Abstract
Transcriptomic analyses have identified tens of thousands of intergenic, intronic, and cis-antisense long noncoding RNAs (lncRNAs) that are expressed from mammalian genomes. Despite progress in functional characterization, little is known about the post-transcriptional regulation of lncRNAs and their half-lives. Although many are easily detectable by a variety of techniques, it has been assumed that lncRNAs are generally unstable, but this has not been examined genome-wide. Utilizing a custom noncoding RNA array, we determined the half-lives of ∼800 lncRNAs and ∼12,000 mRNAs in the mouse Neuro-2a cell line. We find only a minority of lncRNAs are unstable. LncRNA half-lives vary over a wide range, comparable to, although on average less than, that of mRNAs, suggestive of complex metabolism and widespread functionality. Combining half-lives with comprehensive lncRNA annotations identified hundreds of unstable (half-life < 2 h) intergenic, cis-antisense, and intronic lncRNAs, as well as lncRNAs showing extreme stability (half-life > 16 h). Analysis of lncRNA features revealed that intergenic and cis-antisense RNAs are more stable than those derived from introns, as are spliced lncRNAs compared to unspliced (single exon) transcripts. Subcellular localization of lncRNAs indicated widespread trafficking to different cellular locations, with nuclear-localized lncRNAs more likely to be unstable. Surprisingly, one of the least stable lncRNAs is the well-characterized paraspeckle RNA Neat1, suggesting Neat1 instability contributes to the dynamic nature of this subnuclear domain. We have created an online interactive resource (http://stability.matticklab.com) that allows easy navigation of lncRNA and mRNA stability profiles and provides a comprehensive annotation of ∼7200 mouse lncRNAs.
High-throughput studies of mammalian genomes have revealed that most of the genome is transcribed in a complex manner, including the production of tens of thousands of long non-protein-coding RNAs (lncRNAs) (Carninci et al. 2005; Birney et al. 2007; Guttman et al. 2009, 2010). Despite rapid progress, only a small proportion of lncRNAs have been functionally characterized (Amaral et al. 2011), and little is known about most lncRNAs. One particularly poorly understood aspect of lncRNAs is their post-transcriptional regulation and metabolism in the cell. Despite an absence of genome-wide studies to examine lncRNA stability, it has been a general expectation that lncRNAs will be less stable than protein-coding mRNAs, both due to their lower average level of expression and the existence of known unstable classes of lncRNAs (Dinger et al. 2009a). These classes, including cryptic unstable transcripts (CUTs) in yeast (Wyers et al. 2005), upstream noncoding transcripts (UNTs) in Arabidopsis (Chekanova et al. 2007), and promoter upstream transcripts (PROMPTs) in humans (Preker et al. 2008), have supported the suggestion that many, if not most, lncRNA transcripts are highly unstable (Houseley and Tollervey 2009). However, the stability of the small number of functional lncRNAs that have been determined (Sheardown et al. 1997; Seidl et al. 2006; Sone et al. 2007; Askarian-Amiri et al. 2011), suggest that lncRNAs, like mRNAs, have a wide diversity of half-lives. However, genome-wide methods, which provide a more objective view of lncRNAs as a class, are required to test this hypothesis.
Previous studies have investigated mRNA stability genome-wide using microarray technology and RNA sequencing (Raghavan et al. 2002; Yang et al. 2003; Narsai et al. 2007; Friedel et al. 2009; Sharova et al. 2009; Rabani et al. 2011; Schwanhausser et al. 2011) but have not considered lncRNAs. These studies revealed a wide variation in stability between different mRNAs and its importance in combining with transcription to determine the steady-state levels of transcripts and also to control the speed and timing of changes in gene expression (Elkon et al. 2010; Rabani et al. 2011). For example, the low stability of groups of mRNAs, including those that encode transcription factors, is integral to their rapid dynamic regulation (Yang et al. 2003; Narsai et al. 2007; Friedel et al. 2009). Although changes in transcription are responsible for most dynamic changes in gene expression, changes in RNA stability also provide a mechanism to regulate gene expression levels (Blattner et al. 2000; Sharova et al. 2009; Rabani et al. 2011).
Here, we perform a genome-wide analysis of lncRNA stability using custom microarrays that examine ∼7200 lncRNAs together with ∼20,000 protein-coding transcripts. Although the average turnover of lncRNAs is higher than mRNAs, we find that lncRNAs display a wide range of stabilities comparable to that of protein-coding transcripts. The functionally characterized lncRNA Neat1 was also found to be highly unstable, highlighting the notion that high turnover is not refractory to function and rather can promote the dynamic nature of lncRNAs and the cellular constituents with which they are associated.
Results
Genome-wide determination of RNA half-lives
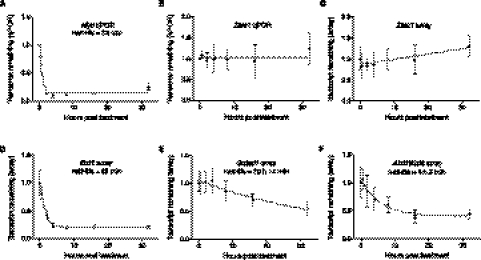
To examine the stability of lncRNAs, we inhibited transcription in mouse Neuro-2a (N2A) neuroblastoma cells with actinomycin D (Hurwitz et al. 1962) and measured RNA levels over a 32-h time course. In the absence of transcription, the percentage of an RNA remaining is determined by its rate of decay. Use of an extended time course, similar to that conducted for mRNAs in Arabidopsis (Narsai et al. 2007), allows improved half-life measurements for transcripts with slow decay rates (Friedel et al. 2009) compared to previous transcriptional inhibition experiments in mammals that were continued to only 8 h (Raghavan et al. 2002; Yang et al. 2003; Sharova et al. 2009). We validated the transcriptional blocking methodology by determining the stability of the Myc oncogene mRNA, which is known to have a very short half-life (Dani et al. 1984; Raghavan et al. 2002), by qPCR. Consistent with previous observations, Myc mRNA was highly unstable (Fig. 1A) with a half-life of 33 min (95% confidence interval 25–48 min).
Figure 1.
RNA half-life determination following transcription inhibition. (A,B) Transcript decay curves after blocking transcription in N2A with actinomycin D and measuring transcript remaining relative to a control gene by qPCR. Error bars show standard deviation. (A) Myc decay relative to Gapdh. Gapdh is a suitable control gene for transcripts that are not highly stable. Results are from four biological replicates, which were subsequently used for microarray analysis. The fitted curve was modeled by one-phase decay using nonlinear least squares regression. Myc expression was also tested in mock treated time courses, which did not show evidence of transcript decay. (B) Transcript decay curve for Zfas1 relative to Atp5e. Results from three biological replicates. No degradation is observed, and nonlinear regression supports a horizontal line fit. (C) Zfas1 expression over 32-h time course following transcription inhibition from microarray. Four biological replicates; error bars show standard deviations. Nonlinear regression was used to test model fits and supports a linear fit with a positive slope showing apparent up-regulation of expression by 32 h. (D–F) Decay curves and half-lives determined for two random mRNAs (D,E) and one lncRNA (F) transcript from microarrays. All were modeled using one-phase exponential decay. Error bars represent standard deviations.
Genome-wide analysis of lncRNA stability was conducted using NCode (Life Technologies) microarrays, which contain probes that target ∼7200 distinct lncRNA transcripts as well as probes that target most annotated protein-coding genes. As reported previously (Sharova et al. 2009), the most stable transcripts increased in relative abundance in samples from later time points and so give the artifactual appearance of up-regulation. Therefore, we normalized the scale of the array expression data to a pool of ultra-stable control transcript(s). Gapdh had been used to normalize shorter time courses in a previous study (Raghavan et al. 2002), but it is not stable over 32 h (Supplemental Fig. S1), and scaling to a single mRNA over a long time course could introduce noise. We recently identified the lncRNA Zfas1 (1500012F01Rik) as highly stable with no degradation over a 16-h time course (Askarian-Amiri et al. 2011) and were able to confirm by qPCR that its relative expression was virtually unchanged after 32 h (Fig. 1B). As expected, the microarray data showed a relative increase in Zfas1 expression by 32 h (Fig. 1C). Therefore, we selected six genes showing similar profiles to Zfas1 that also had previous evidence of high stability in mouse cells (Friedel et al. 2009). This high stability was confirmed by qPCR analysis of two of these mRNAs, Atp5e and Gstm1 (Supplemental Fig. S1), which showed little or no change in abundance over the time course. The average fold change between 0 h and each treated time point for these seven genes was adjusted to zero, and the intensity values of the probes were scaled proportionately to create the normalized time course data set.
We identified 14,987 transcripts that were expressed significantly above background (Supplemental Table S1), from which we calculated transcript half-lives by modeling each transcript with one-phase exponential decay or linear decay. As one-phase exponential decay is not optimal for modeling some highly stable transcripts, we calculated half-lives using both exponential decay and linear decay and selected the best model for each transcript (see Methods). Examination of transcript half-lives revealed that linear decay began to be favored for transcripts with half-lives above 18 h. After filtering to remove transcripts that could not be accurately modeled, we determined the half-lives for 12,670 transcripts (including 823 lncRNAs), (Supplemental Table S2). Examples of half-lives calculated from the microarray data are shown in Figure 1D–F.
To confirm modeled transcripts were within the dynamic range of the arrays, we compared transcript half-lives with their expression values at 0 h. We did not observe increased half-lives at low expression levels, which could have suggested that array background was preventing the decay of signal. This observation held true whether comparing all RNAs, or lncRNAs only (Supplemental Fig. S2). Indeed, there was a significant correlation (r = 0.2045, Spearman correlation) between higher expression and longer half-lives for all RNAs, but no correlation between lncRNA expression and half-life (Supplemental Fig. S2).
We validated that the arrays were sufficiently sensitive to detect unstable transcripts by comparing the calculated half-life for Myc as determined by qPCR to that determined by the microarrays, as well as the array stability of Znfx1, which we recently showed to be unstable in N2A cells by qPCR (Askarian-Amiri et al. 2011). The array-determined half-life of Myc was 34 min, which is in excellent agreement with qPCR, as was that of Znfx1 (65 min by array versus 50 min by qPCR). These data confirmed the accuracy of the array results and our ability to determine the half-lives of short-lived transcripts.
GO analysis of stable and unstable protein-coding genes
Previous studies have shown that unstable protein-coding transcripts are enriched for transcription factors, while mRNAs encoding proteins for various cellular metabolic processes are enriched amongst stable transcripts (Yang et al. 2003; Friedel et al. 2009; Sharova et al. 2009; Rabani et al. 2011). To assess the biological robustness of our data, we performed a GO analysis of protein-coding transcripts found within the unstable (half-life < 2 h) and highly stable (half-life > 12 h) fractions using Fatigo (Medina et al. 2010). Consistent with previous studies, we found that genes involved in transcription and regulation of gene expression were enriched in the unstable fraction, especially proteins with transcription factor activity, which were strongly overrepresented (P = 1.59 × 10−7), while genes involved in numerous cellular metabolic processes and with oxidoreductase activity were enriched amongst the highly stable mRNAs (Supplemental Table S3). Together, these observations suggest our experimental model for RNA decay is applicable for making biologically relevant interpretations.
Stability of lncRNAs
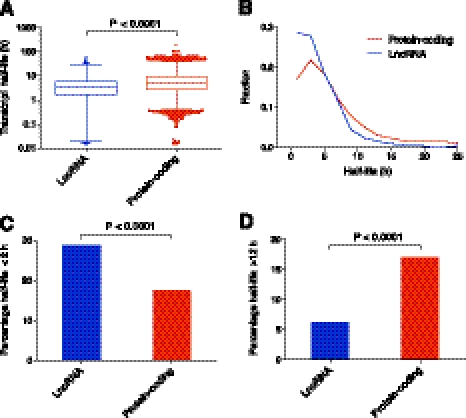
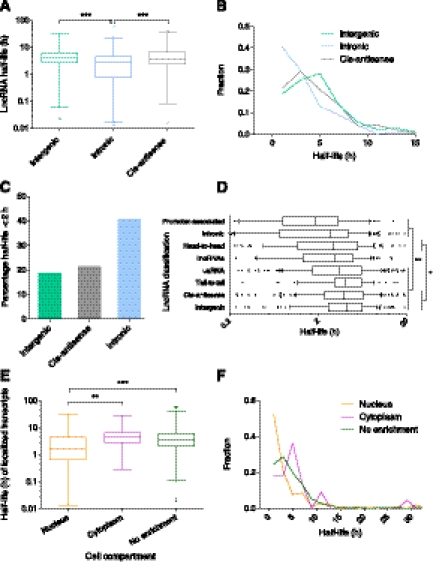
To examine the stability of lncRNAs and how they compare to mRNAs, we graphed the distribution of half-lives. Figure 2A reveals that transcript half-lives ranged from <30 min to >48 h. LncRNAs show a similar range of half-lives to protein-coding transcripts, suggesting that lncRNA stability is a regulated process. The median lncRNA half-life was 3.5 h (mean 4.8 h), whereas the median half-life for protein-coding transcripts was 5.1 h (mean 7.7 h). This result shows that lncRNAs are not unstable as a class, although their half-lives are, on average, shorter than protein-coding RNAs (P < 0.0001) (Fig. 2A). We also find that a higher percentage of lncRNAs are classified as unstable (29% versus 17%) (t½ < 2 h; P < 0.0001) (Fig. 2B,C) and a lower percentage as highly stable (6% versus 17%) (t½ > 12 h; P < 0.0001) (Fig. 2B,D).
Figure 2.
Half-lives of lncRNA and protein-coding transcripts. (A) Box-and-whisker plot of coding and lncRNA transcript half-lives. (Whiskers) 1st–99th percentile, with individual transcripts outside this shown as dots. (Box) 25th–75th percentile. Difference calculated using a nonparametric Mann-Whitney t-test. (B) Frequency distribution showing the fraction of protein-coding and lncRNA transcripts in 2-h bins. Plotted points are at the center of the 2-h bin. Only time points with 1% or more of transcripts are plotted. (C) Percentage of unstable (half-life under 2 h) lncRNA and protein-coding transcripts. Significant difference calculated using a χ2 test. (D) Percentage of highly stable (half-life over 12 h) lncRNA and protein-coding transcripts. Significant difference calculated using a χ2 test.
In total, we identified ∼240 unstable lncRNA transcripts (t½ < 2 h). Although most lncRNAs are yet to be annotated, this group contains a number of known and functionally validated lncRNAs. These include Neat1 (see below), the probable mouse Adapt33 (5430416N02Rik) homolog, a number of lncRNAs that associate with chromatin-binding proteins (Guttman et al. 2011) (see below), small RNA host transcripts including Mir17hg, an isoform of Rmst, and several snoRNA host genes including Snhg3, Snhg5, and one Snhg8 isoform, as well as the imprinted lncRNAs Kcnq1ot1 and Peg13. The low stability of several snoRNA host transcripts is in stark contrast to the extreme stability of Zfas1, which is also a snoRNA host gene.
Fifty-one lncRNAs had half-lives of over 12 h and were, therefore, classified as highly stable. These lncRNAs corresponded to a variety of genomic context classifications including intergenic, bidirectional with other transcripts, and intronic to protein-coding loci. Few have been characterized in detail with the exception of 4933436C20Rik (Linc1399), identified as involved in maintaining pluripotency in embryonic stem cells, where it associates with the chromatin-binding protein CBX3 (Guttman et al. 2011). Given that we have characterized the stability of only a subset of the total number of lncRNAs, these results suggest that there are a large number of highly stable lncRNAs. Although the function of these transcripts is generally unknown, genes with enzymatic and housekeeping functions are enriched amongst stable protein-coding RNAs, so we hypothesize that housekeeping lncRNAs will also show high stability.
Clustering of RNA decay profiles
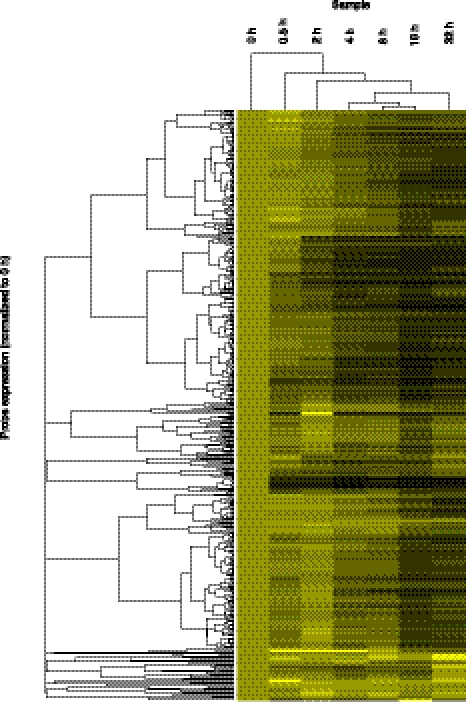
Transcripts of different stabilities provide a rich variety of decay profiles. To visualize these, we clustered all lncRNAs using unsupervised hierarchical clustering (Fig. 3). These results further reinforce the notion that lncRNAs are not generally unstable but rather show a wide range of stability profiles.
Figure 3.
Hierarchical clustering of transcript decay rates. Unsupervised hierarchical clustering of all lncRNA transcripts above the expression cut-off. Clustering was performed using cluster3 (de Hoon et al. 2004) and visualized in Java Treeview (Saldanha 2004). All transcripts were set to an expression level of 1 at 0 h, so clustering is determined only by decay rate. Transcripts that decay quickly turn black during the early time points; transcripts that show no degradation remain bright yellow. As clustering was performed on all transcripts above the expression cut-off, transcripts whose half-lives could not be determined are also included.
Previous analyses of coding transcript stability have revealed relationships between stability and gene function and that mRNAs that act in common biological pathways (same GO function) (Sharova et al. 2009) or proteins found within the same complexes (Friedel et al. 2009) often have similar stabilities. On this basis, we hypothesize that the stability of lncRNAs may be similarly related to their functional classes and that lncRNAs with related functions may tend to cluster together according to their stability profiles.
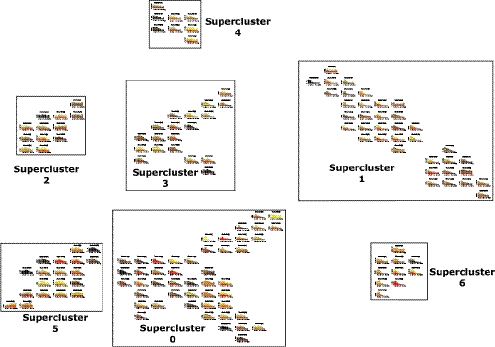
To reveal potential relationships between stability and function, we reclustered all expressed transcripts by their decay profiles using QAPgrid (Inostroza-Ponta et al. 2011). The decay profile for each individual cluster is shown in Figure 4. As a result of this optimization-based approach to visualization, the distance between each cluster in the final layout is related to the difference between the decay profiles. From our data set of 14,987 decay profiles, the method produced 152 clusters, which, in turn, aggregate into seven “super-clusters” containing related decay profiles. The 152 clusters contain from 2 to 4447 transcripts. Most transcripts are found in a relatively small number of clusters, with 29.7% present in the single most populous cluster (Cluster 0), which is part of the largest super-cluster (Super-cluster 0). Indeed, the ten most populous individual clusters contain 79.9% of all transcripts, from the generally highly unstable transcripts in Cluster 1 (1087 transcripts) through to the extremely stable transcripts in Clusters 15 (196 transcripts) and 27 (797 transcripts). Hence, it appears that a reasonably small number of clusters and super-clusters can define the dynamics of RNA degradation.
Figure 4.
Distance-based clustering of transcript decay rates. Transcripts are automatically clustered; with those showing indistinguishable decay profiles over the time course present in the same cluster, while transcripts with similar profiles are found in nearby clusters. The physical distance between individual clusters and between super-clusters represents the degree of difference in the decay profile. Clusters of clusters or super-clusters are created by applying the method used to form the clusters to the clusters themselves. Title gives the cluster number and the number of probes in the cluster. (X-axis) Cumulative expression of all probes in the cluster. Decay profiles are stacked bar graphs with every stack representing a separate transcript; when there are many transcripts in a cluster, the expression level of some transcripts cannot be individually visualized and are seen as areas of black (representing many transcripts). An interactive version of this figure can be found at http://stability.matticklab.com/.
To facilitate the browsing and visualization of transcript stabilities, we developed an interactive, web-accessible version of the QAPgrid representation of the clusters (http://stability.matticklab.com/). Each decay cluster is linked to a table of the transcripts it contains, together with their associated half-lives and further annotations including transcript classification, structural features, conservation, subcellular localization and expression information.
Comparing the annotated transcripts present in different super-clusters (i.e., independent of modeled half-lives) revealed a number of features overrepresented within individual super-clusters. For example, Super-cluster 1 showed an enrichment for genes encoding nuclear proteins (P < 5.72 × 10−60) involved in the regulation of transcription (P < 4.40 × 10−56) as estimated by g:profiler (Reimand et al. 2011), while a MEDLINE-based analysis using GATHER (Chang and Nevins 2006) revealed a significant enrichment of genes associated with the term “oncogene” (P < 0.0003) (Supplemental Table S4). Super-cluster 1 also had the highest proportion of lncRNAs (13.5%). Taken together these analyses demonstrate how QAPgrid clustering facilitates both the extraction of biologically meaningful data from, and visualization of, genome-wide data sets.
Combining clustering and RNA half-lives to identify potentially independent transcripts
The identification of transcripts from common loci with different half-lives or occurrence within different super-clusters provides a potential means to disentangle independent transcripts from complex loci. This is useful as such loci can contain numerous overlapping transcripts with alternative splice sites and varying start and end positions, which can be very challenging to distinguish.
We searched the four most populous super-clusters (0, 1, 3, and 5, containing 94% of clustered probes) for Unigene genes with multiple probes that clustered into more than two super-clusters. This analysis identified 21 genes (Supplemental Fig. S3; Supplemental Table S4). One gene identified was Tcf4 (transcription factor four). Two probes to constitutive splice junctions showed half-lives of 14.5 and 6.7 h, respectively, whereas three probes distributed across the 5-kb Tcf4 3′UTR (Supplemental Fig. S4) all identified a half-life of 3.4–3.5 h, The high concordance and divergent half-life of the 3′UTR suggests its regulation is independent of the common coding isoform, potentially due to processing of the 3′UTR (Mercer et al. 2011) or due to specific use of the 3′UTR by an alternative Tcf4 isoform. This differential stability across the gene locus would be difficult to detect by standard expression arrays or next-generation sequencing, but regulation of this nature is readily identifiable from transcript decay.
Another example that illustrates the value of stability profiling for disentanglement of transcripts is the noncoding snoRNA host gene Snhg4, which terminates ∼4 kb upstream of the coding gene Matr3. Both cDNA cloning (Bortolin and Kiss 1998) and next-generation sequencing (Guttman et al. 2010) suggest Snhg4 is commonly a 5′ upstream start site for Matr3 (Supplemental Fig. S4). However, our data show that Snhg4 is unstable (and found in Super-cluster 1), whereas Matr3 is highly stable (and found in Super-cluster 5), suggesting that these transcripts can be distinct and independently regulated.
Genomic location impacts lncRNA stability
Previous studies have defined a number of sequence elements that influence mRNA stability (Yang et al. 2003; Narsai et al. 2007; Sharova et al. 2009). To further understand how lncRNAs differ in stability and why they are, on average, less stable than mRNAs, we investigated the relationship between lncRNA stability and their corresponding sequence elements and features.
To analyze various features that could affect lncRNA stability, we created a comprehensive set of genome-wide lncRNA annotations (Supplemental Table S2), which represent the most detailed annotations of mouse lncRNAs to date. An important feature of lncRNAs is the genomic location they are transcribed from in relation to protein-coding genes (Mercer et al. 2009). On this basis, lncRNAs can be broadly classified as intronic, where they originate from within coding gene loci, cis-antisense, where they are transcribed from the opposite stand to other transcripts, or intergenic, where they are transcribed outside of, or between, coding genes (Supplemental Fig. S5). Comparison of the stability of intergenic, intronic and cis-antisense lncRNAs revealed significant differences between the groups, with intronic lncRNAs less stable than those from intergenic regions or antisense to other transcripts (P < 0.001) (Fig. 5A) with a much greater percentage of intronic lncRNAs classified as unstable (t½ < 2 h) (P < 0.0001) (Fig. 5B,C), although there was no difference between the number classified as highly stable (t½ > 12 h) (P = 0.34, χ2 test).
Figure 5.
Effect of lncRNA features on stability. (A) Comparison of the stability of intronic versus intergenic and cis-antisense lncRNAs. Box-and-whisker plot. (Whiskers) 1st–99th percentile, with individual transcripts outside this shown as dots. (Box) 25th–75th percentile. Difference calculated using one-way ANOVA with Kruskal-Wallis nonparametric test and Dunn's post-test to compare individual annotations. (B) Frequency distribution showing the fraction of lncRNA transcripts in 2-h bins. Plotted points are at the center of the 2-h bin. Only time points with 1% or more of transcripts are plotted. (C) Percentage of unstable (half-life under 2 h) lncRNA intergenic, cis-antisense, and intronic transcripts. Significant difference calculated using χ2 test. (D) Comparison of the stability of all lncRNA genomic classifications. Box-and-whisker plot. To focus on the center of the distribution, whiskers show 10th–90th percentile, with individual transcripts outside this shown as dots, and only half-lives between 0.2 h and 20 h are shown. Significant differences found between stability of classes using one-way ANOVA with Kruskal-Wallis nonparametric test and Dunn's post-test to compare individual annotations. (*) Level of significance common to all comparisons. (E) Comparison of the stability of localized transcripts. Box-and-whisker plot and statistical testing as per A. (F) Frequency distribution showing the fraction of nuclear, cytoplasmic, and nonenriched transcripts in 2-h bins. Plotted points as per B. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
LncRNAs can be transcribed from other genomic loci including bidirectionally (either head-to-head or tail-to-tail) with other transcripts (Engstrom et al. 2006), 3′ from coding genes (uaRNAs) (Mercer et al. 2011) and from promoter regions (promoter-associated) (Preker et al. 2008) (Supplemental Fig. S5). Another subset of lncRNAs are large intergenic noncoding RNAs (lincRNAs) (Guttman et al. 2009, 2010), many of which have critical roles in differentiation and development (Guttman et al. 2011). We identified the half-lives for 90 transcripts from lincRNA regions, including LincRNA-p21 (Gm16197), which acts downstream from p53 (also known as TP53) to repress many genes (Huarte et al. 2010) and which was quite stable with a half-life of over 6 h.
Comparing all classes of lncRNAs revealed that intergenic, cis-antisense, tail-to-tail bidirectional and uaRNA transcripts are significantly more stable (on average) than intronic and promoter associated lncRNAs (P < 0.01) (Fig. 5D). Intergenic and cis-antisense transcripts were also more stable than head-to-head bidirectional transcripts (P < 0.05). Hierarchical clustering of lncRNA stability profiles supported this result. The most stable classes (intergenic, cis-antisense, tail-to-tail and uaRNAs) clustered together, as did the unstable group of intronic, promoter-associated and head-to-head bidirectional lncRNAs, while lincRNAs showed intermediate characteristics (Supplemental Fig. S6).
Comparison of cellular localization with stability
Functional studies of lncRNAs have shown that many act as repressors or activators of gene expression through various mechanisms, including modification of epigenetic state, alternative splicing, and RNA polymerase binding efficiency (Dinger et al. 2008; Khalil et al. 2009; Zhao et al. 2010; Amaral et al. 2011). Proteins with similar regulatory roles, such as transcription factors, typically have short half-lives and are nuclear-localized. Therefore, to identify lncRNAs with potential regulatory roles in the nucleus, we interrogated total RNA isolated from nuclear and cytoplasmic fractionated N2A cells with the NCode microarrays. Successful isolation of pure nuclei and cytoplasmic fractions was confirmed by qPCR and Western blot (Supplemental Fig. S7). The array data were validated by qPCR for a number of genes ranging from nuclear-specific to cytoplasmically enriched (Supplemental Fig. S8) and also by the high nuclear enrichment of known nuclear lncRNAs, such as Xist and Bace1as. Array analysis revealed 499 lncRNAs that were significantly enriched in the cytoplasm and 191 in the nucleus. Intersection with lincRNAs and lncRNAs listed in lncRNAdb revealed a number of known and uncharacterized localizations of lncRNAs, including nuclear enrichment of Adapt33, Zfas1, and several other snoRNA host genes. Taken together, these results support the widespread trafficking of lncRNAs to different subcellular locations.
Among the subset of lncRNAs for which a half-life could be determined, we identified 22 that were significantly enriched in the cytoplasm and 105 that were enriched in the nucleus. Nuclear-localized transcripts displayed a significantly lower stability (Fig. 5E) with a greater proportion (52%) of the nuclear-enriched transcripts classified as highly unstable (P < 0.0001, χ2 test) (t½ < 2 h) (Fig. 5F; Supplemental Fig. S9). In contrast, stable transcripts (t½ > 6 h) were not enriched in either compartment (P = 0.26, χ2 test).
We considered whether the unstable nature of nuclear-localized transcripts was due to an overabundance of unstable lncRNA genomic classes (intronic, promoter-associated, and head-to-head). Head-to-head transcripts were, instead, underrepresented in the nucleus (P = 0.0154, Fisher's exact test). An increased prevalence of exosome-targeted transcripts similar to CUTs and PROMPTs could also help explain the low stability of nuclear transcripts; however, there was no enrichment for promoter-associated lncRNAs in the nucleus (P = 0.74, Fisher's exact test) or among the unstable nuclear fraction (P = 1.0, Fisher's exact test). Although intronic transcripts were enriched in the nuclear fraction (P = 0.0005, Fisher's exact test), nuclear-localized intronic lncRNAs were less stable than those intronic lncRNAs not found in the nucleus (P = 0.015, Mann Whitney t-test). Taken together, these results suggest the impact on stability is due to the cellular location of these lncRNAs, rather than resulting from their genomic classification.
The relatively high turnover of nuclear-localized lncRNAs is consistent with the notion that these lncRNAs are acting as regulatory molecules, which, like their protein-coding counterparts, are turned over rapidly to provide a high degree of dynamism to the processes they regulate. We find preliminary evidence to support this hypothesis by investigating the stability of RNAs identified as interacting with chromatin-binding proteins (Amaral et al. 2011; Guttman et al. 2011), which are less stable than lncRNAs (P = 0.049, Mann Whitney t-test), more likely to be unstable (P = 0.017, Fisher's exact test), and more likely to be nuclear-localized (P = 0.028, Fisher's exact test).
LncRNA decay elements
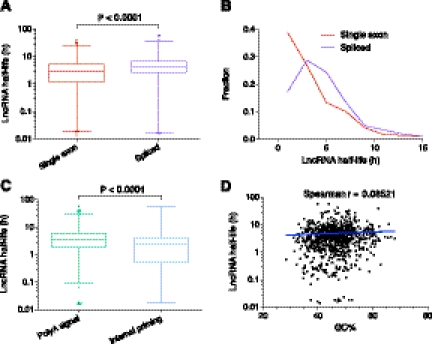
A number of decay elements have been detected, predominantly in the 3′UTR, of mRNAs, including AU-rich elements (AREs) (Bakheet et al. 2001) and Puf family protein-binding sites (Xie et al. 2005). Spliced mRNA transcripts are more stable than single exon mRNAs (Narsai et al. 2007), and there are positive correlations between both the number of introns and the density of splice junctions per kb of ORF length and stability (Sharova et al. 2009). To investigate whether these factors known to affect coding transcript stability also impact the stability of noncoding RNAs, which would suggest common regulatory and decay pathways, we analyzed the splicing status and putative AREs and Puf-binding sites of lncRNAs in the context of their stability. Similar to mRNAs, we find that spliced lncRNAs are more stable than single exon transcripts (P < 0.0001) (Fig. 6A) and that single exon transcripts are overrepresented among unstable transcripts (t½ < 2 h) (P < 0.0001) (Fig. 6B; Supplemental Fig. S11). However, unlike mRNAs (Supplemental Fig. S10), no correlation was found between the stability of spliced transcripts and the number of introns or density of splice junctions (Supplemental Fig. S11). Furthermore, no significant relationship was found between lncRNA stability and predicted ARE or Puf-binding sites (Supplemental Fig. S11), although the closeness to statistical significance for ARE sites (P = 0.091, Spearman correlation), suggests such a relationship might be present with a larger sample size. As expected, we did observe a negative relationship between the presence of these motifs and mRNA stability (Supplemental Fig. S10).
Figure 6.
LncRNAs and decay elements. (A) Comparison of the stability of single exon versus spliced lncRNAs. Box-and-whisker plot. (Whiskers) 1st–99th percentile, with individual transcripts outside this shown as dots. (Box) 25th–75th percentile. Difference calculated using a nonparametric Mann-Whitney t-test. (B) Frequency distribution showing the fraction of single exon versus spliced lncRNAs in 2-h bins. Plotted points are at the center of the 2-h bin. Only time points with 1% or more of transcripts are plotted. (C) Comparison of the stability of lncRNAs containing a major or minor polyA signal versus those with evidence of internal priming. Box-and-whisker plot and statistical testing as per A. (D) Correlation between GC% and lncRNA half-life. Spearman correlation = 0.0852 (P = 0.0145) indicates a small positive relationship between increased half-life and lncRNAs with higher GC%. Spearman correlation utilized because data is non-Gaussian. Trend line shows a semilog fit from nonlinear regression. Axes are log10-linear.
Utilizing our genome-wide lncRNA annotations, we examined other properties of lncRNAs that could impact stability. LncRNAs with a polyadenylation signal had higher stability than those transcripts that appeared internally primed (P < 0.0001) (Fig. 6C; Supplemental Fig. S11). We also identified positive correlations between the stability of lncRNAs and increases in transcript conservation and GC% percentage (Fig. 6D; Supplemental Fig. S11). These relationships were also identified in mRNAs (Supplemental Fig. S10). Although current long RNA structure prediction approaches are limited, GC% is a simple proxy for the potential of a RNA to fold into secondary structures due to the higher stability of GC base pairs. The positive relationship between GC% and stability suggests that lncRNAs with more structural elements are, perhaps not surprisingly, more stable.
Finally, we also investigated whether there were any characteristics of lncRNAs that could explain why they were less stable than mRNAs. Unexpectedly, we observed similar proportions of mRNAs and lncRNAs localized to the nucleus, with nuclear-enriched mRNAs also displaying lower stability than cytoplasmic or nonenriched transcripts (P < 0.001, one-way ANOVA). These results suggest it is not the frequent localization of lncRNAs to the nucleus that results in their lower stability relative to mRNAs; instead, the differential stability may be due to other RNA features. We find that many RNA characteristics that positively correlate with stability in lncRNAs and/or mRNAs, such as whether or not a transcript is spliced, the density of splice junctions, GC%, and degree of evolutionary conservation, have much lower values in lncRNAs compared to mRNAs (all P < 0.0001, Mann Whitney t-tests and χ2 tests). For example, the average intron density in lncRNAs is much lower than for mRNAs, which may explain why the correlation for lncRNAs was positive but not significant; i.e., too few lncRNAs have sufficient intron density to provide measureable stabilization, as is the case for mRNAs. Taken together, we find evidence that a number of sequence features known to affect mRNA stability also impact lncRNAs and provide some insight into why lncRNAs are, on average, less stable.
Comparison of lncRNA stability between mouse and human
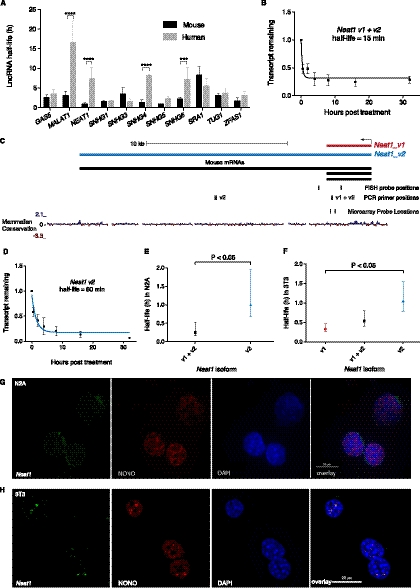
We investigated the conservation and variability in lncRNA half-lives between mouse and human. Searching the stability microarrays performed by Friedel et al. (2009) on human B cells and mouse 3T3 cells for probes corresponding to the genomic locations of lncRNAs from lncRNAdb (Amaral et al. 2011), we identified 11 lncRNAs with measured half-lives in both species (Supplemental Table S5). While most lncRNAs showed similar half-lives between the two species, four were significantly different, showing higher stability in human (all P < 0.001) (Fig. 7A). For example, MALAT1, which is found in SC35 splicing speckles and plays a role in the regulation of splicing (Tripathi et al. 2010) was highly stable in human B cells, with a half-life of 16.5 h, compared to 3 h in mouse 3T3 cells. Investigation of Malat1 in mouse N2A cells by qPCR identified a half-life of 4 h (Supplemental Fig. S13), in good agreement with Friedel et al. (2009) but significantly shorter than determined previously in mouse embryonic fibroblasts (Bernard et al. 2010). NEAT1 was also stable in human cells but unstable in mouse (investigated further below).
Figure 7.
Neat1 stability. (A) Comparison of lncRNA stabilities in mouse and human from Friedel et al. (2009). Error bars are standard deviations. Significant differences determined by two-way ANOVA using Bonferroni multiple comparisons. (***) P < 0.001, (****) P < 0.0001. (B) Transcript decay curve for Neat1 (both v1 and v2 isoforms) in N2A cells after blocking transcription with actinomycin D and measuring transcript remaining relative to Gapdh by qPCR. Four biological replicates. Error bars are standard deviations. Fit modeled by one-phase decay using nonlinear least squares regression. (C) Neat1 genomic locus showing v1 and v2 isoforms plus positions of PCR amplicons, FISH, and microarray probes. (D) Neat1_v2/ long isoform half-life in N2A cells, qPCR as per B. (E) Comparison of Neat1 (both v1 and v2 isoforms) (95% CI = 10–32 min) and Neat1_v2/ long isoform (95% CI = 40 min–1 h, 57 min) in N2A cells. Error bars show 95% confidence intervals. Unpaired t-test. (F) Comparison of stability of Neat1 isoforms in 3T3 cells. qPCR from three biological replicates. Error bars show 95% confidence intervals. Unpaired t-test. (G,H) Combined RNA protein FISH on N2A cells (G) and 3T3 cells (H). (Left panel) Neat1 RNA; (second panel) localization of paraspeckle protein NONO; (third panel) DAPI nuclear stain; (final panel) overlay.
We recently reported on the extreme stability of the Zfas1 lncRNA, which is expressed from a bidirectional promoter with the Znfx1 protein-coding gene (Askarian-Amiri et al. 2011) and showed no evidence of degradation after 16 h of transcriptional inhibition. Using the improved stable control genes, we extended this analysis to find that Zfas1 levels remain unchanged even after 32 h (Fig. 1B). This remarkably high level of stability may explain its very high expression level in N2A cells (top 0.5% of all expressed transcripts). In contrast, the stability of Zfas1 in mouse 3T3 cells and in human revealed much shorter half-lives of 1.7 and 3 h, respectively (Friedel et al. 2009). Friedel et al. (2009) did not block transcription to determine RNA half-lives; however, this result appears independent of the method used, as another study of mRNA half-lives in mouse embryonic stem cells using actinomycin D found the Zfas1 half-life to be 63 min (Sharova et al. 2009). Rather, these results suggest there can be large changes in lncRNA stability both within and between species and highlights the dynamic nature of lncRNA post-transcriptional regulation.
The architectural lncRNA Neat1 is highly unstable
Neat1 is a structural RNA involved in the formation and integrity of nuclear paraspeckles (Bond and Fox 2009; Sunwoo et al. 2009; Mao et al. 2011). Surprisingly, given its identification as a functional “architectural” RNA and similar to Friedel et al. (2009), Neat1 was very unstable with a half-life of <30 min by array and 15 min by qPCR (Fig. 7B). As Neat1 is unspliced and has short, ∼3.2 kb (Neat1_v1/Men epsilon) and long, ∼20 kb (Neat1_v2/Men beta) isoforms transcribed from a single promoter (Guru et al. 1997; Sunwoo et al. 2009), both the array and qPCR were quantifying the combined stability of the short and long isoforms (Fig. 7B,C). Neat1_v2, but not Neat1_v1, is able to rescue a Neat1 knockdown (Sasaki et al. 2009) and is required for paraspeckle formation in mice (Nakagawa et al. 2011), suggesting this isoform is more central to Neat1 function. Although qPCR specific to this isoform revealed it was significantly more stable than the combined short and long isoforms (P < 0.05), the long isoform is still unstable with a half-life of 60 min (Fig. 7D,E).
To investigate whether the high instability of Neat1 affected the formation of paraspeckles, we performed RNA-protein FISH in N2A cells. This revealed only a few, small nuclear speckles within a minority of cells by RNA FISH, which was consistent with the lack of localization of the paraspeckle protein NONO to nuclear speckle structures (Fig. 7G; Supplemental Fig. S12). N2A cells are “undifferentiated,” and the up-regulation of Neat1 (and formation of paraspeckles) upon differentiation has been reported in other cell-type specific differentiation systems (Chen and Carmichael 2009; Sunwoo et al. 2009). To examine the possibility that Neat1 up-regulation may be partially post-transcriptional and that low stability may prevent proper paraspeckle formation, we investigated Neat1 stability in 3T3 cells, which have abundant paraspeckles (Fig. 7H; Supplemental Fig. S12). The half-lives of Neat1 (v1 and v2) and Neat1_v2 were calculated as 32 min (95% CI = 24 min–47.8 min) and 63 min (95% CI = 47.3 min–93 min), respectively, indicating that there was no difference in Neat1 stability between cells with and without paraspeckles. Subtracting the expression of Neat1_v2 from Neat1 (v1 and v2) revealed that Neat1_v1 was significantly less stable than the long isoform, with a half-life of 19.6 min (95% CI = 15.22 min–27.59 min) (Fig. 7F). Therefore, we conclude that the presence/absence of paraspeckles in these cells is not related to Neat1 stability. Furthermore, the highly unstable nature of Neat1 demonstrates that instability is not a barrier to lncRNA function and that, instead, the high turnover of Neat1 could contribute to the highly dynamic nature of paraspeckles (Mao et al. 2011).
Discussion
Contrary to some expectations, examination of lncRNA stability and comparison to that of protein-coding genes has revealed that lncRNAs are not generally unstable but rather show a wide variation in their stability profiles in a manner similar to mRNAs. In light of several studies that show remarkably specific expression profiles for lncRNAs in diverse tissues and development systems (Dinger et al. 2008; Guttman et al. 2010; Cabili et al. 2011), as well as a recent large-scale screen showing functions for many lncRNAs in embryonic stem cell differentiation (Guttman et al. 2011), the large variation in lncRNA stability is consistent with their functional diversity and is likely a reflection of their complex post-transcription regulation. Indeed, post-transcriptional regulation is particularly important for lncRNAs because, unlike protein-coding genes, they do not have any further translational and post-translational opportunities for regulation. We find that a number of lncRNA characteristics correlate with stability, including genomic location, subcellular localization, splicing, and GC percentage. Other characteristics, such as the density of exon junctions, had no measurable impact on lncRNAs despite providing the strongest correlation with the mRNA stability of those tested. LncRNA expression level also was not correlated with stability, suggesting that lncRNAs below the expression cutoff will not be generally unstable either, although this possibility cannot be completely discounted. Overall, we find that analysis of lncRNA sequence features is an effective means of gaining insight into factors underlying stability.
A large number of post-transcriptional regulatory pathways act by regulating RNA stability and degradation. Some, such as the nuclear and cytoplasmic exosomes, have been found to degrade lncRNAs (including CUTs, UNTs, and PROMPTS) and regulate their function (Camblong et al. 2007; Chekanova et al. 2007; Berretta et al. 2008; Preker et al. 2008; van Dijk et al. 2011). How other pathways, such as miRNAs and nonsense mediated decay (NMD), act on lncRNAs is poorly understood. In Arabidopsis, putative lncRNAs can be regulated by NMD (Tycowski et al. 1996; Kurihara et al. 2009), a process thought to require a pioneer round of translation (Maquat et al. 2010), which could suggest “noisy” translation of lncRNAs. Recently, the first example of miRNA regulation of lncRNAs was identified (Hansen et al. 2011), suggesting this process may be widespread. RNA-binding proteins can also affect RNA stability; for example, the presence of the exon-junction complex has been hypothesized to be responsible for the increased stability of mRNAs with a higher density of splice junctions (Sharova et al. 2009). Indeed, we find this appears to apply to lncRNAs, as spliced lncRNAs are more stable than those that are unspliced.
We find that a larger proportion of lncRNAs than mRNAs are unstable. Both our results and previous findings from mammals and yeast (Seidl et al. 2006; Berretta et al. 2008; van Dijk et al. 2011) have shown that low stability does not mean lncRNAs are nonfunctional. Instead, low stability can be important for regulating lncRNA function. For example, stability can determine where and when a lncRNA can perform its function, as exemplified by PHO84 antisense in yeast (Camblong et al. 2007). Furthermore, by not requiring translation to produce a functional gene product, lncRNAs, especially those acting in the nucleus, can function almost immediately after transcription (Dinger et al. 2009a) and so may not require a long half-life. In addition, unstable transcripts are very sensitive to changes in the level of transcription and respond quickly when transcription changes (Elkon et al. 2010; Rabani et al. 2011). Low stability, such as that observed in many transcription factor mRNAs, allows dynamic gene expression in response to environmental signals (both intra- and extracellular) that can activate or repress genes. A growing number of lncRNAs, have been found to interact with chromatin and chromatin modification complexes, and some have been shown to regulate gene expression (Dinger et al. 2008; Khalil et al. 2009; Zhao et al. 2010; Guttman et al. 2011; Wang et al. 2011). It has previously been hypothesized that unstable lncRNA would be suitable for regulating gene expression (Dinger et al. 2009a), and indeed, we find that RNAs which associate with chromatin binding proteins exhibit lower stability, suggesting that lncRNA “transcription factors” with analogous properties to their coding mRNA counterparts may exist.
Consistent with our finding that low stability does not mean lack of function, we identify the nuclear paraspeckle component Neat1 as among the least stable lncRNAs. Nuclear paraspeckles are highly dynamic nuclear subdomains (Mao et al. 2011). Transcription of Neat1 is required to both nucleate and maintain paraspeckles at the Neat1 locus, with paraspeckles quickly disassembling upon inhibition of Neat1 transcription (Mao et al. 2011). The unstable nature of the Neat1 isoforms could contribute to this dynamic regulation, as paraspeckles formed around a highly stable RNA would likely be less susceptible to such quick degradation. In human, NEAT1 appears to be more stable than in mouse (Friedel et al. 2009; Sasaki et al. 2009; Sunwoo et al. 2009), with unknown consequences for NEAT1 function and paraspeckle dynamics.
Although occurring at a lower incidence than mRNAs, we also identified a number of highly stable lncRNAs. Apart from the recently identified Zfas1 (Askarian-Amiri et al. 2011), which was too stable for array-based half-life modeling but the stability of which has been confirmed by qPCR, few of these lncRNAs have been characterized. Highly stable mRNAs often encode highly stable proteins with “housekeeping” functions, such as those in central metabolism, which do not require dynamic regulation for their proper function (Schwanhausser et al. 2011). Similarly, highly stable lncRNAs may serve “housekeeping” roles. The existence of highly stable lncRNAs also suggests that some lncRNAs have evolved to avoid degradation through various mechanisms, such as secondary structure and interactions with RNA-binding proteins, about which little is known.
Despite the large impact actinomycin D treatment has on cells, previous genomic scale studies have shown good agreement between actinomycin D treatment and other methods, which do not require transcriptional blockade (Dolken et al. 2008; Friedel et al. 2009; Rabani et al. 2011; Schwanhausser et al. 2011), with the proviso that the actinomycin D time course is of sufficient duration (>6 h) to determine the half-life of stable transcripts (Friedel et al. 2009). Although actinomycin D treatment can stabilize transcripts globally, the rank order of transcript stabilities appears relatively unchanged (Rabani et al. 2011; Schwanhausser et al. 2011), supporting the validity of our conclusions regarding lncRNA stability.
The increasing availability of genome-wide technologies such as microarrays, high-throughput proteomics and next-generation sequencing has led to an explosion of studies featuring massive data sets. However, there has been minimal innovation with respect to the visualization of and accessibility to genome-scale data sets, and it is typically a complex process to extract and navigate the data sets associated with such studies. To address this issue, here we have presented our data set using QAPgrid clustering and combined this with an interactive navigation system based on Google Maps and an easily searchable database. As a result, users can become oriented within the data at various levels, such as by browsing the stability profiles or by searching for clusters that contain a particular transcript. In addition to providing convenient access to the data presented here, such an interface can be readily adapted to other genome-scale data sets. We also suggest that such advances in data dissemination are important to provoke further innovation in this important area, which we anticipate will become an increasingly essential component in any study presenting genome-scale data.
Attempts to dissect the regulatory circuitry of a biological system have increasingly led to the application and integration of various “omics” approaches. Our data and that of others suggest that there are significant correlations between stability profile and function (Sharova et al. 2009; Rabani et al. 2011; Schwanhausser et al. 2011). Furthermore, transcripts clustered into the same decay profiles (despite large differences in actual transcript expression levels) may be regulated by the same post-transcriptional regulatory pathways and/or contain similar regulatory sequences. Given the substantial complexity of the mammalian transcriptome, which continues to grow with increasingly sensitive technology, there is an increasing need for techniques to aid in the functional annotation of novel transcripts and variant isoforms, which now by far exceed the number of transcripts for which a function has been assigned. We suggest that stability profiling provides an important facet in the characterization of the transcriptome in a given biological system and is an important consideration in the ranking of candidate transcripts to pursue for more detailed functional studies.
Methods
Cell culture and RNA extraction
Mouse Neuro-2a (N2A) neuroblastoma cells (ATCC number CCL-131) and NIH-3T3 cells were cultured at 37°C, 5% C02, in DMEM plus 10% newborn calf serum and penicillin/streptomycin. For stability experiments, cells were grown to ∼50% confluency before RNA polymerase activity was blocked with 10 μg/mL actinomycin D (Sigma) in DMSO. Control cells were treated with DMSO alone. Transcriptional inhibition of N2A cells was conducted for 32 h, with cells harvested at time zero (0 h) and after 30 min, 2, 4, 8, 16, and 32 h. Inhibition of 3T3 cells was conducted for 24 h with harvesting at time zero (0 h) and after 30 min, 2, 4, 8, and 24 h. Cells were collected by trypsinization and total RNA extracted using RNeasy columns (Qiagen) and treated with DNase. RNA samples were confirmed DNA-free by performing PCR for a genomic target on the purified RNA. Integrity of RNA for microarrays was confirmed using the Agilent 2100 Bioanalyzer (Agilent Technologies). High quality, intact rRNA was found at all time points, although after 32 h of transcriptional inhibition, the first indications of rRNA degradation were visible despite continued high RNA integrity number (RIN) values. Success of transcriptional inhibition was confirmed by testing the stability of Myc mRNA (Fig. 1A; Supplemental Fig. S13).
Stability microarrays
Stability microarrays were conducted using the NCode Mouse Noncoding RNA Microarray (Life Technologies), which contain 27,281 probes targeted against protein-coding transcripts and 7228 probes targeted against noncoding transcripts. Two color arrays were performed where transcriptionally inhibited time points were compared to the untreated 0-h sample. Four biological replicates were performed with two dye swaps. Control DMSO-treated samples were not analyzed by microarray. 0.5 μg of total RNA was used for cRNA production using the Quick Amp Labeling Kit (Agilent). Equal micrograms were used because total RNA is comprised mainly of rRNA, which is highly stable, so the amount of rRNA, and hence total RNA, stays essentially the same even though mRNAs are decaying. Array hybridization was performed as per the Two-color Microarray Protocol (Version 5.7) (Agilent) and scanned using a G2565BA Microarray Scanner (Agilent).
Stability array normalization
Array data was processed using LIMMA (Smyth and Speed 2003; Smyth 2004; Ritchie et al. 2007) via the R Project for Statistical Computing (www.r-project.org). Background correction was performed via the minimum method. Between array normalization of 0-h control samples was performed with G-quantile or R-quantile normalization (depending on the channel of the 0-h sample), while T-quantile normalization was performed for each transcriptionally inhibited time point to better retain transcript degradation signals. Scaling normalization was conducted using the previously identified highly stable gene Zfas1 (Askarian-Amiri et al. 2011) and six other stable genes. These were identified by comparing the top 100 most stable mouse mRNAs from NIH-3T3 cells (Friedel et al. 2009) against the present data and selecting those appearing to be up-regulated during later time points similar to Zfas1. The average fold change of this group of seven genes between each time point was calculated, set to zero, and the probe intensity data was scaled proportionately. Summary statistics for each probe including expression values, fold change, adjusted P-values, and Bayesian (B) statistics were calculated using LIMMA. An eBayes (B statistic) prior of 25% was used. An appropriate expression cutoff was empirically determined by examining the ability of the microarrays to detect transcriptional decay as the 0-h expression level decreased and selecting a 0-h value which was sufficiently above background to allow accurate half-lives to be calculated. Raw and normalized microarray data is available at the ArrayExpress Data Warehouse (EMBL-EBI; ArrayExpress accession number E-MTAB-961).
Half-life calculations
Half-lives were calculated for probes above the expression cut-off of 25. Probe responses ranged from fast degradation to unchanged. Decay profiles were modeled by one-phase exponential decay (constraints: plateau ≥ 0, K > 0) and/or linear decay using nonlinear least squares regression using R (www.r-project.org). Probes showing statistically decreased expression were modeled with both one-phase decay and linear decay. Only fits with an R2 > 0.7 were utilized. Where both models produced good fits, the model producing the better R2 value was chosen. The remaining probes were modeled with linear decay only. Half-lives were calculated from a filtered set of probes shown by an F-test to have a significantly negative slope and with R2 > 0.7.
Clustering
Unsupervised hierarchical clustering was performed using cluster3 software (de Hoon et al. 2004). Clustering of all lncRNAs above the expression cut-off used the command line parameters -g 4 -e 2 -m m, which specifies the pairwise complete linkage method for the clustering and Pearson correlation for both the microarray and probe clustering. Normalized probe intensities are expressed relative to 1 at time 0 h. Clustering of lncRNA genomic location classes used the command line parameters -cg a -g 0 -e 5 -m m, which centers each time point by subtracting the mean, utilizing the pairwise complete linkage method for the clustering and Spearman's rank correlation for the lncRNA classes only. The resulting clusters were visualized using Java TreeView (Saldanha 2004).
The unsupervised graph-based clustering and visualization of decay profiles was performed using QAPgrid. The clustering approach and several illustrative examples on different data sets have been published previously (Inostroza-Ponta et al. 2011). The clustering algorithm receives as input the time expression values of 14,987 probe sets. After computing the Pearson correlation values for all pairs of probe sets (x,y)—which we denote as r(x,y)—a distance matrix is calculated according to the formula d(x,y) = 1−r(x,y). The clustering algorithm (MSTkNN) does not require user-defined parameters such as the number of clusters or number of elements per cluster. According to its built-in stopping criteria, it has returned 152 clusters. In turn, another matrix of distances is computed between each of these 152 clusters (for details of calculations, see Inostroza-Ponta et al. [2011]). With the input of these two matrices, we use a metaheuristic (a memetic algorithm) to solve a quadratic assignment problem defined over a grid. We obtain a layout in which, at least globally, highly similar expression patterns among clusters and super-clusters are likely to be in close proximity in the final figure produced by the method. For the figures, each of the cluster's profile graphs was produced using stacked bar plots using R (www.r-project.org). To allow integration between half-lives, clustering, and transcript annotations, each cluster was linked to a list of the probes contained in the cluster, along with half-life and annotation information. Cluster enrichment analysis was performed with g:profiler and GATHER (Chang and Nevins 2006; Medina et al. 2010; Reimand et al. 2011).
Annotation of coding and noncoding RNA transcripts
Protein-coding gene structures were based on UCSC Known Genes (Fujita et al. 2011) and Refseq (Pruitt et al. 2009) as of April 2011. Noncoding transcripts were derived from the “All mRNA” track in the UCSC Genome Browser (Fujita et al. 2011) and were defined as noncoding if they had <5% overlap with protein-coding exons and an ORF less than 100 amino acids. PhyloCSF (Lin et al. 2011) was used to filter putative lncRNAs, and any transcripts with a score of 50 (or above) were defined as protein coding. A classification as protein coding was overridden if the transcript was found in lncRNAdb (Amaral et al. 2011) or had been identified as binding to chromatin regulatory proteins (Guttman et al. 2011). Genomic locations of lncRNAs were defined as follows: Head-to-head transcripts from bidirectional promoters were defined as transcripts that originate from the antisense strand within 1 kb upstream of or 0.5 kb downstream from the 5′ end of another RNA transcriptional start site. Tail-to-tail transcripts terminate on the antisense strand within 1000 nt downstream or 1000 nt overlapping the termination site of another transcript. Cis-antisense transcripts were defined as those lncRNAs where at least 90% of their exonic length overlapped with a transcript on the opposite strand. Exclusively intronic ncRNAs were defined as those that initiated and terminated within introns of protein-coding genes in the sense orientation. The broader definition of intronic ncRNAs included any sense noncoding transcript where at least 75% of its exonic region covers the introns of a protein-coding gene. Intergenic transcripts were defined as ncRNAs not transcribed within 2 kb upstream of a protein-coding region on either strand or 30 kb downstream in the sense direction or 2 kb downstream in the antisense direction. 3′UTR-associated transcripts (uaRNAs) were defined as any ncRNA that initiates within 30 kb downstream from a stop codon on the same strand [to allow for the potential of 3′UTRs that extend beyond their annotations (Moucadel et al. 2007; Mercer et al. 2011)]. Promoter-associated transcripts were defined as single exon transcripts whose transcription was found within 3 kb upstream of and 1 kb downstream from transcription start sites on either strand. Note that some genomic categories are mutually exclusive, but others are not. LincRNAs were from Guttman et al. (2009, 2010), lncRNAdb (Amaral et al. 2011), and Guttman et al. (2011) (chromatin protein-binding lincRNAs only). We identified the half-lives for 144 transcripts from lincRNA regions. However, we only classified 90 of these as noncoding. This apparent discrepancy is primarily a consequence of the conservative nature of our lncRNA classification but can also be attributed to updated Refseq annotations that class some lincRNAs as probable coding genes. There was no difference between the stability of those lincRNAs we classified as coding or noncoding (P = 0.93).
Puf family-binding sites were predicted using the UGUANAUA consensus sequence (Xie et al. 2005). AU-rich elements were searched for using the AUUUA motif. The number of major and minor polyadenylation signals was calculated for the last 35 nt of transcripts. Major signals: AATAAA and ATTAAA. Minor signals: AATTAA, AAATAA, AGTAAA, AATATA, CATAAA, TAATAA, and AATAAT. Transcripts with 12 or more A nt in the final 20 nt of a transcript or first 20 genomic nt 3′ from the transcript were identified as candidates for internal priming.
GO analysis was performed with Fatigo (Medina et al. 2010; Reimand et al. 2011). Statistical analyses of lncRNA and mRNA features that define stability were performed using GraphPad Prism (GraphPad Software).
N2A fractionation arrays
N2A nuclear and cytoplasmic fractions were isolated using a variation of a procedure described previously (Andersen et al. 2002). Cells were trypsinized, washed twice in ice cold PBS, and resuspended in low salt buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT) on ice to swell (swelling was confirmed using a phase contrast microscope). Cells were transferred to an ice cold Dounce homogenizer and homogenized until at least 95% of cells were burst, but nuclei were still intact. Homogenate was centrifuged at 220g for 5 min at 4°C to separate the cytoplasmic fractions and nuclear-enriched pellets. The pellets were resuspended in 0.25 M sucrose, 10 mM MgCl2 buffer and overlayed on 0.88 M sucrose, 0.5 mM MgCl2 buffer, then were centrifuged at 3000g for 15 min at 4°C to obtain pure nuclei. RNA was extracted, and three biological replicate microarrays performed and normalized as described previously (Askarian-Amiri et al. 2011). An eBayes (B-statistic) prior of 15% was used. Data was uploaded and analyzed using NRED (Dinger et al. 2009b). Probes with expression of log2 ≥ 7 were defined as expressed. Enrichment in either compartment was defined as ≥twofold expression enrichment, P-value < 0.05, B-statistic > 0. Raw and normalized microarray data is available at the ArrayExpress Data Warehouse (EMBL-EBI; ArrayExpress accession number E-MTAB-952).
qPCR
Reverse transcription was carried out with the SuperScript III cDNA synthesis kit (Invitrogen) using random hexamers. Quantitative real-time PCR (qPCR) was performed using SYBR Green PCR Master Mix and real time cyclers (Applied Biosystems). Relative quantification to a stability control (Gapdh or Atp5e) was performed for stability qPCRs. At least three biological replicates were performed for all transcriptionally inhibited time courses. Decay profiles were graphed and half-lives calculated using GraphPad Prism (GraphPad Software) as per Askarian-Amiri et al. (2011). The 0-h time point expression level was set to 100%, and the percentage of transcript remaining at each time point calculated. Half-lives were calculated by nonlinear regression with a least squares fit (plateau > 0, k > 0), using one-phase exponential decay, or, when no decay was observed, a linear line. Primer sequences are provided in Supplemental Table S6.
Absolute quantification was performed for fractionation samples. Equivalent micrograms of nuclear and cytoplasmic RNAs used for qPCR were converted to cell equivalents by multiplying the cytoplasmic expression level by the cytoplasmic/nuclear RNA ratio calculated from the RNA extraction yields. Nuclear:cytoplasmic enrichment ratios were calculated from three biological replicates.
Western blotting
Protein lysate concentrations were measured using the Bradford assay and then run on 12.5% polyacrylamide gels, which were Coomassie-stained to confirm equal loadings. Proteins from SDS-PAGE gels were transferred to PVDF membrane using a semi-dry procedure. Membranes were blocked overnight in 5% milk powder. Primary and secondary antibody incubations were carried out for 2 h. GAPDH (1:4000 dilution) (R&D systems), HISTONE H3 (1:1000), anti-rabbit HRP (1:5000) (Cell Signaling Technologies). Proteins were visualized using ECL (GIBCO).
FISH
N2A and NIH-3T3 cells were seeded onto microscope cover slips and grown until ∼80% confluent. Cover slips were incubated in Extraction buffer (100 mM NaCl, 300 mM sucrose, 10 mM PIPES, 3 mM MgCl2, 0.5% Triton, and 10 mM vanadyl ribonucleoside complex, VRC) for 1 min and then fixed with 4% PFA/PBS at 4°C for 40 min, followed by overnight incubation in 70% ethanol at 4°C. Biotinylated oligos against mouse Neat1 (5′-aaactgttattcccatcaacccacggttccaggcacatt-3′biotin and 5′-aatgacacaccactcaactaaattcagggcaggttggct-3′biotin) or a scrambled control oligo (5′-gacatcctcatcggaatttagggcatatcgcaatcgcgc-3′biotin) were used at a final concentration of 1 μM and denatured at 80°C for 10 min in 70% formamide. Oligos were made up to 20 μL with hybridization buffer (0.4% BSA, 20% dextran sulphate, 4× SSC buffer, 2 U/μL RNase out) and incubated on cover slips at 37°C overnight. Cover slips were washed once in 15% Formamide/2× SSC, then in 2× SSC, and then in 1× SSC. Streptavidin Alexa Fluor488 (Invitrogen) was diluted 1:500 in 4× SSC/0.4% BSA and incubated on each cover slip at 37°C for 1 h, followed by washing in 4× SSC, then 4× SSC/0.1% Triton, then again in 4× SSC. NONO mouse monocolonal antibody (Souquere et al. 2010) was diluted 1:500 in PBS/0.05% Tween-20 and incubated on cover slips at room temperature for 1 h, followed by three washes with PBS/0.05% Tween-20, then probed with anti-mouse-TRITC secondary antibody (Jackson Immuno Research Laboratories) diluted 1:250 in PBS/0.05% Tween-20 for 1 h, followed by three washes in PBS/0.05% Tween-20. Cover slips were stained with DAPI (Sigma), mounted with Vectashield (Vector Laboratories), and z-stacks obtained using a Nikon TiE fluorescent microscope.
Paraspeckle quantification
Maximum projections of deconvolved z-stacks of NIH-3T3 cell nuclei (N = 18) and N2A cell nuclei (N = 31) from Neat1 FISH with NONO and DAPI costaining were converted to grayscale in each of the three fluorescent channels used. The images were then analyzed using CellProfiler (www.cellprofiler.org) using a modification of the “Speckle counting” pipeline (available on request) that is given on the CellProfiler website (http://www.cellprofiler.org/examples.shtml).
Data access
Raw microarray data is available at the ArrayExpress Data Warehouse (EMBL-EBI; ArrayExpress accession numbers E-MTAB-952 and E-MTAB-961). All normalized stability and localization array data is available online at http://stability.matticklab.com and/or in Supplemental Tables S1 and S2.
Competing interest statement
M.E.D. and J.S.M. are license holders of the NCode ncRNA microarrays (Life Technologies).
Acknowledgments
We thank Dr. Matt Ritchie from the Walter and Eliza Hall Institute of Medical Research for helpful advice on microarray normalization and analysis, Prof. Caroline Friedel from the Ludwig-Maximilians-Universität München for sharing half-life data, Dr. Nedda Cecchinato from the Queensland University of Technology for statistical advice, Dr. Sven Hennig from the Western Australian Institute for Medical Research, and the rest of the Mattick Laboratory for useful discussions. This work was supported by a National Health and Medical Research Council (NHMRC) Australia Fellowship (631668; J.S.M.), an NHMRC Career Development Award (631542; M.E.D.), and a Queensland Government Department of Employment, Economic Development and Innovation Smart Futures Fellowship (M.E.D.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.131037.111.
References
- Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS 2011. lncRNAdb: A reference database for long noncoding RNAs. Nucleic Acids Res 39: D146–D151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI 2002. Directed proteomic analysis of the human nucleolus. Curr Biol 12: 1–11 [DOI] [PubMed] [Google Scholar]
- Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, Ru K, Mercer TR, Thompson ER, Lakhani SR, et al. 2011. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA 17: 878–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS 2001. ARED: Human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res 29: 246–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, et al. 2010. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 29: 3082–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta J, Pinskaya M, Morillon A 2008. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev 22: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C, Kannouche P, Litfin M, Bender K, Rahmsdorf HJ, Angulo JF, Herrlich P 2000. UV-Induced stabilization of c-fos and other short-lived mRNAs. Mol Cell Biol 20: 3616–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond CS, Fox AH 2009. Paraspeckles: Nuclear bodies built on long noncoding RNA. J Cell Biol 186: 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolin ML, Kiss T 1998. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA 4: 445–454 [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F 2007. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131: 706–717 [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Chang JT, Nevins JR 2006. GATHER: A systems approach to interpreting genomic signatures. Bioinformatics 22: 2926–2933 [DOI] [PubMed] [Google Scholar]
- Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. 2007. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131: 1340–1353 [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA. Mol Cell 35: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani C, Blanchard JM, Piechaczyk M, El Sabouty S, Marty L, Jeanteur P 1984. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci 81: 7046–7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S 2004. Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. 2008. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Mattick JS 2009a. Pervasive transcription of the eukaryotic genome: Functional indices and conceptual implications. Brief Funct Genomics Proteomics 8: 407–423 [DOI] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS 2009b. NRED: A database of long noncoding RNA expression. Nucleic Acids Res 37: D122–D126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, et al. 2008. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14: 1959–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Zlotorynski E, Zeller KI, Agami R 2010. Major role for mRNA stability in shaping the kinetics of gene induction. BMC Genomics 11: 259 doi: 10.1186/1471-2164-11-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, Lavorgna G, Brozzi A, Luzi L, Tan SL, Yang L, et al. 2006. Complex loci in human and mouse genomes. PLoS Genet 2: e47 doi: 10.1371/journal.pgen.0020047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel CC, Dolken L, Ruzsics Z, Koszinowski UH, Zimmer R 2009. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res 37: e115 doi: 10.1093/nar/gkp542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. 2011. The UCSC Genome Browser database: Update 2011. Nucleic Acids Res 39: D876–D882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guru SC, Agarwal SK, Manickam P, Olufemi SE, Crabtree JS, Weisemann JM, Kester MB, Kim YS, Wang Y, Emmert-Buck MR, et al. 1997. A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus. Genome Res 7: 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. 2010. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J 2011. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 30: 4414–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2009. The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. 2010. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz J, Furth JJ, Malamy M, Alexander M 1962. The role of deoxyribonucleic acid in ribonucleic acid synthesis. III. The inhibition of the enzymatic synthesis of ribonucleic acid and deoxyribonucleic acid by actinomycin D and proflavin. Proc Natl Acad Sci 48: 1222–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inostroza-Ponta M, Berretta R, Moscato P 2011. QAPgrid: A two level QAP-based approach for large-scale data analysis and visualization. PLoS ONE 6: e14468 doi: 10.1371/journal.pone.0014468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, Morosawa T, Tanaka M, Kaminuma E, Mochizuki Y, Matsushima A, et al. 2009. Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc Natl Acad Sci 106: 2453–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M 2011. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–i282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL 2011. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 13: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Tarn WY, Isken O 2010. The pioneer round of translation: Features and functions. Cell 142: 368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina I, Carbonell J, Pulido L, Madeira SC, Goetz S, Conesa A, Tarraga J, Pascual-Montano A, Nogales-Cadenas R, Santoyo J, et al. 2010. Babelomics: An integrative platform for the analysis of transcriptomics, proteomics, and genomic data with advanced functional profiling. Nucleic Acids Res 38: W210–W213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long non-coding RNAs: Insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Wilhelm D, Dinger ME, Solda G, Korbie DJ, Glazov EA, Truong V, Schwenke M, Simons C, Matthaei KI, et al. 2011. Expression of distinct RNAs from 3′ untranslated regions. Nucleic Acids Res 39: 2393–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moucadel V, Lopez F, Ara T, Benech P, Gautheret D 2007. Beyond the 3′ end: Experimental validation of extended transcript isoforms. Nucleic Acids Res 35: 1947–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Naganuma T, Shioi G, Hirose T 2011. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol 193: 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Millar AH, O'Toole N, Small I, Whelan J 2007. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19: 3418–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854 [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Klimke W, Maglott DR 2009. NCBI Reference Sequences: Current status, policy, and new initiatives. Nucleic Acids Res 37: D32–D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, et al. 2011. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol 29: 436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, Krathwohl M, Bohjanen PR 2002. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res 30: 5529–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Arak T, Vilo J 2011. g:Profiler—a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res 39: W307–W315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK 2007. A comparison of background correction methods for two-colour microarrays. Bioinformatics 23: 2700–2707 [DOI] [PubMed] [Google Scholar]
- Saldanha AJ 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T 2009. MENɛ/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci 106: 2525–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Seidl CI, Stricker SH, Barlow DP 2006. The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. EMBO J 25: 3565–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS 2009. Database for mRNA half-life of 19,977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res 16: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheardown SA, Duthie SM, Johnston CM, Newall AE, Formstone EJ, Arkell RM, Nesterova TB, Alghisi GC, Rastan S, Brockdorff N 1997. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell 91: 99–107 [DOI] [PubMed] [Google Scholar]
- Smyth GK 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: 1–25 [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T 2003. Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S 2007. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci 120: 2498–2506 [DOI] [PubMed] [Google Scholar]
- Souquere S, Beauclair G, Harper F, Fox A, Pierron G 2010. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell 21: 4020–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL 2009. MEN ɛ/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Steitz JA 1996. A mammalian gene with introns instead of exons generating stable RNA products. Nature 379: 464–466 [DOI] [PubMed] [Google Scholar]
- van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, et al. 2011. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475: 114–117 [DOI] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. 2005. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE Jr 2003. Decay rates of human mRNAs: Correlation with functional characteristics and sequence attributes. Genome Res 13: 1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT 2010. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40: 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]