaller than −0.85 were considered as indicating a significantly reproducible dilution trend.
For low-abundance peaks, where signals for the highest dilutions were below the limit of detection, correlation values were calculated for the detectable consecutive measurements (at least 3 dilution points were required).
All statistical analyses and graphical routines were handled in R (R Development Core Team, R: A Language and Environment for Statistical Computing, Austria: 2011; http://www.R-project.org).
Raw data files in mzXML format, R code containing the complete data processing pipeline, as well final peak tables are available for download at http://mzmatch.sourceforge.net/metabolomics.html.
Results and discussion
Our study was carried out in two steps. First we wanted to validate our filtering method by applying it to the data sets of the mixtures of analytical standards. The resulting numbers of detected peaks are shown in Table 2. Data for both chromatographic columns are shown: even for relatively simple samples (39 compounds in the mix of standards) a huge amount of the peaks were detected (2831 peak sets for C18 data, and 11169 for HILIC). Only about 20–30% of these signals can be identified in chemical databases or assigned to known contaminants. A significant amount of the uninformative signals could be removed after application of the dilution trend filter. For example, in the unfiltered data set for HILIC data 28 unique standard compounds were matching 409 features within 5 ppm mass accuracy window. After application of the dilution trend filter, this number decreased to 91 features matching 26 unique standard compounds. In other words, the number of detected compounds is not significantly changing, while the number of total peaks in the data set is decreasing by almost 5 times and the number of unambiguous matches is substantially increased (Fig. 1a). Manual inspection showed that the two putative standard compounds removed by application of the filter were artefacts, i.e. these two compounds were not really detectable. Also, a very large amount of the signals matching the ScoCyc database (which should not be present in samples of analytical standards) was removed by the trend filter, as were most of the unidentifiable compounds, which also do not match the expected composition of the samples. Overall the fraction of correctly identifiable compounds is dramatically increased.
Table 2.
Comparison of number of the peaks extracted for the standard mixtures samples
| C18 | HILIC | |||||||
|---|---|---|---|---|---|---|---|---|
| Before filtering | Filtered | Before filtering | Filtered | |||||
| 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | |
| Detected as standards (bp) | 49 | 12 | 11 | 10 | 409 | 28 | 91 | 26 |
| Detected as standards (rp) | 30 | 27 | 20 | 20 | 256 | 30 | 99 | 28 |
| Detected as contaminants (bp) | 69 | 23 | 1 | 1 | 227 | 28 | 13 | 8 |
| Detected as contaminants (rp) | 40 | 17 | 5 | 4 | 147 | 22 | 22 | 9 |
| Detected in ScoCyc (bp) | 94 | 17 | 6 | 2 | 516 | 68 | 70 | 29 |
| Detected in ScoCyc (rp) | 72 | 37 | 24 | 23 | 383 | 92 | 115 | 58 |
| Unidentified (bp) | 1335 | 106 | 4745 | 493 | ||||
| Unidentified (rp) | 1142 | 348 | 4486 | 1337 | ||||
The fraction of features uniquely identified as standard compounds is significantly increased after application of trend filtering
bp labelled as base peaks by mzMatch software, rp labelled as derivative peaks, 1 number of peaks, 2 number of unique identifiers
Fig. 1.
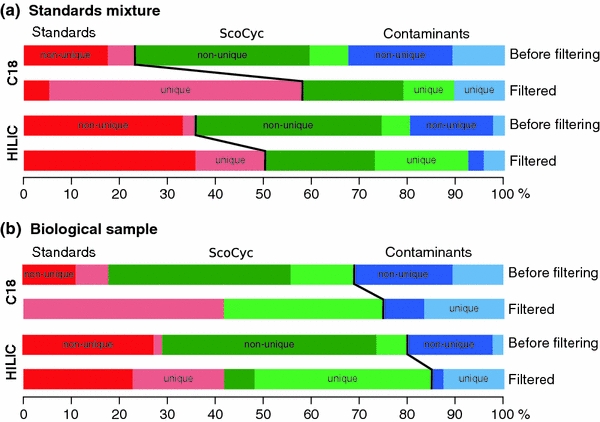
Proportional relationship between identified compounds before and after filtering on dilution trend. Compounds labelled as base peaks by the mzMatch software are shown. For the standards mixture (a) where only matches to the standard compounds are expected, a clear increase of the fraction of identified peaks can be seen after filtering. Importantly, the fraction of uniquely identified compounds (lighter shade of the color) is also strongly increasing. In other words, after filtering more compounds with unambiguous, unique identifications are retained. The same trend can be also seen in the data for the biological samples (b), where matches to the standard compounds and the ScoCyc data base are expected. Matches to the contaminant compounds decrease in the filtered data, and the number of unique identifications increases substantially (Color figure online)
A list of the standard compounds detected on both C18 and HILIC columns is shown in Table 3. The following structural isomers could not be distinguished: l-alanine, l-sarcosine and β-alanine; γ-amino-N-butyric acid, d,l-β-aminoisobutryic acid and L-α-amino-n-butyric acid. For l-isoleucine/l-leucine and 1-methyl-l-histidine/3-methyl-l-histidine two peaks eluting close to each other were observed. Ammonium chloride was not detected on either column (because of its low molecular weight), and l-ornithine was not detected on the HILIC column. Almost no separation was achieved on the C18 column (most of the signals eluted within the first minute of the analytical run). Surprisingly high quantification accuracy (correlation value is close to −1, i.e. a linear relationship between intensity and sample dilution) can be observed for almost all analytical standards on both chromatographic columns.
Table 3.
Identified compounds in the analytical mixture
| Metabolite (KEGG compound ID) | Molecular formula | Monoisotopic mass | C18 | HILIC | ||
|---|---|---|---|---|---|---|
| Corr. | RT | Corr. | RT | |||
| Urea (C00086) | CH4N2O | 60.03240 | −0.88 | 0 min 40 s | −0.99 | 8 min 33 s |
| Ethanolamine (C00189) | C2H7NO | 61.05280 | −0.94 | 0 min 32 s | −0.99 | 20 min 56 s |
| Glycine (C00037) | C2H5NO2 | 75.03200 | −0.89 | 0 min 35 s | −1 | 17 min 58 s |
| l-Alanine (C00041) | C3H7NO2 | 89.04770 | −0.97 | 0 min 35 s | −0.98 | 14 min 58 s |
| γ-Amino-N-butyric acid (C00334) | C4H9NO2 | 103.06330 | −0.91 | 0 min 37 s | −0.90 | 13 min 56 s |
| l-Serine (C00065) | C3H7NO3 | 105.04260 | −0.88 | 0 min 35 s | −1 | 18 min 05 s |
| l-Creatinine (C00791) | C4H7N3O | 113.05890 | −0.97 | 0 min 34 s | −0.85 | 14 min 44 s |
| l-Proline (C00148) | C5H9NO2 | 115.06330 | −0.92 | 0 min 38 s | −0.98 | 14 min 26 s |
| l-Valine (C00183) | C5H11NO2 | 117.07900 | −0.99 | 0 min 48 s | −0.92 | 13 min 18 s |
| l-Threonine (C00188) | C4H9NO3 | 119.05820 | −0.95 | 0 min 35 s | −0.89 | 18 min 16 s |
| Taurine (C00245) | C2H7NO3S | 125.01470 | −0.86 | 0 min 36 s | −0.99 | 15 min 01 s |
| Hydroxy-l-proline (C01157) | C5H9NO3 | 131.05820 | −0.96 | 0 min 36 s | −0.85 | 15 min 23 s |
| l-Isoleucine (C00407) | C6H13NO2 | 131.09460 | −0.99 | 1 min 34 s | −1 | 11 min 48 s |
| l-Ornithine (C00077) | C5H12N2O2 | 132.08990 | −0.95 | 0 min 28 s | ||
| l-Aspartic acid (C00049) | C4H7NO4 | 133.03750 | −0.90 | 0 min 36 s | −1 | 16 min 39 s |
| l-Lysine (C00047) | C6H14N2O2 | 146.10550 | −0.95 | 0 min 28 s | −1 | 30 min 7 s |
| l-Glutamic acid (C00025) | C5H9NO4 | 147.05320 | −0.92 | 0 min 36 s | −0.90 | 15 min 41 s |
| l-Methionine (C00073) | C5H11NO2S | 149.05100 | −0.99 | 1 min 02 s | −1 | 12 min 48 s |
| l-Histidine (C00135) | C6H9N3O2 | 155.06950 | −0.89 | 0 min 29 s | −1 | 29 min 19 s |
| δ-Hydroxylysine (C01211) | C6H14N2O3 | 162.18700 | −0.95 | 0 min 28 s | −1 | 30 min 18 s |
| l-Phenylalanine (C00079) | C9H11NO2 | 165.07900 | −0.99 | 3 min 42 s | −1 | 11 min 16 s |
| 1-Methyl-l-histidine (C01152) | C7H11N3O2 | 169.08510 | −0.91 | 0 min 31 s | −1 | 29 min 38 s |
| l-Arginine (C00062) | C6H14N4O2 | 174.11170 | −0.95 | 0 min 32 s | −1 | 30 min 10 s |
| l-Citrulline (C00327) | C6H13N3O3 | 175.09570 | −0.93 | 0 min 36 s | −1 | 18 min 35 s |
| l-Tyrosine (C00082) | C9H11NO3 | 181.07390 | −0.99 | 1 min 40 s | −1 | 13 min 42 s |
| l-Tryptophan (C00078) | C11H12N2O2 | 204.08990 | −0.99 | 4 min 39 s | −0.99 | 12 min 01 s |
| l-Cystathionine (C02291) | C7H14N2O4S | 222.06740 | −0.87 | 0 min 35 s | −1 | 26 min 00 s |
| l-Carnosine (C00386) | C9H14N4O3 | 226.10660 | −0.88 | 0 min 28 s | −1 | 31 min 01 s |
| l-Cystine (C00491) | C6H12N2O4S2 | 240.02380 | −0.88 | 0 min 35 s | −1 | 25 min 17 s |
| l-Anserine (C01262) | C10H16N4O3 | 240.12220 | −0.98 | 0 min 30 s | −1 | 30 min 53 s |
| l-Homocystine (C01817) | C8H16N2O4S2 | 268.05510 | −1 | 0 min 44 s | −1 | 24 min 02 s |
Corr. Pearson’s correlation coefficient between sample number and the logarithm of the signal intensity, RT retention time
The resulting numbers of detected peaks after processing of biological samples are shown in Table 4 . Surprisingly, the amount of detected peaks is comparable to the numbers seen for the analytical standards, both in the filtered and unfiltered data sets. For the HILIC data set, 639 features were putatively identified in the ScoCyc database (78 unique compounds), but only 28 peaks (24 unique identifiers) were retained after application of the dilution trend filtering. Clear trends in improvement of the data set quality are shown in Fig. 1b. Interesting compounds that were identified (and expected) only in the biological samples on both chromatographic columns are the osmoregulator compound ectoine and hypoxanthine. In Fig. 2, an example of dilution trends and chromatographic peaks for the biological sample (Fig. 1a) and the standard mixture (Fig. 1b) is given. In both data sets, a peak was identified as matching the mass of ectoine with an apparent mass error less than 1 ppm, but in the standard mixture (which does not contain ectoine), this peak was successfully discarded by the trend filter, as the signal intensity patterns (shown in the left panel of the plot) are not following the sample dilution trend.
Table 4.
Comparison of the number of the peaks extracted for the biological samples before and after trend filtering
| C18 | HILIC | |||||||
|---|---|---|---|---|---|---|---|---|
| Before filtering | Filtered | Before filtering | Filtered | |||||
| 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | |
| Detected as standards (bp) | 34 | 13 | 5 | 5 | 366 | 22 | 32 | 15 |
| Detected as standards (rp) | 16 | 12 | 8 | 8 | 208 | 23 | 20 | 10 |
| Detected as contaminants (bp) | 59 | 20 | 3 | 2 | 254 | 28 | 10 | 8 |
| Detected as contaminants (rp) | 29 | 16 | 4 | 4 | 129 | 29 | 16 | 12 |
| Detected in ScoCyc (bp) | 97 | 25 | 4 | 4 | 639 | 78 | 28 | 24 |
| Detected in ScoCyc (rp) | 36 | 22 | 7 | 7 | 362 | 78 | 46 | 33 |
| Unidentified (bp) | 1235 | 19 | 4962 | 146 | ||||
| Unidentified (rp) | 632 | 123 | 3053 | 359 | ||||
The fraction of compounds with putative identifications is significantly increased after application of the trend filter
bp Labelled as base peaks by mzMatch software, rp labelled as derivative peaks, 1 number of peaks, 2 number of unique identifiers
Fig. 2.
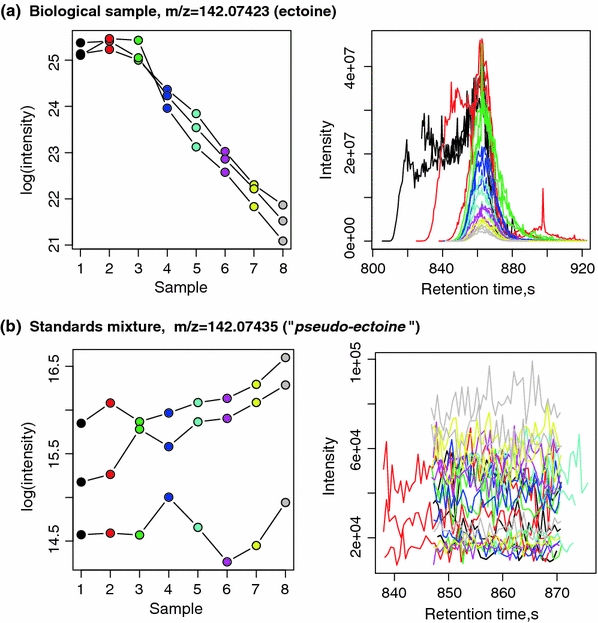
Example of the dilution trends (on the left) and extracted mass chromatograms (on the right) for a metabolite putatively identified as ectoine. For the biological samples, which are expected to contain ectoine (Kol et al. 2010), three technical replicates show clearly identifiable dilution trend (trend correlation value −0.97). For the standard mixture, which does not contain ectoine, a random trend is seen in all replicates for the signal putatively identified as ectoine (mass error 0.86 ppm); this putative technical artefact can thus be removed by the trend filtering (Color figure online)
The biological samples used in this illustrative example are particularly challenging, due to a large number of peaks with low signal intensities. Our results show that even for such difficult data, the dilution trend filter can be applied with no real danger of losing information of interest. It is also quite obvious that sample dilution factors should be adjusted according to the expected overall metabolite levels in the analysed samples, to avoid over-dilution and loss of signals of interest. To avoid the problem of large correlations occurring by chance when the number of observations is low, the statistical significance of the observed correlation can be examined and the obtained p-values can be used to determine the threshold for peak selection. This method can also be integrated with a quality control sample approach (Sangster et al. 2006), where repeated injection of a pooled randomized sample throughout the analysis serves as a reference for quality control; this approach is commonly used in large populations studies (Zelena et al. 2009). This control sample can be replaced with injections of pooled dilution samples in randomized order. Thereby, without increasing the number of injections for a typical analytical sample batch, it will be possible to simultaneously assess machine stability (as the dilution trend should stay constant) and do a filtering of the data set on highly reproducible signals.
The method suggested here is therefore a useful complement to the commonly used relative standard deviation (RSD) filters (Shah et al. 2000; Scheltema et al. 2008) and the CoDA-DW filters, (Windig 2004), allowing automatic retrieval of signals of interest, reducing the complexity of the data and consequently speeding up the interpretation process.
The dilution filtering approach can be easily integrated in a complete data processing pipeline (based on mzMatch and XCMS software tools) and used in a semi-automated manner. This is illustrated in the R script provided as supplementary material for this study (http://mzmatch.sourceforge.net/metabolomics.html).
Concluding remarks
We have been able to demonstrate the effectiveness and reliability of a relatively simple data filtering strategy. The proposed trend correlation filter significantly decreases the amount of non-informative signals in the data sets and makes metabolite identification much easier. We could show that even very stringent filtering of the data is not causing a loss of informative signals.
Our illustrative application to biological samples demonstrates that our approach can also be applied to assess the performance of metabolite extraction from the samples. This allows a more reliable estimate of the true metabolomic complexity observed in a particular experiment.
Acknowledgments
The authors gratefully acknowledge the contributions of Richard Scheltema (Max Planck Institute for Biochemistry, Germany), Ruben t’Kindt (Metablys, Belgium) and Darren Creek (University of Glasgow, UK) during many discussions on data processing and mass spectroscopy-related topics. The authors have declared that no competing interests exist. AJ is supported by an NWO-Vidi award to RB. MEM is funded by a 4 × 4 Ubbo Emmius scholarship and ET by a Rosalind Franklin Fellowship, both from the University of Groningen. RJV was supported by an investment grant from NWO.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Arita M. What can metabolomics learn from genomics and proteomics? Current Opinion in Biotechnology. 2009;20:610–615. doi: 10.1016/j.copbio.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeño-Tárraga AM, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrometry Reviews. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Molecular Biology. 2002;48:155–171. doi: 10.1023/A:1013713905833. [DOI] [PubMed] [Google Scholar]
- Kamleh A, Barrett MP, Wildridge D, Burchmore RJS, Scheltema RA, Watson DG. Metabolomic profiling using Orbitrap Fourier transform mass spectrometry with hydrophilic interaction chromatography: a method with wide applicability to analysis of biomolecules. Rapid Communications in Mass Spectrometry. 2008;22:1912–1918. doi: 10.1002/rcm.3564. [DOI] [PubMed] [Google Scholar]
- Karp PD, Ouzounis CA, Moore-Kochlacs C, et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Research. 2009;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Paley S, Romero P. The Pathway Tools software. Bioinformatics. 2002;18:S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.S225. [DOI] [PubMed] [Google Scholar]
- Keller BO, Sui J, Young AB, Whittal RM. Interferences and contaminants encountered in modern mass spectrometry. Analytica Chimica Acta. 2008;627:71–81. doi: 10.1016/j.aca.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Kind T, Fiehn O. Advances in structure elucidation of small molecules using mass spectrometry. Bioanalytical Reviews. 2010;2:23–60. doi: 10.1007/s12566-010-0015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kol S, Merlo ME, Scheltema RA, et al. Metabolomic characterization of the salt stress response in Streptomyces coelicolor. Applied and Environmental Microbiology. 2010;76:2574–2581. doi: 10.1128/AEM.01992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Clasquin MF, Melamud E, et al. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone Orbitrap mass spectrometer. Analytical Chemistry. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieselt K, Battke F, Herbig A, et al. The dynamic architecture of the metabolic switch in Streptomyces coelicolor. BMC Genomics. 2010;11:10. doi: 10.1186/1471-2164-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldiges M, Lütz S, Pflug S, et al. Metabolomics: current state and evolving methodologies and tools. Applied Microbiology and Biotechnology. 2007;76:495–511. doi: 10.1007/s00253-007-1029-2. [DOI] [PubMed] [Google Scholar]
- Sangster T, Major H, Plumb R, Wilson AJ, Wilson ID. A pragmatic and readily implemented quality control strategy for HPLC-MS and GC-MS-based metabonomic analysis. Analyst. 2006;131:1075–1078. doi: 10.1039/b604498k. [DOI] [PubMed] [Google Scholar]
- Scalbert A, Brennan L, Fiehn O, et al. Mass-spectrometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics. 2009;5:435–458. doi: 10.1007/s11306-009-0168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltema R, Decuypere S, Dujardin J, et al. Simple data-reduction method for high-resolution LC-MS data in metabolomics. Bioanalysis. 2009;1:1551–1557. doi: 10.4155/bio.09.146. [DOI] [PubMed] [Google Scholar]
- Scheltema R, Jankevics A, Jansen RC, Swertz MA, Breitling R. PeakML/mzMatch: A file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Analytical Chemistry. 2011;83:2786–2793. doi: 10.1021/ac2000994. [DOI] [PubMed] [Google Scholar]
- Scheltema R, Kamleh A, Wildridge D, et al. Increasing the mass accuracy of high-resolution LC-MS data using background ions: a case study on the LTQ-Orbitrap. Proteomics. 2008;8:4647–4656. doi: 10.1002/pmic.200800314. [DOI] [PubMed] [Google Scholar]
- Shah VP, Midha KK, Findlay JW, et al. Bioanalytical method validation—a revisit with a decade of progress. Pharmaceutical Research. 2000;17:1551–1557. doi: 10.1023/A:1007669411738. [DOI] [PubMed] [Google Scholar]
- Smith CA, O’Maille G, Want EJ, et al. METLIN: A metabolite mass spectral database. Therapeutic Drug Monitoring. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Takano E, Chakraburtty R, Nihira T, Yamada Y, Bibb MJ. A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2) Molecular Microbiology. 2001;41:1015–1028. doi: 10.1046/j.1365-2958.2001.02562.x. [DOI] [PubMed] [Google Scholar]
- Tautenhahn R, Böttcher C, Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics. 2008;9:504. doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windig W. The use of the Durbin–Watson criterion for noise and background reduction of complex liquid chromatography/mass spectrometry data and a new algorithm to determine sample differences. Chemometrics and Intelligent Laboratory Systems. 2004;77:206–214. doi: 10.1016/j.chemolab.2004.10.008. [DOI] [Google Scholar]
- Zelena E, Dunn WB, Broadhurst D, et al. Development of a robust and repeatable UPLC-MS method for the long-term metabolomic study of human serum. Analytical Chemistry. 2009;81:1357–1364. doi: 10.1021/ac8019366. [DOI] [PubMed] [Google Scholar]


