Abstract
Mitochondrial DNA (mtDNA) lineages of macro-haplogroup L (excluding the derived L3 branches M and N) represent the majority of the typical sub-Saharan mtDNA variability. In Europe, these mtDNAs account for <1% of the total but, when analyzed at the level of control region, they show no signals of having evolved within the European continent, an observation that is compatible with a recent arrival from the African continent. To further evaluate this issue, we analyzed 69 mitochondrial genomes belonging to various L sublineages from a wide range of European populations. Phylogeographic analyses showed that ∼65% of the European L lineages most likely arrived in rather recent historical times, including the Romanization period, the Arab conquest of the Iberian Peninsula and Sicily, and during the period of the Atlantic slave trade. However, the remaining 35% of L mtDNAs form European-specific subclades, revealing that there was gene flow from sub-Saharan Africa toward Europe as early as 11,000 yr ago.
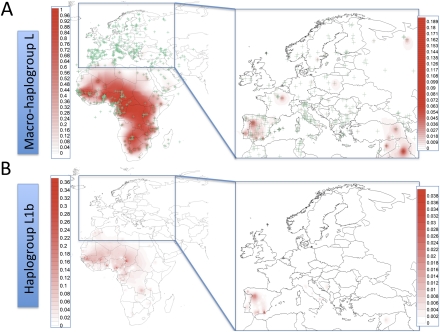
Analysis of the mitochondrial DNA (mtDNA) variation has been successfully used for reconstructing ancestral and modern human migrations. Most of the sub-Saharan African mtDNAs fall into one of the multiple subclades of macro haplogroup L (excluding M and N that derived from L3 and represent the branches from which the entire non-sub-Saharan mtDNA diversity arose). Previous studies (Salas et al. 2004) showed that L haplotypes account for <1% of mtDNAs in Europe (Fig. 1), with L1b being the most common haplogroup (Fig. 2). However, frequencies vary widely, ranging from 3% in Southern Europe (with a peak of 4%–6% in Iberia) to 0.7% in Central Europe and 0.5% in Northern Europe. Moreover, it has been proposed that their arrival into Europe most likely occurred in recent times, during the Romanization period, the Arab/Berber conquest of Iberia and South Italy, and more recently during the Atlantic slave trade. This hypothesis mainly relies on the observation that most of the L-mtDNAs in Europe represent patchy members of the African phylogeny as randomly taken from different African source populations (Fig. 3), without evidence indicating accumulation of molecular divergence within the European continent.
Figure 1.
Spatial haplogroup distribution of sub-Saharan African lineages in Europe based on control-region data. (A) Macro-haplogroup L; (B) haplogroup L1b. Green crosses in A indicate the sampled regions (see also Supplemental Data S1).
Figure 2.
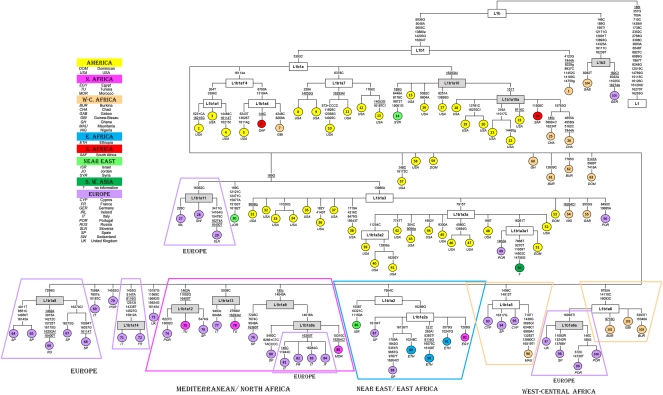
Maximum parsimony tree of L1b mtDNA genomes. The mutations are displayed along the branches; numbering is according to the revised Cambridge Reference Sequence (Andrews et al. 1999); uppercase letters (A, C, G, T) indicate transitions, whereas lowercase letters refer to transversions. (+) Insertions; (d) deletions. An additional suffix “!” indicates a back mutation. Population codes are indicated in the legend within the figure. Branches shaded in gray indicate subclades that are reported for the first time in the present study.
Figure 3.
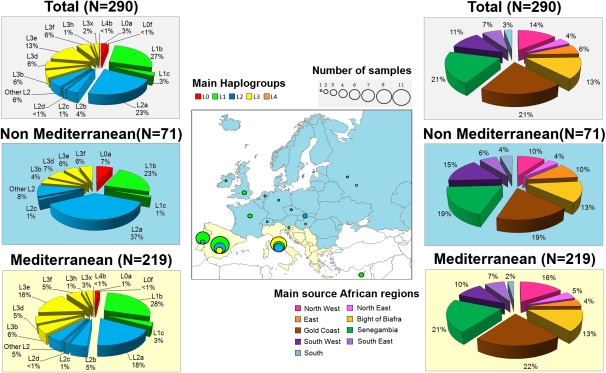
The pie charts on the left indicate the frequency distribution of African haplogroups in Europe; the color circles within the map indicate the distribution of entire genomes in Europe clustered in main haplogroups. The pie charts on the right show the admixture components of L-European lineages in Africa.
The mitochondrial genetic imprint of the African trans-Atlantic slave routes to the Americas received substantial attention in the literature (Salas et al. 2004, 2005; Veeramah et al. 2010; Stefflova et al. 2011), but its impact on the European gene pool was poorly evaluated, probably due to the scantiness of the data. Salas et al. (2004) surveyed the mtDNA hypervariable segment I (HVS-I) of about 15,000 individuals from Europe and found 113 mtDNAs of African ancestry (including haplogroups U6, M1, and L). According to these investigators, African mtDNAs in Eurasia can be broadly attributed to gene flow from Eastern, Western, and Southeastern Africa, but neither the proportions nor their arrival time in Europe were estimated. Based on the analysis of control-region data compiled from the literature, Malyarchuk and Czarny (2005) proposed that most of the European L-lineages might have arrived in Europe very recently, with the exception of two monophyletic clusters of L1b and L3b that could be older (no more than 6.5 thousand years ago [kya]) (Malyarchuk and Czarny 2005). In a follow-up study, the initial findings were not replicated, but a new subclade was identified (L2a1k; initially named as L2a1a) that could have specifically evolved within Europe ∼10 kya (Malyarchuk et al. 2008). Other studies undertaken in populations from Iberia tried to explain the origin of the observed L lineages (Pereira et al. 2000; Larruga et al. 2001; A´lvarez-Iglesias et al. 2009), concluding that virtually all of these probably arrived in recent times, the majority as a result of the slave trade between the 15th and 19th Centuries. It is worth mentioning that genetic links between Europe and North Africa are well established, supporting ancient migratory event from the Franco-Cantabrian refuge toward the Mediterranean regions of Africa ∼9 kya (Achilli et al. 2005; Pereira et al. 2010).
In this study, we have conducted the largest analysis of complete mtDNA genomes belonging to haplogroup L from Europe to date (n = 69) in order to unravel their temporal and geographic origin, under the null hypothesis that most of these lineages could have arrived in Europe very recently. Given that the targeted L mtDNAs are of undisputed sub-Saharan origin, these lineages signal the migrations of peoples moving from sub-Saharan locations to European destinations, but not in the opposite direction. Analysis of a panel of ancestry informative markers (AIMs) was used to investigate the presence of undetected individuals with recent genomic African ancestry.
Results
The phylogeny of European haplogroup L mitogenomes
The phylogenetic relationships of the 56 novel and 13 previously reported L mitochondrial genomes from Europe are reported in Supplemental Figure S1. Several novel minor L haplogroups that are not present in Africans (or in African-Americans) were detected. This finding, together with the fact that these haplogroups show a certain level of molecular divergence, suggests that these branches could have evolved within Europe. One of the most singular European haplogroup L subclades is L3d1b1a, defined by the stable diagnostic transversion A8014C (Soares et al. 2009). We found L3d1b1a exclusively in Italy (five subjects from Tuscany, Umbria, and Campania) (Supplemental Table S1), thus most likely it evolved in situ. All individuals carried different haplotypes suggesting local divergence dating back to 3.6 kya (95% C.I.: 2.2–5.1) (Supplemental Table S2).
Malyarchuk et al. (2008) recently proposed that the subclade L2a1k, defined by the mutational motif G6722A-T12903C-C16218T-T16519C, could have originated in Europe ∼10,300 ± 5150 yr ago. We have built a maximum parsimony tree using all available L2a1 entire mtDNA sequences (Supplemental Fig. S2; Supplemental Table S3). Our data provide further support to the finding of Malyarchuk et al. (2008) because we did not observe representatives of L2a1k in our collection of African sequences (n = 2426). We also searched L2a profiles carrying the diagnostic control-region variant C16218T in a large survey of African control-region sequences (>13,700), but no potential L2a1k candidates were observed. However, it is important to note that variant C16218T could be a misleading diagnostic marker for L2a1k. Two of our samples from Benin and Cameroon carrying this variant were completely sequenced and attributed to different clades, L2a1c8 and L2a1d (Supplemental Data S1; Supplemental Fig. S2). The TMRCA for L2a1k of 10.6 kya (95% C.I.: 9.9–11.3) is very similar to the previous estimate (Malyarchuk et al. 2008).
L1b is by far the most common L-African lineage in Europe, 49% according to complete mitogenomes (a total of 34, of which 30 are newly reported here), and 23% according to control-region data. Previous studies mainly based on control-region sequences (Salas et al. 2002) showed that haplogroup L1b is most frequent and diverse in West-central Africa (Fig. 1).
To further evaluate haplogroup L1b mtDNAs, we collected 73 L1b entire genomes (mainly from Africans and African-Americans) from the literature and GenBank, which together with the 30 novel sequences from Europe sum to 103 (Supplemental Table S4).
The phylogeny of the 103 L1b mitogenomes is provided in Figure 2. The vast majority of the non-European lineages were sampled in North America (African-Americans and Hispanics; n = 40) and West-central Africa (n = 13), the latter being the most likely source for the majority of the L1b mtDNAs in America (Salas et al. 2005). Control-region data indicate that L1b haplotype diversity is highest in East Africa, but the values of nucleotide diversity and the average number of nucleotide differences are highest in Western Africa (Bight of Biafra) (Supplemental Table S5). Demographic movements from sub-Saharan Africa could have spread L1b to the North (∼1% and ∼5% in Northeast and Northwest Africa, respectively).
Given the phylogeny, the frequency, and the diversity patterns observed in Africa for L1b, it is likely that this haplogroup arose in West Africa, from where it moved to other African and non-African locations. There is a subclade of L1b defined by the transition A16289G (Fig. 2) and named here L1b1a2a, which could have originated later in East Africa (represented by three divergent sequences from Ethiopia: GenBank accession numbers EU092952, EU092942, and EU092950). L1b1a2a could have moved from East Africa to the North downstream the Nile shores toward Egypt (represented by the complete genome EU092775). The immediate ancestral node, L1b1a2 (Fig. 2), is represented by a single mitogenome observed in Israel (the Bedouin sequence EU092672) (Behar et al. 2008). There are two representatives of L1b1a2a in Spain (one of them in Galicia; Northwest Spain), which could have arrived during the period of the Atlantic slave trade or the Arab invasion of the Iberian Peninsula.
We have also identified a new subclade of L1b1a, here named L1b1a9, characterized by the transversion G185C and the transition T14040C (Fig. 2). In contrast to most of the L1b subclades, L1b1a9 has a clear North African and Mediterranean distribution. It perhaps originated in Northwest Africa (as represented by the Moroccan Jew sequence EU092667) and afterward moved to different European Mediterranean locations (mainly Iberia and Italy). Two L1b1a9 sequences were found in Iberia (Galicia and Catalonia), three in the Italian Peninsula, and one in France.
Another new L1b1a subclade, called L1b1a8, most likely evolved exclusively within Europe. Two alternative phylogenies are possible (Supplemental Fig. S3) given the phylogenetic apparent homoplasmy observed in sequence #66. We favored the phylogeny given in Figure 2 by using the weighting scheme provided by Soares et al. (2009) on positional mutation rates. This clade is defined by the transition A7298G. Figure 2 shows five European members of the L1b1a8 clade—three Andalusians, one Galician (present study), and one Russian (Malyarchuk et al. 2008). The L1b1a8 status was investigated in the mtDNA coding region SNP data analyzed by us (M Cerezo, L Gusmão, V Černý, A Carracedo, P Schneider, A Salas, unpubl.). Here, 43 mtDNAs were found to belong to L1b1a from a total of 2426 profiles (∼1.7%) representing different African and African-American donors. These L1b1a sequences were observed in America (African-American, African-Caribbean, Colombia, and Argentina) and Africa (Angola, Ghana, Morocco, Nigeria, Sierra Leone, Ivory Coast, Togo, Tanzania, and Mozambique), but none of them carried the transition A7298G, thus further supporting a European origin of L1b1a8.
Apart from L1b1a8, there are other minor new clades of L1b that might have originated in Europe. Members of haplogroup L1b1a11 were only found in North-central Europe (Ireland, Switzerland, and Slovenia), while L1b1a12 has representatives only in Iberia (Portugal and Catalonia). L1b1a6a is defined by a reversion at position 16093 (T to C, which is likely to be mutationally more stable than the common 16093 C-to-T transition), and it is present in two Portuguese, one Spaniard, and one individual from Wales. Finally, the immediate ancestor of L1b1a6a seems to have evolved in West-central Africa (as represented by two entire genomes from Burkina Faso and Guinea Bissau), and from there it could have spread into Europe through the Atlantic facade.
Admixture analysis
Admixture proportions were calculated based on control-region data as done previously (Supplemental Table S6; Salas et al. 2009; Sandoval et al. 2009). The main source populations for the European L lineages were from the West-central African coast (P0: Gold Coast + Senegambia + Bight of Biafra ≅ 54%), and North Africa (P0: North East + North West ≅ 18%) (Fig. 3; Supplemental Table S6). When estimating the admixture proportions of the L-European lineages observed in the Mediterranean Basin versus non-Mediterranean, the contribution of North Africa just increases slightly (Northwest: ∼21%) (Fig. 2). Supplemental Table S7 also indicates that Europe shares the highest amount of haplotypes with West-central Africa. These results suggest that a proportion of the ancient L lineages that arrived to Europe from sub-Saharan Africa most likely traveled following a coastal Atlantic route, while other lineages arrived to Mediterranean Europe through North Africa, previously crossing the Sahara desert, but without leaving any detectable imprint in present-day North African populations.
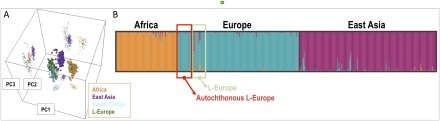
A panel of AIMs was genotyped in the same samples whose mtDNAs were completely sequenced (Supplemental Table S8) in order to estimate their three-way continental admixture genetic proportions (Africa, Asia, and Europe) and identify recent migrants whose presence in our data set would affect our results. Principal Component Analysis of AIM profiles identifies three clear-cut clusters (Fig. 4A), one of them grouping all Europeans (including L-European carriers with only one outlier; see comments below), and two other clusters representing East Asians and sub-Saharan Africans. Structure analysis on AIM profiles showed that the full sample of L-European donors analyzed in the present study harbored an average membership in the European cluster of ∼91% and reached ∼97% for the carriers of autochthonous L-European lineages (a value that is comparable with the average membership of other European populations in the same analysis, ∼98%). On the other hand, structure identifies only one European donor belonging to haplogroup L1b2 (the German individual #105 of Fig. 2) with a membership value of ∼98% in the sub-Saharan cluster (Fig. 4B: see the prominent orange bar in the European cluster of the structure bar plot). In addition, ancestry prediction was also performed based on maximum likelihood and the AIMs genotyping profiles (Supplemental Table S9). The results were again concordant with those obtained from PCA and structure (Pritchard et al. 2000): All of the L-European subjects were classified as Europeans, whereas the L1b2 German outlier was the only individual assigned to the African cluster (sample ID #32 in Supplemental Table S9).
Figure 4.
(A) 3D principal component analysis performed on samples analyzed in the present study (L-European carriers), Africans (n = 102; CEPH panel), East Asians (n = 229; CEPH panel), and South Europeans (n = 160; data set from Spain and Italy taken from SNPforID (Sánchez et al. 2006; Phillips et al. 2007), all genotyped for a panel of 34 AIMs. The genotyping data were compiled using SPSmart (Amigo et al. 2008). (B) Structure bar plot indicating ancestral components of L-European mtDNA carriers based on profiles derived from a set of 34 AIMs.
Discussion
In this study, we investigated the most likely origin of haplogroup L lineages of African origin in Europe. Given the criteria of autochthony used coupled with the results yielded by the analysis of ancestry based on AIMs, we could rule out (at least in our samples) the bias that recent immigrants could have introduced in the results.
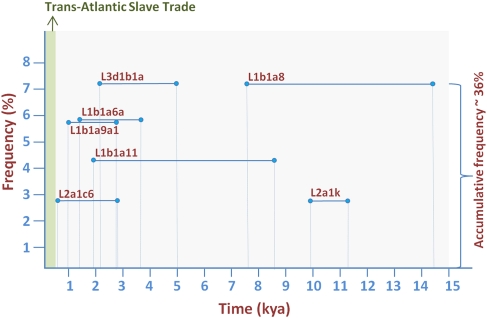
A large proportion (65%) of the African-European mtDNAs investigated could be attributed to modern and well-documented demographic routes that existed during the Romanization period, the Arab conquest, and the trans-Atlantic slave trade. However, there is strong evidence pointing to the fact that the remaining 35% of the L-European mtDNAs stand as modern witnesses of sporadic population movements occurring between the two continents that might have begun as early as 11,000 yr ago (Fig. 5). These contacts were not only restricted to North Africa, but connected sub-Saharan regions to Europe directly via coastal routes or first crossing North African territories toward the Mediterranean Sea.
Figure 5.
Diagram showing the coalescence ages of L-European lineages and their 95% C.I. (see also Supplemental Table S2) and the estimated frequencies in Europe over the total number of existing L mitochondrial genomes from Europe.
Previous studies (Achilli et al. 2005; Ottoni et al. 2010; Pereira et al. 2010) provided evidences of mtDNA flow from Europe toward North Africa during the Holocene; thus, North Africa would represent the Southern African edge of post–Last Glacial Maximum expansions spreading from European refugee. Here, we show, for the first time, genetic evidence signaling prehistorical movements in the opposite direction, from sub-Saharan Africa toward Europe. It is likely that most of the signals in the nuclear genome of this ancestral gene admixture between African immigrants and local Europeans had been erased by historical recombination and genetic drift. Therefore, as demonstrated in the present study, the mtDNA genome (and perhaps the Y chromosome) (Capelli et al. 2009) is the source to rescue the echoes of prehistorical sub-Saharan movements into Europe.
Methods
Sampling and sequence analysis
We analyzed a total of 69 haplogroup L-mitochondrial genomes from Europe, 56 of which were newly sequenced here. Two additional African genomes belonging to haplogroup L2a1 are also included in the study (see above and Supplemental Data S1). Most of the donors are from European Mediterranean regions, mainly from Iberia (Portugal and Spain) and the Italian Peninsula. Detailed information concerning the geographical origin of the grandparents was collected in order to exclude individuals whose ancestors arrived from Africa just recently (see Supplemental Data S1 for more details on the geographic origin of the donors). DNA extraction, PCR, and sequencing were performed as described in Cerezo et al. (2009).
In addition to the sequence of entire mtDNA genomes, all of the samples were genotyped for a panel of 34 AIMs according to Phillips et al. (2007) in order to investigate their autosomal genomic ancestry.
Genome data mining
A database of more than 9400 mtDNA genomes was compiled from the literature and GenBank in order to search for worldwide haplogroup L mitogenomes and perform interpopulation comparisons. For this purpose, we used the information provided by Phylotree Build 13 (van Oven and Kayser 2009).
In addition, almost 46,000 HVS-I control-region profiles of European (n = 27,494), Near Eastern (n = 4806), and African (n = 13,705) origin were collected following literature survey (Supplemental Data S1); 225 of those from Europe (∼0.8%) belonged to different sublineages of macro-haplogroup L (Fig. 1A). Several European haplogroup L sequences with dubious assignment into haplogroup L3 (n = 82; 27%) were disregarded because they lacked solid diagnostic markers.
Statistical analysis and molecular dating
Several maximum parsimony trees were built in this study for complete L genomes. The time to the most recent common ancestor (TMRCA) for each cluster was calculated by computing the averaged distance (ρ) of all the haplotypes in a clade to the respective root haplotype. Heuristic estimates of the standard error (σ) were calculated from an estimate of the genealogy (Saillard et al. 2000). Calculations were obtained using the entire mtDNA genomes but excluding hotspot mutations such as 16182C, 16183C, and 16519. Mutational distances were converted into years using the corrected evolutionary rate proposed by Soares et al. (2009). Howell et al. (2004) reported that African L-haplogroup sequences do not evolve in a clock-like fashion; however, the impact on our TMRCA estimates are probably negligible given that the oldest European L-haplogroup is ∼11,000 years old.
Principal Component Analysis (PCA) was performed on population frequencies of 34-plex SNP genotypes using R (http://www.r-project.org/). Analysis of ancestry was carried out using structure v.2.5 0. Group membership was assessed using runs of 200,000 Markov Chain steps after a burn-in of length 200,000 with five replicates for each value of K in the range 2–4. Posterior probabilities of K were calculated using the values of ln P(X|K), where X denotes the genotypes of the sampled individuals.
Spatial geographical representations of mtDNA haplogroup frequencies were obtained using Surfer 8.0 software (http://www.goldensoftware.com). We used the inverse-squared distance method for interpolating frequency values. The haplogroup frequencies are presented in a regular grid covering the region of interest (Africa and Europe).
Data access
GenBank accession numbers of mitochondrial genomes analyzed in the present study are JN214430–JN214454, JN214457–JN214480, JN225464–JN225468, and JN858955–JN858956.
Acknowledgments
This research received support from the Ministerio de Ciencia e Innovación (SAF2008-02971 and SAF2011-26983) and the European project “A European Initial Training Network on the History, Archaeology, and New Genetics of the Trans-Atlantic Slave Trade (EUROTAST)” (EU project: 290344) (to A.S.), Progetti Ricerca Interesse Nazionale 2009 (Italian Ministry of the University) (to A.A. and A.T.), FIRB-Futuro in Ricerca (Italian Ministry of the University) (to A.A. and A.O.), and the Fondazione Alma Mater Ticinensis (to A.T.). We thank Norman Angerhofer of the Sorenson Molecular Genealogy Foundation for data mining and queries of their mtDNA database. We are grateful to all of the donors for providing blood samples. This study was approved by the ethical committee of the University of Santiago de Compostela, and it conforms to the Spanish Law for Biomedical Research (Law 14/2007- 3 of July).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.134452.111.
References
- Achilli A, Rengo C, Battaglia V, Pala M, Olivieri A, Fornarino S, Magri C, Scozzari R, Babudri N, Santachiara-Benerecetti AS, et al. 2005. Saami and Berbers—an unexpected mitochondrial DNA link. Am J Hum Genet 76: 883–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- A´lvarez-Iglesias V, Mosquera-Miguel A, Cerezo M, Quintáns B, Zarrabeitia MT, Cuscó I, Lareu MV, García O, Pérez-Jurado L, Carracedo A´, et al. 2009. New population and phylogenetic features of the internal variation within mitochondrial DNA macro-haplogroup R0. PLoS ONE 4: e5112 doi: 10.1371/journal.pone.0005112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amigo J, Salas A, Phillips C, Carracedo A´ 2008. SPSmart: Adapting population based SNP genotype databases for fast and comprehensive web access. BMC Bioinformatics 9: 428 doi: 10.1186/1471-2105-9-428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N 1999. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23: 147 doi: 10.1038/13779 [DOI] [PubMed] [Google Scholar]
- Behar DM, Villems R, Soodyall H, Blue-Smith J, Pereira L, Metspalu E, Scozzari R, Makkan H, Tzur S, Comas D, et al. 2008. The dawn of human matrilineal diversity. Am J Hum Genet 82: 1130–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelli C, Onofri V, Brisighelli F, Boschi I, Scarnicci F, Masullo M, Ferri G, Tofanelli S, Tagliabracci A, Gusmão L, et al. 2009. Moors and Saracens in Europe: Estimating the medieval North African male legacy in southern Europe. Eur J Hum Genet 17: 848–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerezo M, Bandelt H-J, Martin-Guerrero I, Ardanaz M, Vega A, Carracedo A´, Garcia-Orad A, Salas A 2009. High mitochondrial DNA stability in B-cell chronic lymphocytic leukemia. PLoS ONE 4: e7902 doi: 10.1371/journal.pone.0007902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell N, Elson JL, Turnbull DM, Herrnstadt C 2004. African Haplogroup L mtDNA sequences show violations of clock-like evolution. Mol Biol Evol 21: 1843–1854 [DOI] [PubMed] [Google Scholar]
- Larruga JM, Diez F, Pinto FM, Flores C, Gonzalez AM 2001. Mitochondrial DNA characterisation of European isolates: The Maragatos from Spain. Eur J Hum Genet 9: 708–716 [DOI] [PubMed] [Google Scholar]
- Malyarchuk BA, Czarny J 2005. African DNA lineages in the mitochondrial gene pool of Europeans. Mol Biol 39: 703–709 [PubMed] [Google Scholar]
- Malyarchuk BA, Derenko M, Perkova M, Grzybowski T, Vanecek T, Lazur J 2008. Reconstructing the phylogeny of African mitochondrial DNA lineages in Slavs. Eur J Hum Genet 16: 1091–1096 [DOI] [PubMed] [Google Scholar]
- Ottoni C, Primativo G, Hooshiar Kashani B, Achilli A, Martinez-Labarga C, Biondi G, Torroni A, Rickards O 2010. Mitochondrial haplogroup H1 in north Africa: An early holocene arrival from Iberia. PLoS ONE 5: e13378 doi: 10.1371/journal.pone.0013378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Prata MJ, Amorim A 2000. Diversity of mtDNA lineages in Portugal: Not a genetic edge of European variation. Ann Hum Genet 64: 491–506 [DOI] [PubMed] [Google Scholar]
- Pereira L, Černý V, Cerezo M, Silva NM, Hájek M, Vašíková A, Kujanová M, Brdička R, Salas A 2010. Linking the sub-Saharan and West Eurasian gene pools: Maternal and paternal heritage of the Tuareg nomads from the African Sahel. Eur J Hum Genet 18: 915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C, Salas A, Sánchez JJ, Fondevila M, Gómez-Tato A, A´lvarez-Dios J, Calaza M, de Cal MC, Ballard D, Lareu MV, et al. 2007. Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci Int Genet 1: 273–280 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt H-J, Nørby S 2000. mtDNA variation among Greenland Eskimos: The edge of the Beringian expansion. Am J Hum Genet 67: 718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Richards M, De la Fé T, Lareu MV, Sobrino B, Sánchez-Diz P, Macaulay V, Carracedo A´ 2002. The making of the African mtDNA landscape. Am J Hum Genet 71: 1082–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Richards M, Lareu MV, Scozzari R, Coppa A, Torroni A, Macaulay V, Carracedo A´ 2004. The African diaspora: Mitochondrial DNA and the Atlantic slave trade. Am J Hum Genet 74: 454–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Carracedo A´, Richards M, Macaulay V 2005. Charting the ancestry of African Americans. Am J Hum Genet 77: 676–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Lovo-Gómez J, A´lvarez-Iglesias V, Cerezo M, Lareu MV, Macaulay V, Richards MB, Carracedo A´ 2009. Mitochondrial echoes of first settlement and genetic continuity in El Salvador. PLoS ONE 4: e6882 doi: 10.1371/journal.pone.0006882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez JJ, Phillips C, Børsting C, Balogh K, Bogus M, Fondevila M, Harrison CD, Musgrave-Brown E, Salas A, Syndercombe-Court D, et al. 2006. A multiplex assay with 52 single nucleotide polymorphisms for human identification. Electrophoresis 27: 1713–1724 [DOI] [PubMed] [Google Scholar]
- Sandoval K, Buentello-Malo L, Peñaloza-Espinosa R, Avelino H, Salas A, Calafell F, Comas D 2009. Linguistic and maternal genetic diversity are not correlated in Native Mexicans. Hum Genet 126: 521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P, Ermini L, Thomson N, Mormina M, Rito T, Röhl A, Salas A, Oppenheimer S, Macaulay V, Richards MB 2009. Correcting for purifying selection: An improved human mitochondrial molecular clock. Am J Hum Genet 84: 740–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefflova K, Dulik MC, Barnholtz-Sloan JS, Pai AA, Walker AH, Rebbeck TR 2011. Dissecting the within-Africa ancestry of populations of African descent in the Americas. PLoS ONE 6: e14495 doi: 10.1371/journal.pone.0014495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oven M, Kayser M 2009. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 30: E386–E394 [DOI] [PubMed] [Google Scholar]
- Veeramah KR, Connell BA, Pour NA, Powell A, Plaster CA, Zeitlyn D, Mendell NR, Weale ME, Bradman N, Thomas MG 2010. Little genetic differentiation as assessed by uniparental markers in the presence of substantial language variation in peoples of the Cross River region of Nigeria. BMC Evol Biol 10: 92 doi: 10.1186/1471-2148-10-92 [DOI] [PMC free article] [PubMed] [Google Scholar]