Abstract
Extracting biomedical information from large metabolomic datasets by multivariate data analysis is of considerable complexity. Common challenges include among others screening for differentially produced metabolites, estimation of fold changes, and sample classification. Prior to these analysis steps, it is important to minimize contributions from unwanted biases and experimental variance. This is the goal of data preprocessing. In this work, different data normalization methods were compared systematically employing two different datasets generated by means of nuclear magnetic resonance (NMR) spectroscopy. To this end, two different types of normalization methods were used, one aiming to remove unwanted sample-to-sample variation while the other adjusts the variance of the different metabolites by variable scaling and variance stabilization methods. The impact of all methods tested on sample classification was evaluated on urinary NMR fingerprints obtained from healthy volunteers and patients suffering from autosomal polycystic kidney disease (ADPKD). Performance in terms of screening for differentially produced metabolites was investigated on a dataset following a Latin-square design, where varied amounts of 8 different metabolites were spiked into a human urine matrix while keeping the total spike-in amount constant. In addition, specific tests were conducted to systematically investigate the influence of the different preprocessing methods on the structure of the analyzed data. In conclusion, preprocessing methods originally developed for DNA microarray analysis, in particular, Quantile and Cubic-Spline Normalization, performed best in reducing bias, accurately detecting fold changes, and classifying samples.
Electronic supplementary material
The online version of this article (doi:10.1007/s11306-011-0350-z) contains supplementary material, which is available to authorized users.
Keywords: Metabolomics, NMR, Data normalization, Preprocessing, Classification
Introduction
Nuclear magnetic resonance (NMR) spectroscopy is a powerful and versatile method for the analysis of metabolites in biological fluids, tissue extracts and whole tissues. Applications include the analysis of metabolic differences as a function of disease, gender, age, nutrition, genetic background, and the targeted analysis of biochemical pathways (Klein et al. 2011). Further, metabolomic data derived from individuals with known outcome are used to train computer algorithms for the prognosis and diagnosis of new patients (Gronwald et al. 2011). There are many good reviews available on these topics (Lindon et al. 2007; Dieterle et al. 2011; Clarke and Haselden 2008).
Due to the chemical complexity of biological specimens such as human urine and serum, which contain hundreds to thousands of different endogenous metabolites and xenobiotics (Holmes et al. 1997), NMR spectra contain a correspondingly large number of spectral features. Spectral data are typically analyzed using multivariate data analysis techniques (Wishart 2010), which all exploit the joint distribution of the metabolomic data including the variance of individual metabolite concentrations and their joint covariance structure. Some sources of variation are the target of analysis such as differences in response to treatment or metabolite concentrations between diseased individuals and controls. Other sources of variation are not wanted and complicate the analysis. These include measurement noise and bias as well as natural, non-induced biological variability and confounders such as nutrition and medication. An additional complication arises from the typically large dynamic spectrum of metabolite concentrations. As described by van den Berg et al. (van den Berg et al. 2006), one can expect order-of-magnitude differences between components of metabolite fingerprints of biological specimens, where the highly abundant metabolites are not necessarily more biologically important. Data normalization needs to ensure that a measured concentration or a fold change in concentration observed for a metabolite at the lower end of the dynamic range is as reliable as it is for a metabolite at the upper end. Also variances of individual metabolite concentrations can differ greatly. This can have a biological reason as some metabolites show large concentration changes without phenotypic effects, while others are tightly regulated. Moreover, one observes that the variance of non-induced biological variation often correlates with the corresponding mean abundance of metabolites leading to considerable heteroscedasticity in the data. However, differences in metabolite variance can also have technical reasons, because relative measurements of low abundance metabolites are generally less precise than those of high abundance metabolites. The goal of data preprocessing is to reduce unwanted biases such that the targeted biological signals are depicted clearly.
In accordance with the layout suggested by Zhang et al. (Zhang et al. 2009), methods applicable to NMR spectra may be grouped into (i) methods that remove unwanted sample-to-sample variation, and (ii) methods that are aimed at adjusting the variance of the different metabolites to reduce for example heteroscedasticity. These include variable scaling and variance stabilization approaches. There are methods that attempt both tasks simultaneously.
The first group includes approaches such as Probabilistic Quotient Normalization (Dieterle et al. 2006), Cyclic Loess Normalization (Cleveland and Devlin 1988; Dudoit et al. 2002), Contrast Normalization (Astrand 2003), Quantile Normalization (Bolstad et al. 2003), Linear Baseline Normalization (Bolstad et al. 2003), Li-Wong Normalization (Li and Wong 2001), and Cubic-Spline Normalization (Workman et al. 2002). The second group comprises among others Auto Scaling (Jackson 2003) and Pareto Scaling (Eriksson et al. 2004). These are so-called variable scaling methods that divide each variable by a scaling factor determined individually for each variable. The next tested method of the second group is a non-linear transformation that is aimed at the reduction of heteroscedasticity by use of a Variance Stabilization Normalization (Huber et al. 2002; Parsons et al. 2007; Durbin et al. 2002; Anderle et al. 2011). Several of the aforementioned methods, including Variance Stabilization Normalization, were developed originally for the analysis of DNA microarray data. Since factors complicating the analysis of DNA microarray data also affect the analysis of metabolomics data, it appeared promising to conduct a comprehensive evaluation of these methods for their application to NMR-based metabolite fingerprinting. A similar evaluation, albeit limited to six linear scaling and two heteroscedasticity reducing methods, had been already performed for mass spectrometry based metabolomic data (van den Berg et al. 2006).
For the evaluation of the performance of the different data normalization methods in the identification of differentially produced metabolites and the estimation of fold changes in metabolite abundance, we spiked eight endogenous metabolites at eight different concentration levels into a matrix of pooled human urine following a Latin-square design (Laywine and Mullen 1998) that keeps the total spike-in amount constant while the molar amounts of the individually added metabolites were varied. To investigate the effect of the different normalization methods on sample classification by a support vector machine (SVM) with nested cross validation, a previously published dataset comprising NMR urinary fingerprints from 54 autosomal polycystic kidney disease (ADPKD) patients and 46 apparently healthy volunteers was employed (Gronwald et al. 2011).
Materials and methods
Urinary specimens
As a background for the spike-in data human spot-urine specimens were collected from volunteers at the University of Regensburg. Samples were pooled and immediately frozen at −80°C until preparation for NMR analysis. The classification data had been generated previously employing urine specimens collected at the Klinikum Nürnberg and the University Hospital Erlangen from 54 ADPKD patients and 46 apparently healthy volunteers, respectively (Gronwald et al. 2011).
Latin-square spike-in design
For the generation of the Latin-square spike-in data, eight endogenous metabolites, namely 3-aminoisobutyrate, alanine, choline, citrate, creatinine, ornithine, valine, and taurine, were added in varied concentrations to eight aliquots of pooled human urine keeping the total concentration of metabolites added consistently at 12.45 mmol/l per aliquot of urine. The highest concentration level of an individual metabolite was 6.25 mmol/l and was halved seven times down to a minimum concentration 0.0488 mmol/l, i.e. in each of the 8 aliquots of urine each metabolite was present at a different concentration. In contrast to a dilution series, the overall concentration of the contents remains the same, thus eliminating the impact of differing total concentrations on normalization. The spike-in samples were prepared once.
NMR spectroscopy
To each 400-μl specimen of human urine 200 μl of 0.1 mol/l phosphate buffer, pH 7.4, and 50 μl of deuterium oxide containing 0.75% (w/v) trimethylsilyl-2,2,3,3-tetradeuteropropionic acid (TSP) as a reference [Sigma-Aldrich, Steinheim, Germany] were added. 1D 1H spectra were measured as described previously (Gronwald et al. 2008) on a 600 MHz Bruker Avance III spectrometer [Bruker BioSpin GmbH, Rheinstetten, Germany], which was equipped with a cryogenic probe with z-gradients and a cooled automatic sample changer. A 1D nuclear Overhauser enhancement spectroscopy (NOESY) pulse sequence was used in all cases and solvent signal suppression was achieved by presaturation during relaxation and mixing time. All spectra were measured once. Spectra were Fourier transformed and phase corrected by automated routines. A flat baseline was obtained employing the baseopt option of TopSpin2.1 [Bruker BioSpin] that corrects the first points of the observed signal, i.e. of the free induction decay (FID). All spectra were chemical shift referenced relative to the TSP signal. For subsequent statistical data analysis, bin (feature) tables were generated from the 1D 1H NMR spectra using AMIX 3.9 (Bruker BioSpin).
Signal positions between samples may be subject to shifts due to slight changes in pH, salt concentration, and/or temperature. In addition, the TSP signal used for spectral referencing may also show pH-dependent shifts. Here we chose to use equidistant binning to compensate for these effects, which is still the most widely used method. In order to keep a clear focus on data normalization other parameters of metabolomic data evaluation such as the initial data processing including spectral binning were kept constant. Competing methods include peak alignment (Forshed et al. 2003; Stoyanova et al. 2004), approaches working at full resolution using statistical total correlation spectroscopy (Cloarec et al. 2005a), and orthogonal projection to latent structures (Cloarec et al. 2005b). In another approach termed targeted profiling a pre-selected set of metabolites is quantified from 1D spectra and these values are used for subsequent data analysis (Weljie et al. 2006). Quantitative values may also be obtained from 2D spectra (Lewis et al. 2007; Gronwald et al. 2008). For the data presented here an optimized bin size of 0.01 ppm was applied and bins were generated in the regions from 9.5 to 6.5 ppm and from 4.5 to 0.5 ppm, respectively, to exclude the water artifact and the broad urea signal, leaving 701 bins for further analysis. To correct for variations in urine concentration, all data in the classification data set was linearly scaled to the signal of the CH2 group of creatinine at 4.06 ppm. This can be considered as normalization in itself. Each dataset was arranged in a data matrix X = (x ij) with i = 1…I and I = 701 representing the feature or bin number, and j = 1…J with J = 8 and J = 100 for the spike-in and classification datasets, respectively, representing the number of specimens. For further analysis, tables were imported into the statistical analysis software R version 2.9.1 (Development Core Team 2011).
Basic characteristics of the normalization algorithms employed
For all normalization methods discussed it is assumed that NMR signal intensities scale linearly with metabolite concentration and are mostly independent of the chemical properties of the investigated molecules. The equations describing the different normalization approaches are listed in Supplemental Table S1. The first group of methods evaluated aims to reduce between-sample variations. If not stated otherwise, it is assumed in the following that only a relatively small proportion of the metabolites is regulated in approximately equal shares up and down. The first group includes the following approaches:
Probabilistic Quotient Normalization (Dieterle et al. 2006) assumes that biologically interesting concentration changes influence only parts of the NMR spectrum, while dilution effects will affect all metabolite signals. In case of urine spectra, dilution effects are caused, for example, by variations in fluid intake. Probabilistic Quotient Normalization (PQN) starts, with an integral normalization of each spectrum, followed by the calculation of a reference spectrum such as a median spectrum. Next, for each variable of interest the quotient of a given test spectrum and reference spectrum is calculated and the median of all quotients is estimated. Finally, all variables of the test spectrum are divided by the median quotient.
Cyclic Locally Weighted Regression (Cyclic Loess) is based on MA-plots, which constitute logged Bland-Altman plots (Altman and Bland 1983). The presence of non-linear such as intensity-depended biases is assumed. Briefly, the logged intensity ratio M of spectra j 1 and j 2 is compared to their average A feature by feature (Dudoit et al. 2002). Then, a normalization curve is fitted using non-linear local regression (loess) (Cleveland and Devlin 1988). This normalization curve is subtracted from the original values. If more than two spectra need to be normalized, the method is iterated in pairs for all possible combinations. Typically, almost complete convergence is reached after two cycles. If only a relatively small proportion of the metabolites are regulated, all data points can be taken into account. Otherwise, rank-invariant metabolites can be selected for the computation of the loess lines. Here, all data points were used.
Contrast Normalization also uses MA-plots (Astrand 2003) and makes the same assumptions as Cyclic Loess. The data matrix of the input space is logged and transformed by means of an orthonormal transformation matrix T = (t ij) into a contrast space. This expands the idea of MA-plots to several dimensions and converts the data into a set of rows representing orthonormal contrasts. A set of normalizing curves is then fitted similarly to those in Cyclic Loess Normalization, using a robust distance measure є based on the Euclidean norm that renders the normalization procedure independent of the particular choice of T. The contrasts are then evened out by a smooth transformation, ensuring that features with equal values prior to normalization retain identical values. Subsequently, data are mapped back to the original input space. The use of a log function impedes the handling of negative values and zeros. Therefore, all non-positive values were set beforehand to a residual value (10−11) three orders of magnitude smaller than the smallest value in the original data. Subtracting the 10%-quantile from each spectrum will minimize the bias introduced thereby.
The goal of Quantile Normalization is to achieve the same distribution of feature intensities across all spectra. Similarity of distributions can be visualized in a quantile–quantile plot (Bolstad et al. 2003). If two spectra share the same distribution, all quantiles will be identical and, hence, align along the diagonal. The idea is to bring simply all spectra to an identical distribution of intensities across features (bins). This is achieved by sorting the vector of feature intensities in ascending order separately for each spectrum. In the sorted vector each entry corresponds to a quantile of the distribution. Next the mean of identical quantiles across spectra is calculated, i.e. the mean of the highest abundances, the mean of the second highest abundances, and so on. This mean is assigned to all features that realize the corresponding quantile. For example, the feature with the highest intensity in a spectrum is assigned the average of the highest intensities across spectra irrespectively of their spectral positions. Since different features may display the highest intensity in different samples, this constant average value may be assigned to different features across samples. After Quantile Normalization the vectors of feature intensities consist of the same set of values, however, these values are distributed differently among features.
A completely different normalization approach used in DNA microarray analysis is Baseline Scaling. In contrast to normalizing the data to a measure of the full dataset, here the data is normalized only to a subset of it, the so-called baseline. This can be conducted both linearly and non-linearly. Typically, the spectrum with the median of the median intensities is chosen as baseline, but other choices are possible, too. Alternatively, an artificial baseline can be constructed.
Linear Baseline Scaling uses a scaling factor to map linearly from each spectrum to the baseline (Bolstad et al. 2003). Therefore, one assumes a constant linear relationship between each feature of a given spectrum and the baseline. In the version implemented in this paper, the baseline is constructed by calculating the median of each feature over all spectra. The scaling factor β is computed for each spectrum as the ratio of the mean intensity of the baseline to the mean intensity of the spectrum. Then, the intensities of all spectra are multiplied by their particular scaling factors. However, the assumption of a linear correlation between spectra may constitute an oversimplification.
A more complex approach is to fit a Non-Linear Baseline Normalization relationship between the spectra that are to be normalized and the baseline as implemented by Li and Wong (2001). It is assumed that features corresponding to unregulated metabolites have similar intensity ranks in two spectra, allowing a reliable determination of a normalization curve. In addition, possible non-linear relationships between the baseline and the individual spectra are assumed. The normalization process is based on scatter plots with the baseline spectrum (having the median overall intensity) on the x-axis and the spectrum to be normalized on the y-axis. Ideally, the data should align along the diagonal y = x. As the non-normalized data generally deviates from that, the normalization curve is then fitted to map the data to the diagonal. To make sure that the normalization curve is fitted only on non-differentially expressed features, a set of almost rank-invariant features (invariant set) is calculated and used for finding the normalizing piecewise linear running median line.
Another non-linear baseline method makes use of Cubic Splines (Workman et al. 2002). As in quantile normalization the aim is to obtain a similar distribution of feature intensities across spectra. In this method as well the existence of non-linear relationships between baseline and individual spectra are assumed. A baseline, called target array in the original publication that corresponds to a target spectrum here, is built by computing the geometric mean of the intensities of each feature over all spectra. In this paper, the geometric mean was substituted by the arithmetic mean for reasons of robustness to negative values. For normalization, cubic splines are fitted between each spectrum and the baseline. To that end, a set of evenly distributed quantiles is taken from both the target spectrum and the sample spectrum and used to fit a smooth cubic spline. This process is iterated several times shifting the set of quantiles by a small offset each time. Next, a spline function generator uses the generated set of interpolated splines to fit the parameters of a natural cubic spline (B-spline). Here, for each spectrum five iterations comprising 14 quantiles each were calculated and interpolated to normalize the data.
The second group of methods is aimed at adjusting the variance of different metabolites. These include variable scaling and variance stabilization approaches. The simplest of these approaches uses the standard deviation of the data as a scaling factor. This method is called Auto Scaling or unit variance (uv) scaling (Jackson 2003). It results in every feature displaying a standard deviation of one, i.e. the data is transformed to standard units. Briefly, one centers the data first by subtracting from each feature its mean feature intensity across spectra. This will result in a fluctuation of the data around zero, thereby adjusting for offsets between high and low intensity features. From the centered data the standard deviation of each feature is obtained and data is divided by this scaling factor. Auto Scaling renders all features equally important. However, measurement errors will also be inflated and between-sample variation due to dilution effects, which in case of urine spectra are caused, for example, by variations in fluid intake will not be corrected.
Using the square root of the standard deviation is an alternative used by Pareto Scaling (Eriksson et al. 2004). It is similar to Auto Scaling, but its normalizing effect is less intense, such that the normalized data stays closer to its original values. It is less likely to blow up noisy background and reduces the importance of large fold changes compared to small ones. However, very large fold changes may still show a dominating effect.
Variance Stabilization Normalization (VSN) transformations are a set of non-linear methods that aim to keep the variance constant over the entire data range (Huber et al. 2002; Parsons et al. 2007; Durbin et al. 2002; Anderle et al. 2011). In the VSN R-package used here (Huber et al. 2002), a combination of methods that corrects for between-sample variations by linearly mapping all spectra to the first spectrum followed by adjustment of the variance of the data is applied. Looking at the non-normalized data, the coefficient of variation, i.e. the variance divided by the corresponding mean, does not vary much for the strong and medium signals, implying that the standard deviation is proportional to the mean and, therefore, in VSN it is assumed that the variance of a feature depends on the mean of that feature via a quadratic function. But as values approach the lower limit of detection, variance does not decrease any more, but rather stays constant, thus, the coefficient of variation increases. VSN addresses exactly this problem by using the inverse hyperbolic sine. This transformation approaches the logarithm for large values, therefore removing heteroscedasticity. For small intensities, though, it approaches linear transformation behavior, leaving the variance unchanged. The VSN normalized data is not logged again for comparisons based on logarithmic intensities of the data.
The R-code for performing the different normalization techniques is given in the supplemental material.
Classification of samples using a support vector machine
Classification of samples was performed using the support vector machine (SVM) provided in the R-library e1071 (http://cran.r-project.org/web/packages/e1071). Results were validated by a nested cross-validation approach that consists of an inner loop for model fitting and parameter optimization and an outer loop for assessment of classification performance. From the analyzed dataset of 100 urine specimens two samples were selected arbitrarily and excluded to serve as test data of the outer cross-validation (leave-two-out cross-validation). Then, two of the remaining samples were chosen randomly and put aside to serve as test data of the inner cross-validation. In the inner loop, the SVM was trained on the remaining n − 4 samples in order to find the optimal number of features. For this, the feature number k was increased stepwise within the range k = 10…60. The top k features with the highest t-values were selected and a SVM classifier was trained and applied to the left-out samples of the inner loop.
For each feature number, the SVM was trained (n − 2)/2 times, such that every sample except for the outer test samples was used once as inner test sample. The accuracy on the inner test samples was assessed and the optimal feature number was used to train classifiers in the outer loop. In the outer cross-validation, the SVM was trained on all samples except the outer test samples, using the optimal number of features from the inner loop and the outer test samples were predicted. This was repeated n/2 times, so that all samples were chosen once as outer test data. In all cases a linear kernel was used. In all steps feature selection was treated as part of the SVM training and was redone excluding left out cases for every iteration of the cross validations.
Classification performance was analyzed by evaluating receiver operating characteristic (ROC) plots that had been obtained by using the R-package ROCR (Sing et al. 2005).
Results and discussion
A first overview of the data (Data Overview) was obtained by comparing the densities of the metabolite concentration distributions for each of the 100 urine specimens of the classification dataset. Supplemental Fig. S1 shows the creatinine adjusted-intensity distributions. For comparison, the distribution of the Quantile normalized data that represents an average of the intensity distributions is indicated in red. Roughly similar distributions were obtained for all specimens.
Next we investigated for each normalization method whether comparable profiles were obtained for all samples of the classification dataset (Overall between Sample Normalization Performance). To that end, all preprocessing methods were included, although the variable scaling and variance stabilization methods are not specifically designed to reduce between-sample variation. For all features we calculated the pair-wise differences in intensity between spectra. We argue, that if these differences do not scatter around zero, this is evidence that for one out of a pair of spectra the concentrations are estimated systematically either too high or too low. To assess the performance of methods we calculated for each pair-wise comparison the ratio of the median of differences to the inter-quartile range (IQR) of differences and averaged the absolute values of these ratios across all pairs of samples (average median/IQR ratios). Dividing by the IQR ensures that the differences are assessed on comparable scales. The smaller the average median/IQR ratios are the better is the global between-sample normalization performance of a method. The results for the classification dataset are shown in the first row of Table 1.
Table 1.
Analysis of average inter- and intra-sample differences by means of interquartile ranges
| Crea-normalized/non-normalized | PQN | Cyclic loess | Contrast | Quantile | Linear baseline | Li-Wong | Cubic spline | Auto/pareto scaling | VSN | |
|---|---|---|---|---|---|---|---|---|---|---|
| Average median/IQR ratios | 0.46 | 0.04 | 0.06 | 0.55 | 0.06 | 0.15 | 0.82 | 0.07 | 0.28 | 0.07 |
| Average IQR ratios | 5.12 | 5.12 | 4.13 | 4.69 | 4.31 | 5.12 | 3.37 | 4.46 | 0.82 | 4.95 |
First row: Average ratios of the median to the IQR of the classification data. Lower values are favorable. Second row: Average ratios of the IQR of the spiked features to the IQR of the non-spiked features. Here, higher values are favorable. The two variable scaling methods performed equally and, therefore, are summarized in a single column
Comparing the list of average median/IQR ratios, PQN (0.04), Quantile (0.06), Cyclic Loess (0.06), VSN (0.07), and Cubic Spline (0.07) reduced overall differences between samples the best compared to the creatinine-normalized data only (0.46). The other methods, except for Contrast and Li-Wong Normalization, all improved the comparability between samples, but did not perform as well as the methods mentioned above. Note that the two variable scaling methods performed similarly and, therefore, were summarized as one entry in Table 1. The good performance of the VSN method can be explained by the fact that VSN combines variance stabilization with between-sample normalization. In comparison to the creatinine-normalized data, Auto and Pareto Scaling also showed some improvement.
While good between-sample normalization is desirable, it should not be achieved at the cost of reducing the genuine biological signal in the data. We tested for this in the Latin-square data. By experimental design, all intensity fluctuations except for those of the spiked-in metabolites, which should stand out in each pair-wise comparison of spectra, are caused by measurement imprecision. That is, spike-in features must be variable, while all other features should be constant. We assessed this quantitatively by calculating the IQR of the spike-in feature intensities and dividing it by the IQR of the non-spike-in feature intensities (average IQR ratios). These ratios are given in the second row of Table 1. High values indicate a good separation between spiked and non-spiked data points and, therefore, are favorable.
For the non-normalized data a ratio of 5.12 was obtained, i.e. the spike-in signal stood out clearly. These results were also obtained for the PQN and the Linear Baseline methods. For the Cyclic Loess, Quantile, Cubic Spline, Contrast, VSN, and Li-Wong approaches, the ratio was slightly reduced demonstrating that normalization might affect the true signals to some extent. Nevertheless, the signal-to-noise ratios for these methods were still above 4 and the signals kept standing out.
Importantly, Auto and Pareto Scaling compromised the signal-to-noise ratio severely. As for the classification data, the two variable scaling methods performed comparable and were summarized as one entry.
This prompted us to investigate systematically technical biases in this data (Analysis of intensity-dependent bias). As illustrated in Fig. 1a and b, M versus rank(A)-plots (M-rank(A)-plots) allow the identification of intensity-dependent shifts between pairs of feature vectors. Data in M-rank(A)-plots are log base 2 transformed so that a fold change of two corresponds to a difference of one. For each feature, its difference in a pair of samples (y-axis) is plotted against the rank of its mean value (x-axis). Hence, the x-axis corresponds to the dynamic spectrum of feature intensities, while the y-axis displays the corresponding variability of the intensities.
Fig. 1.
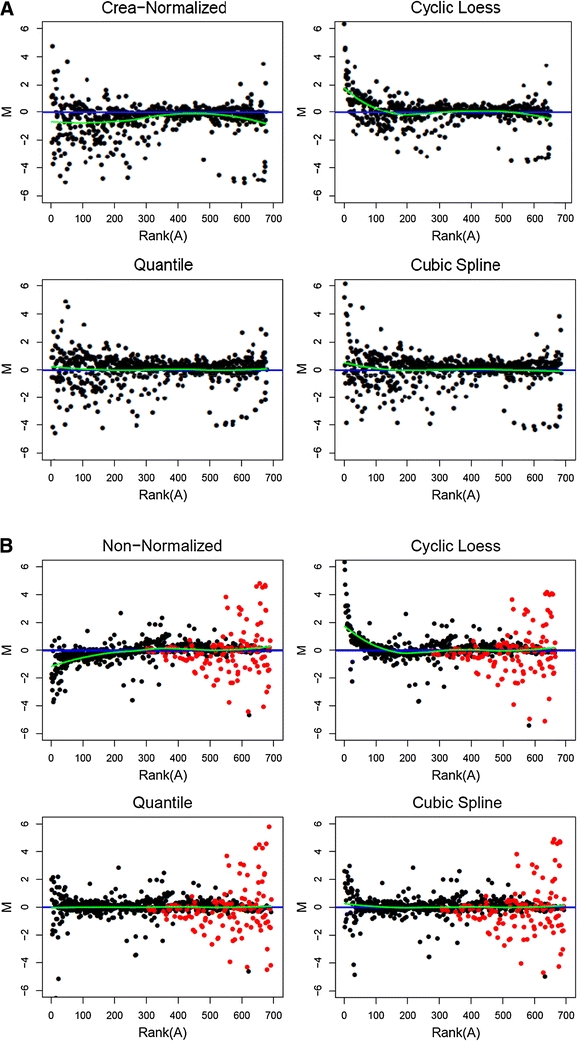
a M-rank(A)-plots comparing the same randomly selected pair of specimens from the classification dataset after creatinine normalization alone (top left) and after additional Cyclic Loess (top right), Quantile (lower left), and Cubic Spline Normalization (lower right). The straight line indicates M = 0, the curved line represents a loess fit of the data points. Deviations of the loess line from M = 0 correlate with bias between samples. The data is log base 2 transformed so that a fold change of two corresponds to a difference of one. b The same methods as above were applied to a pair of samples from the Latin-square spike-in dataset. Note, that for this dataset no prior creatinine normalization was performed and, therefore, in the top left part of the Figure results obtained from non-normalized data are displayed. The black dots represent background features that should not vary, while the differently marked dots, mostly found on the right hand side, represent features for which spike-in differences are expected. Therefore, they preferably stand out from the non-spike-in background
For all possible pair-wise comparisons of spectra and all investigated normalization methods, M-rank(A)-plots were produced from the classification data as well as from the Latin-square data. Representative sets of plots for a randomly selected pair of spectra selected from each of the two datasets are displayed in Fig. 1a and b. Shown are plots for creatinine-normalized classification data, respectively non-normalized Latin-square data and for data after Cyclic Loess, Quantile and Cubic Spline Normalization. In the absence of bias, the points should align evenly around the straight line at M = 0. The additionally computed loess line (curved line) represents a fit of the data and helps to determine how closely the data approaches M = 0.
In the M-rank(A) plots of creatinine-normalized (Fig. 1a) and non-normalized data (Fig. 1b), the curved loess line clearly does not coincide with the straight line at M = 0. The plot of the creatinine-normalized classification data in Fig. 1a suggests that intensities in sample 2 of the pair are systematically overestimated at both ends of the dynamic spectrum but not in the middle. One might want to attribute this observation to a technical bias in the measurements. While we cannot proof directly that the observation originates indeed from a technical bias rather than biological variation, we will show later that correction for the effect improves the estimation of fold changes, the detection of differentially produced metabolites and the classification of samples.
Here, we first evaluated the normalization methods with respect to their performance in reducing such an effect. Looking at Cyclic Loess normalized data in Fig. 1a, the bias is gone for the mid and high intensities, however, in the low-intensity region additional bias is introduced affecting up to 20% of the data points. With Quantile and Cubic Spline Normalization nearly no deviation from M = 0 can be recognized in Fig. 1a, they seem to remove any bias almost perfectly. Similar trends were also observed for the other pair-wise comparisons within the classification data (plots not shown). Application of the other normalization methods to the classification data showed that PQN and VSN evened out most bias well, although they sometimes left the loess line s-shaped. The linear baseline method performed similarly, in that it only partially reduced bias. Contrast, Li-Wong and the two variable scaling methods hardly reduced bias at all.
The M-Rank(A) plots of the Latin-square data, of which 4 examples are shown in Fig. 1b, generally resemble those obtained for the classification data, except for one major difference: Here, we have a large amount of differential spike-in features representing a range of 2- to 128-fold changes. The spike-in differences should not be lost to normalization. Therefore, for better visualization, all empirical data points of the spiked-in metabolites were marked differently, while the non-differential data points were marked in black (Fig. 1b). Ideally, all data points corresponding to the spiked-in metabolites should all be found in the high- and mid-intensity range (A). Moreover, differences (M) should increase with increasing spike-in concentrations, resulting in a triangle-like shaped distribution of the data points corresponding to the spiked-in metabolites and the curved loess line staying close to M = 0. As expected, the spike-ins stood out clearly in the non-normalized data. This was also the case for the PQN, Cyclic Loess, Contrast, Quantile, Linear Baseline, Li-Wong, Cubic Spline and VSN normalized data but not for the variable scaling normalized data.
The performance of all methods with respect to correcting dynamic range related bias can be compared in Loess-Line Plots (Bolstad et al. 2003). In these plots we drew rank(A) (x-axis) against the differences of the average loess line to the baseline at M = 0 (y-axis). The average loess line was computed for each normalization method by a loess fit of the absolute loess lines of the Ranked MA-plots for all pairs of NMR spectra. Our plots are a variation of those used by Bolstad et al. (2003) and (Keeping and Collins 2011) in that we use rank(A) instead of A on the x-axis. Any local offset from zero indicates that the normalization method does not work properly in the corresponding part of the dynamic range.
We calculated these plots for both the classification data (Fig. 2a) and the spike-in data (Fig. 2b). Since in most cases similar trends were obtained for both datasets, the best performing methods will be discussed together if not stated otherwise. In the absence of normalization, an increasing offset with decreasing intensities is observed for the lower ranks of both datasets. Cyclic Loess Normalization reduced the distance for the mid intensities well, but it increased the offset for low intensities. Contrast, Quantile and VSN Normalization all removed the intensity-dependency of the offset well. Regarding the overall distance, Quantile Normalization reduced it best, followed by VSN. Contrast Normalization left the distance at a rather large value. Taken together, this analysis shows that intensity-dependent measurement bias can only be corrected by a few normalization approaches. Not surprisingly, these are methods that model the dynamic range of intensities explicitly.
Fig. 2.
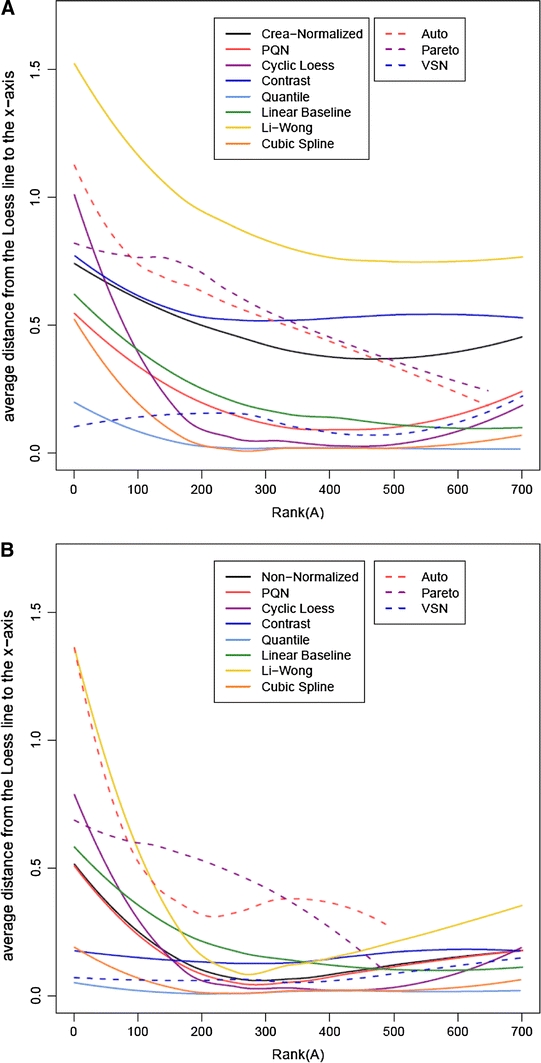
Ranked plot of the averaged loess line versus intensity A of the classification (a) and the spike-in (b) datasets for all normalization approaches. The lines were computed for each normalization method by a loess fit of the absolute loess lines of the M-rank(A)-plots for all sample pairs. Smaller and intensity-independent distances are preferable. The data is log base 2 transformed. For the methods involving centering not all features are well defined after logarithmic transformation leading to shorter average loess lines. Solid lines depict methods that are aimed at reducing sample-to-sample variations, while variable scaling and variance stabilization approaches are marked by dashed lines
M-rank(A)-plots can also detect unwanted heteroscedasticity, which may compromise the comparability of intensity changes across features. Spreading of the point cloud at one end of the dynamic range, as exemplified by the solely creatinine-normalized and non-normalized data, respectively, in Fig. 1a and b, indicates a decrease in the reliability of measurements. In the absence of evidence that these effects reflect true biology or are due to spike-ins (data points corresponding to spiked-in metabolites in Fig. 1b), one should aim at correcting this bias. Otherwise feature lists ranked by fold changes might be dominated by strong random fluctuations at the ends of the dynamic spectrum. Between-feature comparability will only be achieved, if the standard deviation of feature intensities is kept low over the entire dynamic spectrum.
The influence of the different normalization techniques on standard deviation relative to the dynamic spectrum was investigated using plots of the standard deviation for both the classification (Fig. 3a) and the Latin-square dataset (Fig. 3b). For this, the standard deviation of the logged data in a window of features with similar average intensities was plotted versus the rank of the averaged feature intensity, similarly to Irizarry et al. (Irizarry et al. 2003). The plots show for both the creatinine-normalized (Fig. 3a) and the non-normalized data (Fig. 3b), respectively, that standard deviation decreases with increasing feature intensity. The same is true for the PQN normalized data. Further, VSN keeps the standard deviation fairly constant over the whole intensity regime. In contrast, Li-Wong increases the standard deviation compared to the non-normalized data. The two variable scaling approaches increase standard deviation substantially.
Fig. 3.
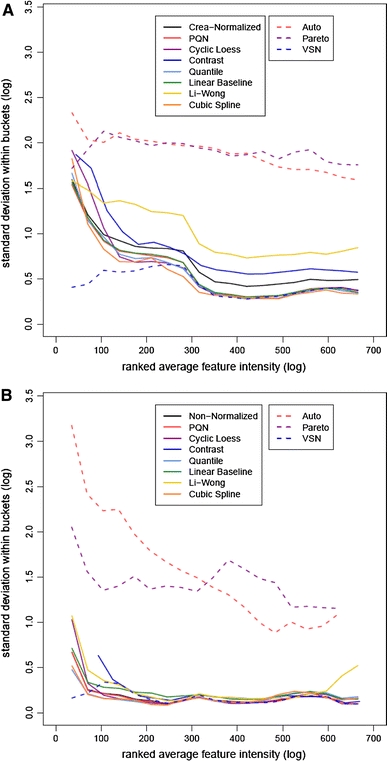
Plot of the logged standard deviation within the features versus the rank of the averaged feature intensity of the classification (a) and spike-in (b) datasets. To make the fit less sensitive to outliers, lines were computed using a running median estimator. The data is log base 2 transformed. Solid lines depict methods that are aimed at reducing sample-to-sample variations, while variable scaling and variance stabilization approaches are marked by dashed lines
Next, we investigated the influence of preprocessing on the detection of metabolites produced differentially, the estimation of fold changes from feature intensities, and the classification of samples based on urinary NMR fingerprints.
In the Latin-square data, we know by experimental design which features have different intensities and which do not. The goal of the following analysis is to detect the spike-in related differences and to separate them from random fluctuations among the non-spiked metabolites (Detection of Fold Changes). To that end, features with expected spike-in signals were identified and separated from background features. Excluded were features that were affected by the tail of spike-in signals, and regions in which several spike-in signals overlaid. As the background signal in the bins containing spike-in signals was, in general, not negligible, it was subtracted to avoid disturbances in the fold change measures.
Then, all feature intensities in all pairs of samples were compared and fold changes were estimated. Fold changes that resulted from a spike-in were flagged. Next, the entire list of fold changes was sorted. Ideally, all flagged fold changes should rank higher than those resulting from random fluctuations. In reality, however, flagged and non-flagged fold changes mix to some degree. Obviously, by design smaller spike-in fold changes tend to be surpassed by random fluctuations. The flagging was performed with three different foci, first flagging all spike-in features, then just low spike-in fold changes up to three and last only high fold changes above ten.
Receiving operator characteristic (ROC) curves with corresponding area under the curve (AUC) values were calculated for each normalization method and are given in Supplemental Table S2. Looking at the AUC values, only four methods yielded consistently better classification results than those obtained with the non-normalized data: Contrast, Quantile, Linear Baseline, and Cubic Spline Normalization. Quantile Normalization reached the highest AUC values in all runs, Cubic Spline and the Linear Baseline method showed comparable results and Contrast Normalization performed slightly better than the non-normalized data.
Differentially produced metabolites may be detected correctly even if the actual fold changes of concentrations are systematically estimated incorrectly. The ROC curves depend only on the order of fold changes but not on the actual values. This can be sufficient in hypothesis generating research but might be problematic in more complex fields such as metabolic network modeling. Therefore, we evaluated the impact of the preprocessing techniques on the accurate determination of fold changes. Based on published reference spectra, for each metabolite a set of features corresponding to the spike-in signals was determined, features with overlapping spike-in signals were removed and the background signal was subtracted. Within this set of features, the feature with the highest measured fold change among all pairs of samples with the highest expected fold change was chosen for evaluating the accuracy of determining the actual fold change for the respective metabolite. Note that the spike-in metabolite creatinine was excluded because of the absence of any non-overlapping spike-in bins. Then, plots of the spike-in versus the measured fold changes between all pairs of samples were computed for each metabolite and each normalization method. For taurine, Fig. 4 shows exemplary results obtained from non-normalized data and from data after Cyclic Loess, Quantile, and Li-Wong Normalization, respectively.
Fig. 4.
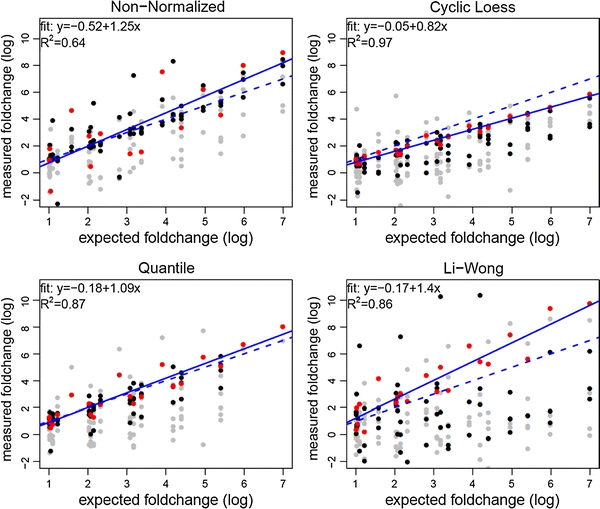
Plot of the reproducibility of determining spike-in fold changes for taurine from the Latin-square spike-in dataset without normalization (upper left), after Cyclic Loess (upper right), Quantile (lower left) and Li-Wong Normalization (lower right). Features at the border of the spike-in signal frequencies are represented by grey dots , whereas features from the inner range of the signals are plotted black. As detailed in the text, for each metabolite one feature was automatically selected. These features (marked differently) were used for fitting a linear model, which is given in the upper left corner of each plot. The solid lines represent the actual models, while the dashed lines represent ideal models with a slope of 1 and an intercept of 0. The data is log base 2 transformed
In analogy to Bolstad et al. (2003) the following linear model was used to describe the observed signal x of a bin i and a sample j:
| 1 |
Here, c 0 denotes the signal present without spike-in, c spike-in the spike-in concentration of the respective metabolite, γ the proportionality between signal intensity and spike-in concentration, which is assumed to be concentration independent within the linear dynamic range of the NMR spectrometer, and εij the residual error.
Comparing two samples j 1 and j 2 leads to the following linear equation, for which we estimate the intercept a and the regression slope b,
| 2 |
In Supplemental Table S3, slope estimates b for the different metabolites and normalizations are given. Again the variable scaling methods were summarized in a single entry. It is obvious that nearly all values exceed one, meaning that the fold changes are overestimated. This can be explained by the choice of features: As one metabolite generally contributes to several features and the feature with the highest fold change between the pair of samples with the highest spike-in difference is selected for each metabolite, features overestimating the fold change are preferred over features underestimating or correctly estimating the fold change. However, we still favored this automated selection algorithm over manually searching for the “nicest looking” bin, to minimize effects of human interference.
Apart from that, it can be seen from an analysis of the slope estimates b that normalization performs quite differently for different metabolites. The methods that showed the most uniform results for all metabolites investigated are Quantile, Contrast, Linear Baseline, and Cubic Spline Normalization.
In Supplemental Table S4, values for the intercept a, the slope b, and the coefficient of determination R 2 are given, averaged over all metabolites. The data shows that the methods that performed best in estimating accurately fold changes are Quantile and Cubic Spline Normalization.
Another common application of metabolomics is the classification of samples. To investigate the degree to which the different normalization methods exerted an effect on this task, the dataset consisting of the ADPKD patient group and the control group was used. Classifications were carried out using a support vector machine (SVM) with a nested cross-validation consisting of an inner loop for parameter optimization and an outer loop for assessing classification performance (Gronwald et al. 2011). The nested cross-validation approach yields an almost unbiased estimate of the true classification error (Varma and Simon 2006). For the nested cross validation, a set of n samples was selected randomly from the dataset. This new dataset was then normalized and classifications were performed as detailed above. Classification performance was assessed by the inspection of the corresponding ROC curves (Supplemental Fig. S2). The classification was conducted five times for every normalization method and classification dataset size n.
In Table 2, the AUC values and standard deviations of the ROC curves are given for all normalization methods and classification dataset sizes of n = 20, n = 40, n = 60, n = 80, and n = 100, respectively. As expected, the classification performance of most normalization methods depended strongly on the size of the training set used for classification. The method with the highest overall AUC value was Quantile Normalization: With 0.903 for n = 100, 0.854 for n = 80, and 0.812 for n = 60, it performed the best among the normalization methods tested, albeit for larger dataset sizes only. For dataset sizes n ≤ 40, its performance was about average. Cubic-Spline Normalization performed nearly as well as Quantile Normalization: It yielded the second highest AUC values for the larger training set sizes of n = 100 (0.892) and n = 80 (0.841). In contrast to Quantile Normalization, it also performed well for smaller dataset sizes: For n = 20 (0.740), it was the best performing method. VSN also showed good classification results over the whole dataset size range, its AUC values were barely inferior to those of the Cubic-Spline Normalization. Cyclic Loess performed almost as well as Quantile Normalization. For small dataset sizes, its classification results were only slightly better than average, but for the larger dataset sizes it was among the best-performing methods. Over the whole dataset size range, the classification results of PQN, Contrast and the Linear Baseline Normalizations and those of the variable scaling methods were similar to results obtained with creatinine-normalized data. Supplemental Table S5 gives the median (first column) and the mean number (second column) of features used for classification with respect to the applied normalization method. As can be seen, the number of selected features strongly depended on the normalization method used. The best performing Quantile Normalization led to a median number of 21 features, while the application of Cubic Spline Normalization and VSN resulted in the selection of 27 and 34 features, respectively. Employment of the PQN approach and the variable scaling methods resulted for the most part in a greater number of selected features without improving classification performance. The third column of Supplemental Table S5 gives the percentage of selected features that are identical to those selected by SVM following Quantile Normalization. As can be seen, PQN yielded about 95% of identical features, followed by Li-Wong and the Linear Baseline method with approx. 90% identical features. This data shows that the ranking of features based on t-values, which was the basis for our feature selection, is only moderately influenced by normalization. The smallest percentage (52.4%) of identical features was observed for Contrast Normalization, which also performed the poorest overall (Table 2).
Table 2.
Classification performance measured on classification dataset
| Normalization | n = 20 | n = 40 | n = 60 | n = 80 | n = 100 |
|---|---|---|---|---|---|
| Crea-normalized | 0.628 ± 0.074 | 0.722 ± 0.037 | 0.776 ± 0.032 | 0.783 ± 0.019 | 0.787 ± 0.003 |
| PQN | 0.710 ± 0.029 | 0.749 ± 0.034 | 0.781 ± 0.018 | 0.802 ± 0.016 | 0.796 ± 0.002 |
| Cyclic loess | 0.683 ± 0.029 | 0.728 ± 0.030 | 0.784 ± 0.027 | 0.797 ± 0.012 | 0.859 ± 0.005 |
| Contrast | 0.611 ± 0.072 | 0.693 ± 0.046 | 0.718 ± 0.036 | 0.764 ± 0.018 | 0.757 ± 0.004 |
| Quantile | 0.688 ± 0.023 | 0.731 ± 0.043 | 0.812 ± 0.033 | 0.854 ± 0.009 | 0.903 ± 0.003 |
| Linear baseline | 0.661 ± 0.034 | 0.728 ± 0.020 | 0.777 ± 0.027 | 0.790 ± 0.019 | 0.756 ± 0.005 |
| Li-Wong | 0.607 ± 0.036 | 0.659 ± 0.024 | 0.723 ± 0.043 | 0.771 ± 0.029 | 0.804 ± 0.005 |
| Cubic spline | 0.740 ± 0.066 | 0.749 ± 0.040 | 0.793 ± 0.018 | 0.841 ± 0.010 | 0.892 ± 0.003 |
| Auto | 0.705 ± 0.032 | 0.703 ± 0.020 | 0.764 ± 0.020 | 0.772 ± 0.011 | 0.789 ± 0.006 |
| Pareto | 0.652 ± 0.037 | 0.717 ± 0.038 | 0.757 ± 0.032 | 0.796 ± 0.010 | 0.785 ± 0.008 |
| VSN | 0.721 ± 0.022 | 0.772 ± 0.013 | 0.790 ± 0.015 | 0.838 ± 0.009 | 0.887 ± 0.003 |
Averaged AUC values and their standard deviation for the classification performance obtained for different sizes of the classification dataset and following different normalization methods. In all cases a SVM with nested cross-validation was employed
We also investigated the impact of the use of creatinine as a scale basis for renal excretion by subjecting the classification data to direct normalization by Quantile and Cubic-Spline Normalization without prior creatinine normalization. For n = 100, AUC values of 0.902 and 0.886 were obtained for Quantile and Cubic-Spline Normalization, respectively. These values are very similar to those obtained for creatinine-normalized data, which had been 0.903 and 0.892 for Quantile and Cubic-Spline Normalization, respectively. However, without prior creatinine–normalization an increase in the average number of selected features was noticed, namely from 21 to 31 and 27 to 36 features, respectively, for Quantile and Cubic-Spline Normalization. In summary one can say that Quantile and Cubic-Spline Normalization are the two best performing methods with respect to sample classification irrespective whether prior creatinine normalization has been performed or not.
Different preprocessing techniques have also been evaluated with respect to the NMR analysis of metabolites in blood serum (de Meyer et al. 2010). Especially, Integral Normalization, where the total sum of the intensities of each spectrum is kept constant, and PQN were tested in combination with different binning approaches. PQN fared the best, but it was noted that none of the methods tested yielded optimal results, calling for improvements in both spectral data acquisition and preprocessing. The PQN technique was also applied to the investigation of NMR spectra obtained from cerebrospinal fluid (Maher et al. 2011).
Several of the preprocessing techniques compared here have been also applied to mass spectrometry-derived metabolomic data and proteomics measurements. Van den Berg et al. (2006) applied 8 different preprocessing methods to GC-MS data. These included Centering, Auto Scaling, Range Scaling, Pareto Scaling, Vast Scaling, Level Scaling, Log Transformation and Power Transformation. They found, as expected, that the selection of the proper data pre-treatment method depended on the biological question, the general properties of the dataset and the subsequent statistical data analysis method. Despite these boundaries, Auto Scaling and Range Scaling showed the overall best performance. For the NMR metabolomic data presented here, the latter two methods were clearly outperformed by Quantile, Cubic Spline and VSN Normalization, all of which were not included in the analysis of the GC-MS data. In the proteomics field, Quantile and VSN normalization are commonly employed (Jung 2011).
Concluding remarks
In this study, normalization methods, different in aim, complexity and origin, were compared and evaluated using two distinct datasets focusing on different scientific challenges in NMR-based metabolomics research. Our main goal was to give researchers recommendations for improved data preprocessing.
A first finding is that improper normalization methods can significantly impair the data. The widely used variable scaling methods were outperformed by Quantile Normalization, which was the only method to perform consistently well in all tests conducted. It removed bias between samples, and accurately reproduced fold changes. Its only flaw was its mediocre classification result for small training sets. Therefore, we recommend it for dataset sizes of n ≥ 50 samples.
For smaller datasets, Cubic Spline Normalization represents an appropriate alternative. We showed that its bias removal and fold change reproduction properties were nearly equal to Quantile Normalization. Moreover, it classified well irrespectively of the dataset size.
VSN also represents a reasonable choice. Concerning the ADPKD data, it showed good results for both classification and bias removal. Concerning the spike-in data it performed less convincingly; however, the spike-in design affects the normalization procedure strongly by inducing additional variance. In conclusion, we found that preprocessing methods originally developed for DNA microarray analysis performed superior.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank Drs. Raoul Zeltner, Bernd-Detlef Schulze and Kai-Uwe Eckardt for providing the urine specimens used for the analysis of classification results. In addition the authors are grateful to Ms. Nadine Nürnberger and Ms. Caridad Louis for assistance in sample preparation. This study was funded in part by BayGene and the intramural ReForM program of the Medical Faculty at the University of Regensburg. The authors who have taken part in this study declare, that they do not have anything to disclose regarding funding from industry or conflict of interest with respect to this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Altman DG, Bland JM. Measurement in medicine: The analysis of method comparison studies. Journal of the Royal Statistical Society: Series D (The Statistican) 1983;32:307–317. [Google Scholar]
- Anderle M, Roy S, Lin H, Becker C, Joho K. Quantifying reproducibility for differential proteomics: Noise analysis for protein liquid chromatography-mass spectrometry of human serum. Bioinformatics. 2011;20:3575–3582. doi: 10.1093/bioinformatics/bth446. [DOI] [PubMed] [Google Scholar]
- Astrand M. Contrast normalization of oligonucleotide arrays. Journal of Computational Biology. 2003;10:95–102. doi: 10.1089/106652703763255697. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Clarke CJ, Haselden JN. Metabolic profiling as a tool for understanding mechanisms of toxicity. Toxicologic Pathology. 2008;36:140–147. doi: 10.1177/0192623307310947. [DOI] [PubMed] [Google Scholar]
- Cleveland WS, Devlin SJ. Locally weighted regression—An approach to regression-analysis by local fitting. Journal of the American Statistical Association. 1988;83:596–610. [Google Scholar]
- Cloarec O, Dumas M-E, Craig A, Barton R, Trygg J, Hudson J, Blancher C, Gauguier D, Lindon JC, Holmes E, Nicholson JK. Statistical total correlation spectroscopy: An exploratory approach for latent biomarker identification from metabolomic 1H NMR data sets. Analytical Chemistry. 2005;77:1282–1289. doi: 10.1021/ac048630x. [DOI] [PubMed] [Google Scholar]
- Cloarec O, Dumas ME, Trygg J, Craig A, Barton RH, Lindon JC, Nicholson JK, Holmes E. Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H NMR spectroscopic metabonomic studies. Analytical Chemistry. 2005;77:517–526. doi: 10.1021/ac048803i. [DOI] [PubMed] [Google Scholar]
- de Meyer T, Sinnaeve D, van Gasse B, Rietzschel E-R, De Buyzere ML, Langlois MR, Bekaert S, Martins JC, Van Criekinge W. Evaluation of standard and advanced preprocessing methods for the univariate analysis of blood serum 1H-NMR spectra. Analytical and Bioanalytical Chemistry. 2010;398:1781–1790. doi: 10.1007/s00216-010-4085-x. [DOI] [PubMed] [Google Scholar]
- Development Core Team, R. (2011). R: A language and environment for statistical computing.
- Dieterle F, Riefke B, Schlotterbeck G, Ross A, Senn H, Amberg A. NMR and MS methods for metabolomics. Methods in Molecular Biology. 2011;691:385–415. doi: 10.1007/978-1-60761-849-2_24. [DOI] [PubMed] [Google Scholar]
- Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dillution of complex biological mixtures. Application to 1H NMR metabolomics. Analytical Chemistry. 2006;78:4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Yang YH, Callow MJ, Speed TP. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Statistica Sinica. 2002;12:111–139. [Google Scholar]
- Durbin BP, Hardin JS, Hawkins DM, Rocke DM. A variance stabilizing transformation for gene-expression microarray data. Bioinformatics. 2002;18 suppl. 1:S105–S110. doi: 10.1093/bioinformatics/18.suppl_1.S105. [DOI] [PubMed] [Google Scholar]
- Eriksson L, Antti H, Gottfries J, Holmes E, Johansson E, Lindgren F, Long I, Lundstedt T, Trygg J, Wold S. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (gpm) Analytical and Bioanalytical Chemistry. 2004;380:419–429. doi: 10.1007/s00216-004-2783-y. [DOI] [PubMed] [Google Scholar]
- Forshed J, Schuppe-Koistinen I, Jacobsson SP. Peak alignment of NMR signals by means of a genetic algorithm. Analytica Chimica Acta. 2003;487:189. doi: 10.1016/S0003-2670(03)00570-1. [DOI] [Google Scholar]
- Gronwald W, Klein MS, Kaspar H, Fagerer S, Nürnberger N, Dettmer K, Bertsch T, Oefner PJ. Urinary metabolite quantification employing 2D NMR spectroscopy. Analytical Chemistry. 2008;80:9288–9297. doi: 10.1021/ac801627c. [DOI] [PubMed] [Google Scholar]
- Gronwald W, Klein MS, Zeltner R, Schulze B-D, Reinhold SW, Deutschmann M, Immervoll A-K, Böger CA, Banas B, Eckardt K-U, Oefner PJ. Detection of autosomal polycystic kidney disease using NMR spectroscopic fingerprints of urine. Kidney International. 2011;79:1244–1253. doi: 10.1038/ki.2011.30. [DOI] [PubMed] [Google Scholar]
- Holmes E, Foxall PJD, Spraul M, Farrant RD, Nicholson JK, Lindon JC. 750 MHz 1H NMR spectroscopy characterisation of the complex metabolic pattern of urine from patients with inborn errors of metabolism: 2-hydroxyglutaric aciduria and maple syrup urine disease. Journal of Pharmaceutical and Biomedical Analysis. 1997;15:1647–1659. doi: 10.1016/S0731-7085(97)00066-6. [DOI] [PubMed] [Google Scholar]
- Huber W, Heydebreck AV, Sültmann H, Poustka A, Vingron M. Variance stabilisation applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18:S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.S96. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JE. A user’s guide to principal components. Hoboken, NJ: Wiley-Interscience; 2003. [Google Scholar]
- Jung K. Statistics in experimental design, preprocessing, and analysis of proteomics data. Methods in Molecular Biology. 2011;696:259–272. doi: 10.1007/978-1-60761-987-1_16. [DOI] [PubMed] [Google Scholar]
- Keeping AJ, Collins RA. Data variance and statistical significance in 2D-gel electrophoresis and DIGE experiments: Comparison of the effects of normalization methods. Journal of Proteome Research. 2011;10:1353–1360. doi: 10.1021/pr101080e. [DOI] [PubMed] [Google Scholar]
- Klein MS, Dorn C, Saugspier M, Hellerbrand C, Oefner PJ, Gronwald W. Discrimination of steatosis and NASH in mice using nuclear magnetic resonance spectroscopy. Metabolomics. 2011;7:237–246. doi: 10.1007/s11306-010-0243-6. [DOI] [Google Scholar]
- Laywine CF, Mullen GL. Discrete mathematics using Latin squares. New York: Wiley; 1998. [Google Scholar]
- Lewis IA, Schommer SC, Hodis B, Robb KA, Tonelli M, Westler WM, Sussman MR, Markley JL. Method for determining molar concentrations of metabolites in complex solutions from two-dimensional 1H–13C NMR spectra. Analytical Chemistry. 2007;79:9385–9390. doi: 10.1021/ac071583z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., & Wong, W. H. (2001). Model-based analysis of oligonucleotide arrays: Model validation, design issues and standard error application. Genome Biology, 2, research0032. [DOI] [PMC free article] [PubMed]
- Lindon JC, Holmes E, Nicholson JK. Metabolomics in pharmaceutical R & D. FEBS Journal. 2007;274:1140–1151. doi: 10.1111/j.1742-4658.2007.05673.x. [DOI] [PubMed] [Google Scholar]
- Maher AD, Cysique LA, Brew BJ, Rae CD. Statistical integration of 1H NMR and MRS data from different biofluids and tissue enhances recovery of biological information from individuals with HIV-1 infection. Journal of Proteome Research. 2011;10:1737–1745. doi: 10.1021/pr1010263. [DOI] [PubMed] [Google Scholar]
- Parsons HM, Ludwig C, Günther UL, Viant MR. Improved classification accuracy in 1- and 2-dimensional NMR metabolomics data using the variance stabilising generalised logarithm transformation. BMC-Bioinformatics. 2007;8:234. doi: 10.1186/1471-2105-8-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: Visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- Stoyanova R, Nicholls AW, Nicholson JK, Lindon JC, Brown TR. Automatic alignment of individual peaks in large high-resolution spectral data sets. Journal of Magnetic Resonance. 2004;170:329–335. doi: 10.1016/j.jmr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC-Genomics. 2006;7:142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma S, Simon R. Bias in error estimation when using cross-validation for model selection. BMC-Bioinformatics. 2006;7:91. doi: 10.1186/1471-2105-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted profiling: Quantitative analysis of 1H NMR metabolomics data. Analytical Chemistry. 2006;78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- Wishart DS. Computational approaches to metabolomics. Methods in Molecular Biology. 2010;593:283–313. doi: 10.1007/978-1-60327-194-3_14. [DOI] [PubMed] [Google Scholar]
- Workman, C., Jensen, L. J., Jarmer, H., Berka, R., Gautier, L., Nielser, H. B., et al. (2002). A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Bioogy, 3, research0048. [DOI] [PMC free article] [PubMed]
- Zhang S, Zheng C, Lanza IR, Nair KS, Raftery D, Vitek O. Interdependence of signal processing and analysis of urine 1H NMR spectra for metabolic profiling. Analytical Chemistry. 2009;81:6080–6088. doi: 10.1021/ac900424c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


