Abstract
Malaria genetic variation has been extensively characterized, but the level of epigenetic plasticity remains largely unexplored. Here we provide a comprehensive characterization of transcriptional variation in the most lethal malaria parasite, Plasmodium falciparum, based on highly accurate transcriptional analysis of isogenic parasite lines grown under homogeneous conditions. This analysis revealed extensive transcriptional heterogeneity within genetically homogeneous clonal parasite populations. We show that clonally variant expression controlled at the epigenetic level is an intrinsic property of specific genes and gene families, the majority of which participate in host–parasite interactions. Intrinsic transcriptional variability is not restricted to genes involved in immune evasion, but also affects genes linked to lipid metabolism, protein folding, erythrocyte remodeling, or transcriptional regulation, among others, indicating that epigenetic variation results in both antigenic and functional variation. We observed a general association between heterochromatin marks and clonally variant expression, extending previous observations for specific genes to essentially all variantly expressed gene families. These results suggest that phenotypic variation of functionally unrelated P. falciparum gene families is mediated by a common mechanism based on reversible formation of H3K9me3-based heterochromatin. In changing environments, diversity confers fitness to a population. Our results support the idea that P. falciparum uses a bet-hedging strategy, as an alternative to directed transcriptional responses, to adapt to common fluctuations in its environment. Consistent with this idea, we found that transcriptionally different isogenic parasite lines markedly differed in their survival to heat-shock mimicking febrile episodes and adapted to periodic heat-shock with a pattern consistent with natural selection of pre-existing parasites.
Adaptation to environmental changes is essential for survival. In the case of Plasmodium falciparum asexual blood stages, which are responsible for all clinical malaria symptoms and complications, the intra-erythrocytic niche provides a relatively stable environment compared with the environments where free-living unicellular organisms reside, but there are still challenges that the parasite must confront (Mackinnon and Marsh 2010). Among the conditions that can change during the course of a blood infection or between different human hosts are immune pressure, drug pressure, host metabolic and nutritional conditions, presence and magnitude of febrile episodes, host genetics, and presence of competing parasites.
Long-term, slow adaptation typically occurs at the genetic level. In P. falciparum, the genetic bases of adaptation have been extensively studied (Mu et al. 2010; Ochola et al. 2010; Van Tyne et al. 2011). However, adaptation to fast-fluctuating environmental pressures requires a much faster time scale and reversibility and operates at the transcriptional level. In the majority of organisms, including bacteria, yeast, and higher eukaryotes, rapid adaptation typically occurs through directed transcriptional responses: An external condition is sensed, which triggers a signaling cascade that results in transcription factor–driven activation or repression of specific genes that mediate adaptation to the new condition. Some evidence suggests that P. falciparum may have a limited capacity to mount directed transcriptional responses: (1) There is a marked paucity of specific transcription factors in the P. falciparum genome (Coulson et al. 2004), despite the recent identification of an expanded family of AP2 domain–containing transcription factors (ApiAP2) (Balaji et al. 2005; Campbell et al. 2010). The specific transcription factors identified seem insufficient to mount a wide range of transcriptional responses. (2) Analysis of transcriptional responses immediately or very soon after environmental perturbations failed to detect large alterations in the levels of transcripts related with the challenge, in contrast to similar studies in other eukaryotes (Gunasekera et al. 2007; Ganesan et al. 2008; Le Roch et al. 2008; Young et al. 2008). This led some investigators to conclude that P. falciparum has an unresponsive, hard-wired transcriptome (Ganesan et al. 2008; Le Roch et al. 2008). This is controversial because other studies detected changes in transcription upon various challenges (Functional Genomics Workshop Group et al. 2007; Oakley et al. 2007; Natalang et al. 2008; Tamez et al. 2008; Hu et al. 2010), but the changes observed were of low amplitude and could be explained by cell cycle arrest or sexual differentiation, or may represent pathways leading to parasite death, rather than being authentic protective transcriptional responses (Functional Genomics Workshop Group et al. 2007; Natalang et al. 2008).
Regardless of the ability of P. falciparum to mount directed transcriptional responses, one source of transcriptional variability in malaria parasites is clonally variant gene expression. Variantly expressed genes can be found in different transcriptional states in genetically identical parasites at the same stage of life cycle progression and grown under homogeneous conditions. The transcriptional state is clonally transmitted by epigenetic mechanisms (Scherf et al. 2008). Clonally variant gene expression has a clear adaptative potential, because spontaneous changes in expression lead to transcriptionally heterogeneous populations. It is well established in the population biology field that heterogeneous populations are fitter than homogeneous populations in changing environments, because diversity provides the grounds for natural selection of individuals with phenotypes that confer maximum fitness upon an environmental change (Kussell and Leibler 2005; Veening et al. 2008). Clonally variant gene expression in P. falciparum was initially described for var genes (Scherf et al. 2008), a family of about 60 genes encoding PfEMP-1, which plays a major role in pathogenesis and immune evasion. Other P. falciparum gene families also show clonally variant expression, including gene families encoding proteins exported to the erythrocyte surface, and families linked to erythrocyte invasion. Variant expression of these gene families may also play a role in immune evasion (Cortés et al. 2007; Lavazec et al. 2007; Petter et al. 2007; Cortés 2008; Scherf et al. 2008; Gomez-Escobar et al. 2010).
Adaptation through clonally variant gene expression would imply that parasite populations spontaneously show transcriptional diversity for genes relevant for adaptation to common environmental pressures. While genetic variation in P. falciparum has been extensively characterized (Jeffares et al. 2007; Mu et al. 2007; Volkman et al. 2007; Van Tyne et al. 2011), the genome-wide extent of clonally variant gene expression remains unknown. Some studies have compared global blood stages transcription between different P. falciparum lines, but their sample size and experimental design resulted in the identification of only a small number of variantly expressed genes: One study compared two isogenic lines selected to display different antigenic and adhesive properties (Mok et al. 2007), whereas another study compared the transcriptome of three genetically different parasite lines, focusing on the comparison of patterns of expression along the life cycle rather than comparing transcript levels (Llinas et al. 2006). Genome-wide transcriptional differences have also been analyzed in field isolates (Daily et al. 2007; Lemieux et al. 2009; Mackinnon et al. 2009), but these studies suffer intrinsic limitations such as the inability to distinguish between spontaneous variation and selection, and the common occurrence of mixed infections. Furthermore, some of the transcript-level differences observed may be attributable to genetic polymorphism. A recent study established that transcriptional diversity in P. falciparum has a strong genetic component (Gonzales et al. 2008).
Here we comprehensively characterized the asexual blood-stage transcriptome of 21 parasite lines and compared transcript levels within sets of parasite lines sharing a common clonal origin. With this approach, transcriptional variation was studied independently of the genetic component. We identified the majority of inherently variant gene families and observed extensive transcriptional heterogeneity among essentially isogenic parasites. Variantly expressed gene families participate in multiple aspects of host–parasite interactions, including but not limited to immune evasion. The characteristics of the genes showing clonally variant expression are suggestive of a central role for variant expression in adaptation, and this was experimentally demonstrated for adaptation to periodic heat-shock mimicking malaria cyclical fever. We also show that functionally diverse variantly expressed gene families are regulated by a common epigenetic mechanism involving heterochromatin formation.
Results
Identification of genes under clonally variant expression
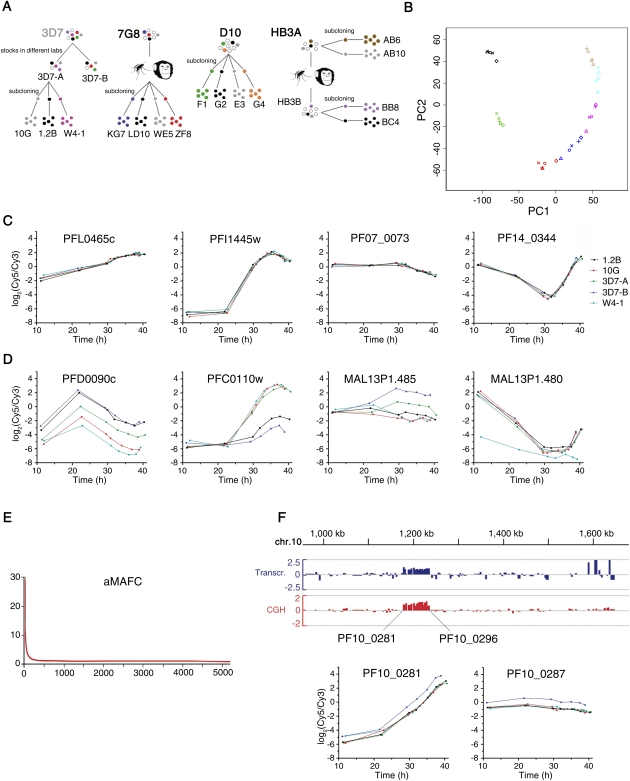
Transcriptional variability was first assessed for two stocks of the clonal parasite line 3D7 maintained in different laboratories for several years, 3D7-A and 3D7-B, and three subclones of 3D7-A (Fig. 1A, 3D7). Subclones were cultured for the minimal number of generations necessary to obtain sufficient material for transcriptional analysis. For genes with stable clonally transmitted expression patterns, this is akin to characterizing transcript levels in single parasites within the clonal population from which the subclones were derived (Cortés et al. 2007). The five parasite lines were tightly synchronized to a 0- to 5-h window, and RNA was obtained at seven time points along the asexual blood cycle. Relative transcript levels were determined by hybridizing samples against a common reference pool in a long-oligonucleotides microarray (Hu et al. 2007). Principal component analysis (PCA) revealed clustering of samples collected at the same time point, as expected for well-synchronized samples. Samples collected at the latest time points approached samples collected at the first time point, reflecting the cyclic nature of asexual blood growth (Fig. 1B).
Figure 1.
Identification of genes under clonally variant expression. (A) Parasite lines used in this study. Parental parasite lines 3D7, 7G8, D10, and HB3A are of clonal origin. 3D7-A and 3D7-B are stocks of 3D7 maintained in different laboratories for some years (Cortés et al. 2004). 7G8 subclones were identified as self-fertilization events after passage through mosquito and splectomized chimpanzee (Hayton et al. 2008). HB3B was derived from self-crossing HB3A (Walliker et al. 1987). Parentals comparison included the three parental parasite lines shown in bold (7G8, D10, and HB3A). (B) Principal component analysis (PCA) of the time-course transcriptional analysis of 3D7 parasite lines. Samples from the same parasite line are represented by the same symbol, whereas samples collected at the same time point are represented by the same color. (C) Time-course expression plots of representative genes not showing clonally variant expression. Expression levels (log2 ratio of expression relative to the reference pool) are plotted against statistically estimated culture age (in hours post-invasion). The first gene has the median aMAFC, whereas the other three genes are single-copy genes commonly used as controls (rhopH2, seryl tRNA synthetase, and ptex150, in this order). (D) Time-course expression plots, as in C, but for genes showing clonally variant expression. The genes belong to the phista, clag, acs, and hrp families, in this order. (E) Distribution of aMAFC. Genes (x-axis) are ranked by their aMAFC in descending order. (F) Duplication of a region of chromosome 10 in 3D7-B. Transcript (blue) and gDNA (red) levels in 3D7-B relative to 3D7-A (log2 ratio) are shown for the second half of chromosome 10. The names of the first and last genes duplicated in 3D7-B are shown. Time-course expression plots for two of the genes within the duplication are shown.
The majority of P. falciparum genes show stage-specific expression, and even small differences in the stage of cycle progression can lead to large differences in observed transcript levels (Bozdech et al. 2003). This is a major confounder for microarray-based studies comparing expression between different parasite lines. To account for this, we estimated the stage of the cycle for each microarray sample using a statistical likelihood-based method (Supplemental Fig. S1; Lemieux et al. 2009) and plotted expression levels against estimated times post-invasion (Fig. 1C,D). To quantify, for each gene, the level of transcriptional variability among the parasite lines compared, we developed a score termed aMAFC (adjusted maximum average fold-change across half the time interval compared), which is an estimate of the expression fold-change between the parasite line that expresses the gene at the highest level and the parasite line that expresses it at the lowest level (see Methods). Only ∼5% of the genes showed substantial transcriptional variation (Fig. 1E). The median aMAFC corresponds to the first gene in Figure 1C.
We generated a high-confidence list of genes showing different transcript levels among 3D7-derived parasite lines, based on criteria dependent on the aMAFC and permutation tests (see Methods). Comparative genome hybridization (CGH) analysis revealed that in six of the 144 genes selected, different transcript levels could be explained by genetic differences (copy number variations, CNVs). Five of these six genes fell within a duplication of ∼56 kb in 3D7-B, which resulted in approximately twofold transcript levels relative to parasite lines without the duplication (Fig. 1F). After excluding these genes to focus only on epigenetic transcriptional variation, we generated a list of 138 genes that we called the “3D7 variantome” (Supplemental Table S1), defined as the set of genes with clonally variant expression in 3D7. Some of these genes (76 of 138) belong to the large hypervariable families var, rif, stevor, and pfmc-2tm (Supplemental Figs. S2, S3; Supplemental Results), for which variant expression has been previously described (Scherf et al. 2008).
Variant expression in parasites of different genetic backgrounds
We followed the same approach to identify genes under clonally variant expression in other sets of parasite lines sharing a clonal origin. Each comparison included the parental parasite line, of clonal origin, and several subclones (Fig. 1A). We also compared transcript levels among the genetically different parental (original) lines 7G8, D10, and HB3A (Fig. 1A, parasite lines in bold) to identify the genes showing top differences in expression among genetically different parasites. In total, we identified genes under variant expression based on five comparisons, four between parasite lines sharing a clonal origin (3D7, 7G8, D10, and HB3 comparisons) and one between genetically different parasites (parentals comparison) (Fig. 1A).
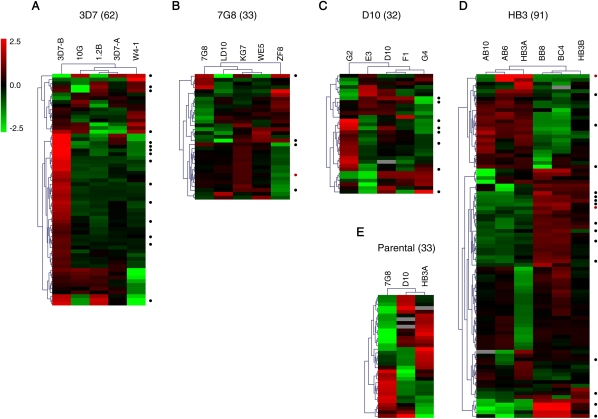
For each non-3D7 genetic background, probes that target polymorphic sequences as inferred from BLAST and CGH analysis were excluded (Supplemental Methods). All genes from the highly polymorphic gene families var, rif, stevor, and pfmc-2tm were excluded from the analysis of non-3D7 parasite lines, because CGH and BLAST analysis revealed that this microarray (designed against the 3D7 reference genome) is unsuitable to study expression of these families in non-3D7 parasite lines (Supplemental Fig. S4). However, it is already well established from our data with 3D7 parasites and by others (Scherf et al. 2008) that these gene families show extensive clonally variant expression. Furthermore, as in the case of the 3D7 comparison, we excluded from the lists of variant genes all the cases in which deletions, duplications, or other genetic alterations (as determined by CGH of individual parasite lines) may account for the differences in transcript levels observed. Genes showing variant expression based only on LD10 (Fig. 1A) were also excluded from the list of 7G8 variant genes because this subclone has DNA sequences of non-7G8 origin (see Methods). Even after excluding all the genes where different transcript levels may be attributable to genetic differences and further filtering the lists of variant genes (Supplemental Figs. S5, S6, S7; Supplemental Table S1; Supplemental Results), we identified a substantial number of variant genes in all comparisons (Fig. 2; Supplemental Tables S1, S2). No two single parasite lines analyzed were transcriptionally identical. Transcriptional differences between isogenic subclones indicate that all of the parental parasite lines from which they were derived, of clonal origin but grown for many generations after cloning, consist of a mixture of single parasites with different transcriptional patterns, despite growing under homogeneous conditions. These results demonstrate that genetically homogeneous clonal populations show high levels of spontaneous epigenetic variation. Importantly, qPCR analysis on cDNA from independent parasite preparations revealed that transcriptional patterns were stable for the majority of genes tested (dots in Fig. 2; Supplemental Fig. S8), implying stable epigenetic heritability, with expression switches occurring at low frequency.
Figure 2.
Overview of genes showing transcriptional variability. Expression patterns are shown for the clonally variant genes identified in the 3D7 (A), 7G8 (B), D10 (C), HB3 (D), and parental lines (E) comparisons. The numbers in parenthesis indicate the number of variant genes in each comparison. Genes from the var, rif, stevor, and pfmc-2tm families are not included in the 3D7 heatmap, to make it comparable with the other parasite lines. Values are the log2 of the expression fold-change relative to the average expression in the parasite lines within each comparison. (Black dots) Genes showing the same transcriptional patterns in qPCR analysis of independent biological preparations; (red dots) genes that showed different patterns (Supplemental Fig. S8; Supplemental Table S3).
Overlap in the variant genes identified in the different comparisons
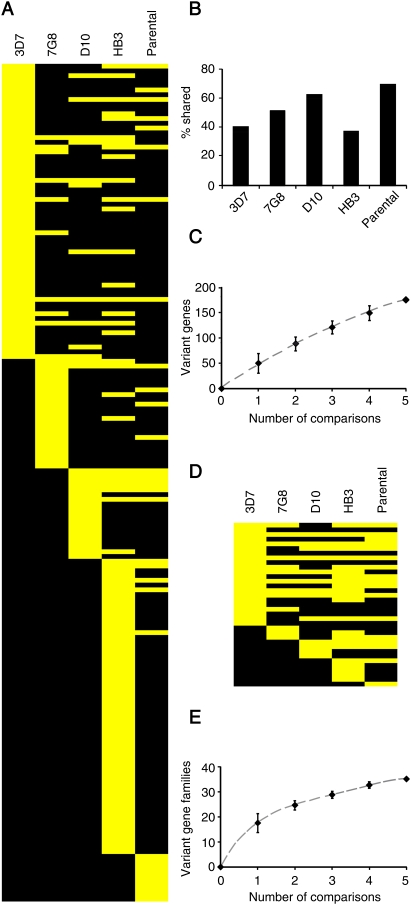
There was extensive overlap in the genes that showed variant expression in the different comparisons (Fig. 3A), indicating that variant expression is an intrinsic property of certain genes. Eight genes showed variant expression in four or five of the comparisons. More than 40% of the genes that showed variant expression in the 3D7 comparison also showed variant expression in at least one other comparison, and the same was true for a similar or higher percentage of the variant genes identified in each comparison (Fig. 3B). Remarkably, transcriptional variability between genetically different parasites seems to affect similar genes as transcriptional variability within clonal parasites populations: 70% of the genes more differentially expressed between the genetically different 7G8, D10, and HB3 parental parasite lines (parentals comparison) were also variantly expressed in at least one of the comparisons involving isogenic parasite lines. The cumulative number of variant genes identified increased with the number of comparisons (Fig. 3C). In total, 176 different variant genes were identified (excluding large gene families that were only analyzed in the 3D7 comparison; 252 variant genes were identified including these families) (Supplemental Table S2).
Figure 3.
Overlap in the variantly expressed genes identified in the different comparisons. (A) Genes (in rows) that showed variant expression in the 3D7, 7G8, D10, HB3, or parental comparisons are shown in yellow in the corresponding lane. Genes from the var, rif, stevor, and pfmc-2tm families were excluded from this analysis, because they were only analyzed in the 3D7 comparison. (B) Percentage of variant genes identified in each comparison that showed variant expression in at least one other comparison. (C) Cumulative number of variantly expressed genes identified with increasing number of comparisons. Values are the average, with 95% ci, of all possible combinations of comparisons. (D,E) Same as panels A and C, but at the gene family level. Gene families are defined in Supplemental Table S4.
The overlap between the gene families with variant expression in the different comparisons was even more extensive (Fig. 3D), and the cumulative number of variant gene families identified clearly approached a plateau (Fig. 3E). This result indicates that we may have captured the majority of P. falciparum gene families under clonally variant expression.
Gene families under clonally variant expression
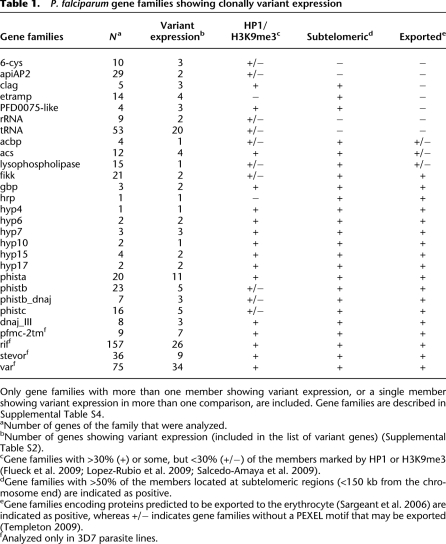
Many of the variantly expressed genes (173 out of 252) are members of gene families, the majority of them known or predicted to participate in host–parasite interactions. In total, we identified 28 gene families with at least two members listed as variantly expressed, or one member listed as variant in two comparisons (Table 1). Even after excluding the large variant gene families var, rif, stevor, and pfmc-2tm from the analysis, 97 of 176 variant genes were members of gene families, and 40% had paralogs according to PlasmoDB/OrthoMCL, compared with only 9% in the full genome (two-tailed Fischer's exact test, P < 0.001). Functional redundancy in some gene families probably results in permissivity for variant expression.
Table 1.
P. falciparum gene families showing clonally variant expression
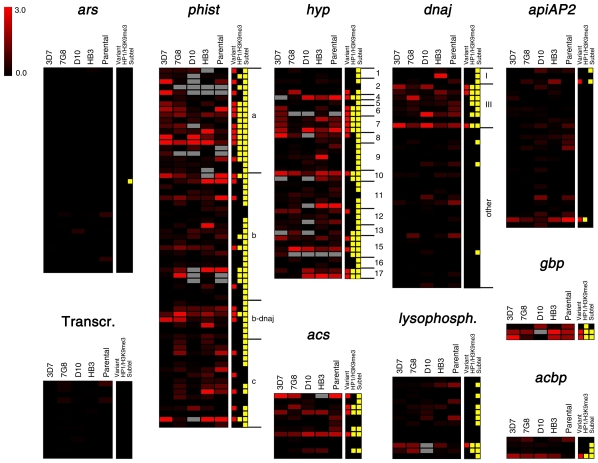
Control gene sets encoding single-copy, essential genes showed minimal levels of transcriptional heterogeneity in all sets of parasite lines compared (Fig. 4). In contrast, we identified several gene families that showed extensive transcriptional variability in all five comparisons, and gene families such as apiAP2 or lysophospholipases that showed stable expression levels for the majority of genes, with substantial variant expression restricted to specific members (Fig. 4). These results indicate again that variant expression is an intrinsic property of some genes and gene families.
Figure 4.
Distribution of variant expression in P. falciparum gene families. The log2 of the aMAFC for each comparison is shown for the genes of selected families. Aminoacyl-tRNA-synthetases (ars) and genes encoding the basal transcription machinery (Transcr.) (Bischoff and Vaquero 2010) provide appropriate controls as gene sets encoding essential genes, for which variant expression is not expected. For phist, hyp, and dnaj families, the specific family identity is indicated at the right of the heatmaps. Gene families are detailed in Supplemental Table S4. The tracks at the right indicate whether a gene was included in the list of variant genes (Supplemental Table S2), whether it was positive for heterochromatin marks HP1 or H3K9me3 in any of the published reports (Flueck et al. 2009; Lopez-Rubio et al. 2009; Salcedo-Amaya et al. 2009), and whether it is subtelomerically located (<150 kb from a chromosome end). Genes that show high aMAFC in some of the tracks but are not in the list of variant genes were excluded because changes in expression were attributable to CNVs, or for other reasons detailed in the text.
Many of the variant gene families are subtelomeric and encode proteins exported to the erythrocyte (Table 1), including exported HSP40-type cochaperones (dnaj) (Sargeant et al. 2006), predicted exported protein kinases (fikk) (Nunes et al. 2007), and exported proteins of unknown function (such as phist, hyp, and gbp families) (Sargeant et al. 2006). Additionally, variant expression was observed in gene families that encode proteins that participate in lipid metabolism, such as acyl-CoA synthetases (acs), acyl-CoA binding proteins (acbp) (Bethke et al. 2006), and lysophospholipases; proteins involved in nutrient import and possibly in erythrocyte invasion (clag) (Cortés 2008; Nguitragool et al. 2011); transcription factors (apiAP2) (Campbell et al. 2010); proteins involved in transmission (6-cys) (Templeton and Kaslow 1999); secreted proteins with functions in hemozoin formation and with procoagulant activity (hrp) (Ndonwi et al. 2011); and proteins located at the parasite—host cell interface (etramp) (Table 1; Spielmann et al. 2003). Surprisingly, many tRNAs were also expressed at different levels between different parasite lines. Other genes were not part of any of the variant gene families included in Table 1, but also showed clonally variant gene expression. Among those, there were 14 genes encoding exported proteins (Sargeant et al. 2006); the gene eba-140 (MAL13P1.60) and the pseudogene PfRh3 (PFL2520w) from families linked to erythrocyte invasion (Cortés 2008); surfin4.1 (PFD0100c) (Winter et al. 2005); three genes encoding putative Zn-finger proteins; genes related with gametocyte formation, for which expression levels in asexual parasites may determine the propensity to enter sexual differentiation (Supplemental Results); two more enzymes linked to lipid metabolism (beta-hydroxyacyl-ACP dehydratase and dephospho-CoA kinase); and 43 genes encoding proteins annotated as “unknown function” (Supplemental Table S2).
Gene set enrichment analysis (GSEA) (Subramanian et al. 2005) did not reveal any additional gene family, metabolic pathway, biochemical process, or protein domain associated with high levels of variant expression (data not shown). However, genes that have paralogs, lack identified orthologs in other species, are located in subtelomeric regions, are pseudogenes, or encode proteins with transmembrane domains showed a significantly skewed distribution toward high levels of transcriptional variability (Supplemental Fig. S9A).
Transcriptional heterogeneity enables adaptation to periodic heat-shock
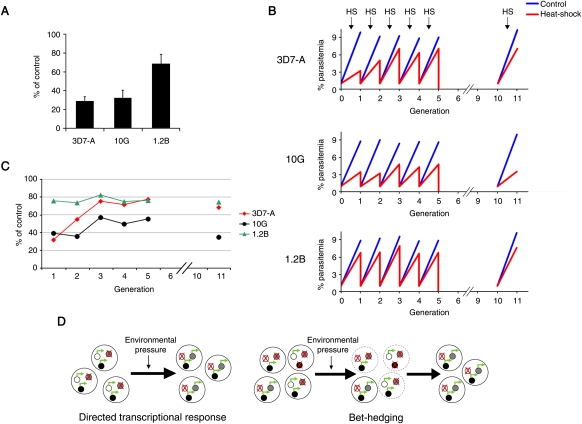
To test the possibility that clonally variant gene expression can mediate adaptation to environmental changes, we measured survival of three parasite lines of the same genetic background (3D7-A and two of its subclones) (Fig. 1A) after a 3-h heat-shock at 41.5°C, mimicking a malaria high-fever episode. Temperature is a major changing condition for malaria parasites that fluctuates between stable 37°C in some human hosts and periodic febrile episodes in others (mainly young children or malaria-naive individuals). One of the recently subcloned parasite lines, 1.2B, showed a markedly higher resistance to heat-shock (Fig. 5A), indicating that transcriptional heterogeneity results in phenotypic differences. We next subjected parasites to periodic heat-shocks for five consecutive generations and observed that the transcriptionally diverse parasite line 3D7-A rapidly adapted and acquired a 1.2B-like heat-shock resistant phenotype (Fig. 5B,C). This is consistent with adaptation by selection of pre-existing parasites with a pattern of expressed and repressed genes that confers low sensitivity to heat-shock. On the other hand, the recently subcloned (transcriptionally homogeneous) line 10G adapted only to a much lower extent and adaptation started later. Adapted 3D7-A parasites maintained the heat-shock resistant phenotype after five generations without heat-shock (Fig. 5B,C, cycle 11), demonstrating stable epigenetic transmission of the transcriptional patterns that confer heat-shock resistance, even in the absence of heat-shock. In summary, the observed pattern of adaptation to heat-shock is exactly as predicted if adaptation occurs via natural selection of pre-existing parasites with specific transcriptional patterns, and not consistent with adaptation via immediate directed transcriptional responses (Fig. 5D).
Figure 5.
Adaptation to heat-shock. (A) Inhibition of parasite growth by a 3-h heat-shock at 41.5°C. Values are the average of three independent experiments, with standard deviation, and represent percentage of growth relative to identical cultures not subjected to heat-shock. Heat-shock was performed when parasites were at the trophozoite stage, and parasitemia was measured by FACS at the next generation. (B) Growth under periodic heat-shock. Parasites at the trophozoite stage were subjected to heat-shock for five consecutive generations. After each cycle, heat-shocked cultures were synchronized, adjusted to 1% parasitemia, and split into two identical dishes to compare again growth between heat-shock and control conditions. Cultures were grown without heat-shock in cycles 6–10. For details, see Supplemental Methods. (C) Percentage of growth under heat-shock relative to normal conditions, calculated from the data in panel B. (D) Schematic model for adaptation through directed transcriptional responses and adaptation through spontaneous clonally variant gene expression (bet-hedging). (Small circles) Individual genes that can be either repressed (crossed) or active (green arrow). In the directed transcriptional response scenario, a change in the environment results in an immediate protective transcriptional response, such that the external change is sensed, and specific genes are activated or repressed to mediate adaptation to the new conditions. In contrast, in the bet-hedging scenario, the population is transcriptionally heterogeneous as a consequence of spontaneous clonally variant gene expression. Upon a change in the environment, pre-existing parasites with combinations of expressed and repressed genes that confer fitness under the new conditions are selected and survive, whereas other parasites die (broken line).
Epigenetic mechanisms regulating clonally variant gene expression
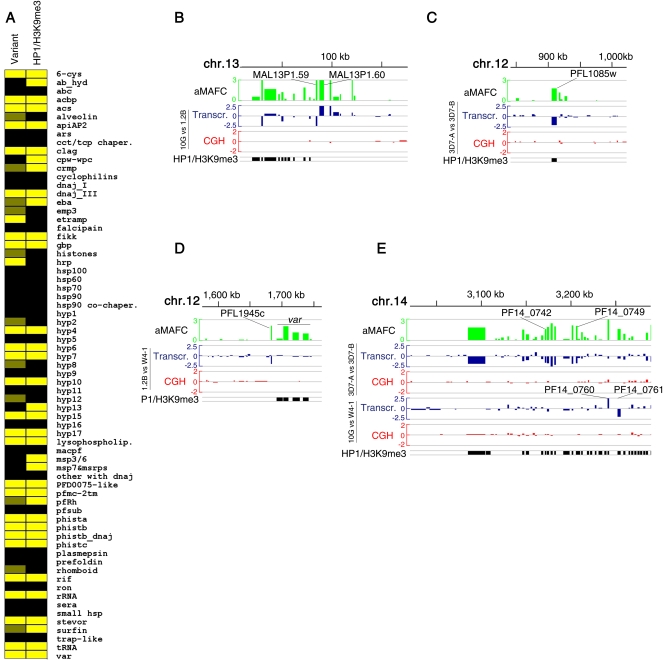
Many of the genes that showed variant expression were found to be positive for the heterochromatin marks H3K9me3 and HP1 in previous chip on ChIP–based mapping studies (Supplemental Table S2; Flueck et al. 2009; Lopez-Rubio et al. 2009; Salcedo-Amaya et al. 2009). Only two of the 28 variant gene families included in Table 1 were HP1- and H3K9me3-negative. Likewise, of 35 gene families reported to be marked by HP1 or H3K9me3, only five small families did not have any member showing clonally variant expression (Fig. 6A). The association between variant expression and heterochromatin marks at the gene family level was significant (two-tailed Fischer's exact test, P < 0.001). The few discrepancies are probably explained by all members of some families being in an active state in the parasite lines used for the HP1/H3K9me3 mapping, or by variant genes being in the same state, either active or repressed, in all our parasite lines. The striking correlation between heterochromatin marks and variant expression at the gene family level was also observed at the individual gene level within some families such as lysophospholipase, apiAP2, acbp (Fig. 4), or clag. The association between variant expression and heterochromatin marks at the gene level was highly significant by a two-tailed Fischer's exact test (P < 0.001), either including only genes from the gene families listed in Table 1 or the full genome, and also excluding the major variant gene families var, rif, stevor, and pfmc-2tm from the analysis. The association was also highly significant when analyzed by GSEA (Supplemental Fig. S9B). These results strongly suggest a general mechanistic link between H3K9me3-based heterochromatin and phenotypic variation. Previous studies showed that variant expression of var genes and genes encoding merozoite proteins is mediated by reversible formation of heterochromatin, with H3K9me3 associated to the repressed state (Chookajorn et al. 2007; Lopez-Rubio et al. 2007; Jiang et al. 2010; Comeaux et al. 2011; Crowley et al. 2011). Our data suggest that this is a general mechanism also controlling variant expression of other functionally unrelated gene families, including members of non-subtelomeric gene families such as apiAP2. However, clonally variant genes that are not members of gene families were generally not marked by H3K9me3 or HP1 and may not be regulated by heterochromatin formation (Supplemental Table S2). Specific transcription factors, such as an ApiAP2 (Campbell et al. 2010) that showed variant expression (PFL1085w) and is HP1/H3K9me3-positive (Fig. 6C), may be master regulators controlling the variant expression of other genes.
Figure 6.
Heterochromatin-based regulation of variant expression. (A) Correlation between variant expression and heterochromatin marks at the gene family level. (Yellow) Gene families (described in Supplemental Table S4) with at least one member showing variant expression (“Variant” column) or positive for heterochromatin marks (“HP1/H3K9me3” column) in any of the published studies (Flueck et al. 2009; Lopez-Rubio et al. 2009; Salcedo-Amaya et al. 2009). (Dark yellow) Gene families with only one gene showing variant expression in only one of the comparisons (gene families not included in Table 1). (B–E) Chromosomal distribution of variantly expressed genes. (Top lane, green) The log2 of the aMAFC in the 3D7 comparison. The other lanes represent the relative transcript (blue) and gDNA (red) levels in pairwise parasite line comparisons, expressed as the log2 ratio of the fold-change. (Bottom lane, black) HP1- or H3K9me3-positive genes.
The stage of expression during the cycle is not coregulated along the chromosomes (Bozdech et al. 2003). However, our results indicate that, at least for gene families involved in host–parasite interactions, and in contrast with developmental regulation, clonally variant expression is regulated by heterochromatin formation. H3K9me3-based heterochromatin has the capacity to spread into adjacent regions (Grewal and Jia 2007), which may result in coordinated repression of neighboring genes. Many of the genes under variant expression cluster together in subtelomeric regions, but we observed little evidence for coordinated expression patterns. Different patterns of expression were often observed for genes within the same variant expression cluster (Fig. 6B), and stand-alone variant genes were observed in pairwise parasite line comparisons (Fig. 6C,D). Possible coordinated repression along a large chromosome region was only observed in a subtelomeric region of chromosome 14, which includes several genes involved in gametocyte formation (Eksi et al. 2005), but further along the chromosome neighboring genes showed opposite expression patterns in some parasite lines (Fig. 6E). The general lack of coordinated expression in clusters of neighboring genes that can be repressed by formation of heterochromatin indicates that subtelomeric regions are organized into small chromatin domains, possibly delimitated by barrier insulators, as previously suggested by us (Crowley et al. 2011). This would allow independent variant expression of neighboring genes, maximizing the number of possible combinations of expressed and repressed genes and consequently the adaptative potential of clonally variant gene expression.
Discussion
Here we present, to our knowledge, the most comprehensive characterization of epigenetic variation in the malaria parasite P. falciparum, and we provide experimental evidence for adaptation of malaria parasites to changes in their environment mediated by clonally variant expression. Our experimental design and analysis resulted in unprecedented accuracy in the transcriptional comparison of different parasite lines, which allowed us to distinguish between authentic epigenetic, clonally transmitted transcriptional differences, and variation attributable to genetic differences or to different relative abundance of parasites at different stages. This approach led to the identification of a large number of clonally variant genes and gene families and demonstrated that genetically homogeneous parasite populations derived from a single parasite (clonal) are epigenetically highly heterogeneous. The extensive overlap in the genes and gene families that show variant expression in comparisons involving different sets of parasite lines indicates that spontaneous clonally variant expression is an intrinsic property of some genes and gene families. Data showing expression variability for each P. falciparum gene and the subclones with defined transcriptional patterns are important resources that will be available through PlasmoDB (http://www.plasmodb.org) and MR4 (http://www.mr4.org).
The critical aspects of our study design and analysis that allowed identification of the majority of inherently variant gene families were (1) transcriptional comparisons involving parasite lines of the same clonal origin, including recently subcloned parasite lines; (2) transcriptional comparison along the full asexual blood cycle for a large number of parasite lines, belonging to four different genetic backgrounds; (3) tight synchronization of parasites to a well-defined narrow age window of 0–5 h post-invasion, completely avoiding contaminating parasites from the previous generation; (4) identification of transcriptional differences based on expression plots with statistically estimated times, and calculation of expression fold-changes from the area underneath the curve of the plots (aMAFC score); (5) exclusion of different probes for the analysis of parasites of different genetic backgrounds, based on CGH and blast analysis; and (6) CGH analysis of all parasite lines, to avoid the potential confounding effect of genetic alterations. This workflow provides the necessary level of accuracy to identify, with high confidence and sensitivity, epigenetic differences between independent parasite lines. However, some clonally variant genes and gene families may have escaped identification by our approach (false negatives). Those may include genes with very fast switching rates (resulting in a mixed population for their expression status even in the limited number of generations from subcloning to analysis), genes that under culture conditions are in the same active or repressed state in the vast majority of parasites, and genes that had to be excluded from the microarray analysis because they are highly polymorphic or have near-identical paralogs. Regarding this last point, only in a small number of gene families did the majority of genes have to be excluded from the analysis (Supplemental Fig. S10).
H3K9me3 is a hallmark of constitutive heterochromatin in higher eukaryotes (Trojer and Reinberg 2007), but in P. falciparum this mark plays a major role in facultative heterochromatin formation at loci involved in host–parasite interactions. The vast majority of gene families that are positive for heterochromatin marks also show variant expression, demonstrating a general association between H3K9me3-based heterochromatin gene repression and phenotypic variation. This observation reveals a scenario in which specific regions of the largely euchromatic P. falciparum genome are organized into bistable chromatin domains. These regions can be found in two alternative clonally transmitted states, euchromatin or heterochromatin, which are associated with transcriptional activity or repression, respectively. Bistable chromatin is prone to infrequent spontaneous switches between its two possible states, as a consequence of molecular noise (Dodd et al. 2007; Crowley et al. 2011). The striking correlation between variant expression and heterochromatin marks allows us to predict that the majority of the genes that were positive for heterochromatin marks in previous studies may be variantly expressed, implying that ∼10% of the genome may be under clonally variant expression. Considering that we observed only very few cases of coregulated variant expression, this represents an unlimited number of possible combinations of expressed and repressed genes, with the exception of families such as var or clag in which specific rules of mutually exclusive expression apply (Cortés et al. 2007; Scherf et al. 2008).
Our results provide a snapshot of transcriptional diversity within clonal parasite populations, revealing that variable expression mainly affects gene families involved in host–parasite interactions. The degree of genetic diversity between isolates in these gene families provides insights into the physiological role of transcriptional variability. Gene families specifically amplified in the P. falciparum lineage can be roughly divided into those that show hypervariability between parasite isolates, such as var, rif, stevor, and pfmc-2tm, and those that are conserved between isolates, including the majority of other amplified families (e.g., phist, dnaj, fikk, etramp, acs, lysophospholipases, and many others) (Jeffares et al. 2007; Mu et al. 2007; Volkman et al. 2007; Templeton 2009). Amplification of these two types of gene families was probably driven by immune evasion and functional diversification, respectively (Templeton 2009). We clearly observed clonally variant expression for both types of gene families, suggesting that transcriptional variability results not only in antigenic variation but also in functional variation.
This idea is further supported by the known or predicted function of the gene families for which transcriptional variability is an intrinsic property and is suggestive of a role for variant expression in P. falciparum adaptation beyond immune evasion. Gene families for which we observed variant expression participate in metabolic processes such as lipid metabolism or nutrient import (acs, acbp, lysophospholipases, and clag), in protein folding/stability (predicted cochaperones of families dnajIII and phistb/dnaj), or in erythrocyte invasion (eba and possibly clag), among other functions. Expression of alternative members of these families, possibly with different functional abilities, has the potential to mediate adaptation to changes in host metabolic/nutritional conditions, changes in temperature associated to febrile episodes, or host genetic polymorphism in erythrocyte receptors, respectively, which are probably the major fluctuating environmental conditions that P. falciparum asexual stages face beyond immune pressure (Merrick and Duraisingh 2010). Importantly, two of the transcriptionally different parasite lines characterized in this study differ in their ability to use alternative invasion pathways or to invade erythrocytes with the cerebral malaria-protective mutation SAO (Cortés et al. 2004), and here we provide evidence that some of the parasite lines also differ in their sensitivity to heat-shock. Also supporting the idea that variant expression can modulate fitness and hence permit adaptation upon challenge, switches in the expression of clag3 genes were recently shown to result in altered sensitivity to chemical inhibitors (Nguitragool et al. 2011) and may determine changes in solute permeability. Another case of special interest is variant expression of the expanded gene family acs. This gene family encodes enzymes that participate in host–parasite interactions by mediating activation of fatty acids scavenged from the host, with different specificities predicted for different members of the family (Tellez et al. 2003; Bethke et al. 2006). In summary, the large number of gene families under clonally variant gene expression and their characteristics indicate that they have the potential to mediate adaptation to the major fluctuating conditions encountered by P. falciparum asexual blood stages, clearly beyond immune evasion.
Adaptation through clonally variant gene expression is fundamentally different from adaptation through directed transcriptional responses (Fig. 5D). We propose that spontaneous, stochastic switches between the active and repressed states for a large number of genes generate transcriptional diversity within clonal parasite populations before any challenge is applied, thus representing a bet-hedging (risk-spreading) strategy (Veening et al. 2008). Upon a change in the environment, transcriptional heterogeneity provides the grounds for natural selection of pre-existing parasites with transcriptional patterns that confer maximum fitness under the new conditions. Diversity confers fitness to a population in changing environments, and mathematical models predict that diversity generated by stochastic variation is favored over sensing followed by transcriptional responses when the environment changes infrequently (Kussell and Leibler 2005), as in the case of the environment where P. falciparum blood stages reside. Adaptation through a stochastic phenotype-switching mechanism, controlled at the epigenetic level, occurs at a much faster time scale than genetic adaptation and is both heritable and reversible. Our results showing extensive transcriptional variation for genes involved in the interaction of the parasite with its environment provide strong support to the idea that bet-hedging strategies play a predominant role in P. falciparum adaptation. This idea is further supported by our experiments studying adaptation to febrile temperatures, one of the common fluctuating conditions faced by malaria parasites. The observed pattern of adaptation is fully consistent with adaptation via selection of pre-existing parasites and epigenetic transmission of the transcriptional patterns that result in adaptation. While adaptation via spontaneous clonally variant expression/selection and adaptation via directed transcriptional responses are not mutually exclusive, the former strategy may compensate for the reported limited ability of P. falciparum to mount immediate directed responses. This is a testable hypothesis that has important implications not only to understand how this devastating parasite adapts to natural fluctuating environmental conditions, but also to predict how it may evolve in front of renewed efforts to control or eradicate malaria (Mackinnon and Marsh 2010). Carefully designed future studies will need to identify the variantly expressed genes that determine survival under specific physiologically relevant conditions. Any studies aimed at identifying genes that mediate adaptation to specific environmental conditions should be designed and interpreted considering that in P. falciparum adaptation may require a time scale consistent with natural selection of pre-existing epigenetic variants.
Methods
Parasites
Clonal parasite lines 3D7, 7G8, D10, and HB3 have been previously described (Anders et al. 1983; Burkot et al. 1984; Walliker et al. 1987). 3D7 stocks 3D7-A and 3D7-B, and 3D7-A subclones have also been described before (Cortés et al. 2004, 2007; Cortés 2005). HB3A and HB3B are stocks of the same HB3 clonal parasite line obtained before (HB3A) or after (HB3B) mosquito and chimpanzee passage (Walliker et al. 1987), kindly provided by O. Kaneko (Nagasaki University, Japan). The 7G8 parental parasite line and 7G8 subclones identified as 7G8 self-fertilization events (MRA-926, MRA-927, MRA-928, MRA-931, and MRA-933, MR4, ATCC; Manassas, VA) in a 7G8 × GB4 genetic cross (Hayton et al. 2008) were obtained from MR4 (http://www.mr4.org). Other subclones were generated in this study by limiting dilution. We cultured subclones for as few generations as possible between subcloning and transcriptional analysis (for subclones generated by us, typically about 20 cycles since limiting dilution). The genetic identity of all parasite lines was corroborated by PCA of CGH data (Supplemental Fig. S11). Sequencing of a highly polymorphic region of the gene ama-1 (nucleotides 450–930, non-3D7 parasite lines) or msp2 restriction fragment length polymorphism (RFLP) (Cortés et al. 2007) confirmed the genetic identity of all parasite lines except for the 7G8 subclone LD10. This subclone had been identified as a 7G8 self-fertilization event by microsatellite typing (Hayton et al. 2008), and our PCA clustered it with other 7G8-derived parasite lines, but ama1 sequencing, performed after completing the study, revealed a GB4 ama-1 sequence. Consequently, genes that were identified as clonally variant based only on transcript levels in LD10 were excluded from the list of 7G8 variant genes (Supplemental Table S1), because in these cases transcriptional differences may be attributable to genetic differences.
Parasites were cultured under standard conditions in media containing Albumax II (Invitrogen) and no human serum, in a 5% CO2, 3% O2, 92% N2 atmosphere. Tight synchronization was achieved by Percoll (3D7 parasite lines) or magnet (all other parasite lines) purification of schizonts followed by sorbitol lysis 5 h later, to obtain a population of a defined age window of 0–5 h post-invasion. Parasites were split into six or seven dishes that were cultured undisturbed for different periods of time (10, 20, 30, 34, 37, 40, or 43 h) before collecting in TRIzol and freezing at −80°C. Details of heat-shock experiments are presented in the Supplemental Methods.
Microarray experiments
The microarray used in this study is based on long oligonucleotides (70 nt) designed against the PlasmoDB 4.4 release of the 3D7 reference P. falciparum genome (Hu et al. 2007). We reannotated the microarray against the 3D7 PlasmoDB 7.0 genome release, which resulted in changes in more than 700 probes, including removal of cross-reactive probes. Additional probes were excluded for the analysis of parasites genetically different from 3D7, because some probes fall within highly polymorphic regions of the genome (Supplemental Figs. S4, S10, S12; Supplemental Table S5; Supplemental Methods).
RNA was purified using the TRIzol method, and cDNA was synthesized by reverse transcription, purified, labeled, and hybridized essentially as previously described (Bozdech et al. 2003). Microarray images were analyzed using GenePix Pro 6.0 (Axon Instruments), with careful visual inspection to refine automatic spot definitions and to eliminate poor quality spots. All test samples (labeled with Cy5) were hybridized against a reference pool (labeled with Cy3), which consisted of a mixture of cDNA from 3D7-A and 3D7-B rings, trophozoites, and schizonts in equal amounts for samples from 3D7-derived parasite lines, or from D10, 7G8, and HB3 for all other samples. 3D7 was not included in the parentals comparison because it was hybridized against a different reference pool. For CGH analysis, sonicated gDNA of test parasite lines was labeled with Cy5 and hybridized against 3D7-A gDNA labeled with Cy3.
Data analysis
All data were processed using Bioconductor in an R environment. Spots with Cy3 and Cy5 intensities lower than 1.5-fold times the microarray background were eliminated. When a spot had intensity higher than 1.5 times the background for only Cy3 or Cy5, the other channel was assigned a value of 1.5 times its microarray background, to avoid extreme ratio values. After background subtraction, the log2 of the Cy5/Cy3 intensity ratio was normalized using locally estimated scatterplot smoothing (LOESS). Gene level log2(Cy5/Cy3) expression ratios were computed from probe level values using median polish. We calculated the most likely time post-invasion along the asexual cycle for each sample using a statistical likelihood method (Lemieux et al. 2009), with the HB3 transcriptome as reference (Bozdech et al. 2003). Expression values for each gene [log2(Cy5/Cy3)] were plotted against estimated times for each parasite line. For genes with values for at least five time points, missing value(s) were estimated using linear model predictors. Estimated values were <1% of the total.
To compare expression levels among a given set of parasite lines and to identify genes under variant expression, the area under the curve of the time-course plots was computed for the time interval between the latest first time point and the earliest last time point among the parasite lines compared (time compared, TC). For each gene, areas were only calculated for parasite lines that had values for all time points (either experimental or estimated). For developmentally regulated genes that are only expressed during part of the asexual cycle (Bozdech et al. 2003), it is expected that potential differences in expression levels between parasite lines would be observed only at stages when the genes are expressed. To prevent a bias against such genes, areas were also computed for the following overlapping half-TC intervals: 0×–0.5× TC, 0.5×–1× TC, 0.25×–0.75× TC, and the complementary to the latter, 0×–0.25× TC + 0.75×–1× TC. The area difference (AD) for a given half-TC interval was the difference between the largest and smallest areas among the parasite lines compared.
We calculated for each gene the maximum average fold-change across half-TC (MAFC). Because expression values were in log2 scale, MAFC was calculated as: MAFC = 2[max(AD)/0.5× TC], where max refers to the maximum of AD among the four half-TC intervals. The MAFC corresponds to the expression fold-change between the parasite line that expressed the gene at the highest level and the parasite line that expressed it at the lowest level, within a given comparison. Rather than corresponding to an expression fold-change at a single time point, the MAFC is the average expression fold-change across a period corresponding to approximately half an asexual cycle. Different sets of parasite lines compared showed different levels of background AD attributable to technical variability, and background AD was also different between the four half-TC intervals (Supplemental Fig. S13). To account for this, we calculated for each gene an adjusted maximum average fold-change across half-TC (aMAFC) as: aMAFC = 2[max(AD-B)/0.5× TC], where max refers to the maximum of AD-B among the four half-TC intervals, and B is the background level of area differences. B was estimated as the 80th percentile of AD for each comparison and each half-TC interval (calculated excluding var, rif, stevor, and pfmc-2tm genes, which were eliminated for the analysis of non-3D7 parasite lines). The 80th percentile was selected based on the assumption that in all comparisons <20% of the genes have authentic variant expression. Because the 80th percentile reflects the high end of area differences that may be attributable to experimental variability, and because the aMAFC is an average-expression fold-change across a long time interval rather than a maximum-expression fold-change at a single time point, the aMAFC is a conservative estimate of the expression fold-change for any given gene. Comparison of the distribution of aMAFC with non-adjusted maximum average fold-changes (MAFC) demonstrates that the adjustment corrects for the different levels of technical variability in the different comparisons (Supplemental Fig. S13).
We set up empirical criteria to produce lists of genes showing variant transcript levels in each comparison. These criteria were based on the aMAFC and permutation tests with 10,000 permutations of parasite line labels within each time point, not allowing two consecutive labels from the same parasite line. The P-values, which correspond to the probability of a random permutation generating a higher value than the observed AD, were corrected for multiple testing using the Benjamini and Hochberg method (Benjamini and Hochberg 1995). For the comparison of sets of parasite lines with the same clonal origin, all genes with aMAFC higher than 2.5 were included in the lists of variantly expressed genes. In all comparisons, an aMAFC of 2.5 was higher than the average plus 3 standard deviations of aMAFC in non-variant genes. Genes with aMAFC between 1.75 and 2.5 were only included in the lists of variant genes if the adjusted P-value of the permutation test was P < 0.05. Genes with aMAFC lower than 1.75 were not included in the lists. Finally, plots for variant genes were visually inspected, and genes with differences in expression based only on data points with low absolute Cy5 signal intensity (less than three times background) were removed from the lists of variantly expressed genes, because near-background values provide lower confidence. For the comparison of non-isogenic parasite lines (parentals comparison), which as expected showed higher levels of transcriptional variability (Supplemental Fig. S13) that may be explained by the effect of genetic differences on transcript levels (Gonzales et al. 2008) or by technical aspects, only genes with aMAFC > 3.5 were retained in the list of variant genes. This represents only the top 2.1% of genes showing highest levels of transcriptional variability in this comparison, with observed expression fold-changes (MAFC) above 6.75.
We also normalized Cy5 intensity values (no ratio) using quantile normalization among samples collected at the same time point. These values provide only a semi-quantitative estimation of the level of expression of a gene, because different oligonucleotides have intrinsically different hybridization efficiencies, and in these arrays each gene is represented by a small number of probes (average 1.9 probes/gene). Gene Cy5 intensity values were calculated as the median of values for the different probes.
Heatmaps and hierarchical clustering were performed using TMEV 4.4 (Saeed et al. 2006). Chromosome plots were constructed using IGV (Robinson et al. 2011). GSEA analysis was conducted using GSEA Preranked (Subramanian et al. 2005).
Data access
The microarray data presented in this study have been submitted to Array Express under accession number E-MTAB-673.
Acknowledgments
This research was funded by Plan Nacional de Investigación Científica, Desarrollo e Innovación Tecnológica (I+D+I), Instituto de Salud Carlos III – Subdirección General de Evaluación y Fomento de la Investigación grant PI07/0891, and Spanish Ministry of Science and Innovation grant SAF2010-20111 to A.C., Singaporean National Medical Research Council grant #IRG07Nov030 to Z.B., and European Union FP7 grant HEALTH-F3-2009-223024 MEPHITIS to L.R.d.P. A.C. acknowledges a travel fellowship from the Catalan Government (Research and Universities Commission), and support from the European Commission (EviMalaR Network of Excellence). We thank D. Rossell for useful discussion on data analysis, J.E. Lemieux for assistance with the statistical estimation of parasite age, and O. Kaneko for providing parasite lines HB3A and HB3B. We also thank MR4 for providing us with malaria parasites contributed by Karen Hayton and Tom Wellems (NIAID, NIH). We are also grateful to A. Casali, A. Vaquero, S. Pagans, and M. Llinas for useful comments on the manuscript.
Authors' contributions: A.C. conceived the study. A.C. and Z.B. designed the experiments. N.R., A.C., V.M.C., and A.P.G. performed the experiments. A.C. and E.P. analyzed the data. S.M., P.R.P., and L.R.d.P. contributed materials/analysis tools/discussion of the data. A.C. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.129692.111.
References
- Anders RF, Brown GV, Edwards A 1983. Characterization of an S antigen synthesized by several isolates of Plasmodium falciparum. Proc Natl Acad Sci 80: 6652–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji S, Babu MM, Iyer LM, Aravind L 2005. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res 33: 3994–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Statist Soc 57: 289–300 [Google Scholar]
- Bethke LL, Zilversmit M, Nielsen K, Daily J, Volkman SK, Ndiaye D, Lozovsky ER, Hartl DL, Wirth DF 2006. Duplication, gene conversion, and genetic diversity in the species-specific acyl-CoA synthetase gene family of Plasmodium falciparum. Mol Biochem Parasitol 150: 10–24 [DOI] [PubMed] [Google Scholar]
- Bischoff E, Vaquero C 2010. In silico and biological survey of transcription-associated proteins implicated in the transcriptional machinery during the erythrocytic development of Plasmodium falciparum. BMC Genomics 11: 34 doi: 10.1186/1471-2164-11-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol 1: E5 doi: 10.1371/journal.pbio.0000005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkot TR, Williams JL, Schneider I 1984. Infectivity to mosquitoes of Plasmodium falciparum clones grown in vitro from the same isolate. Trans R Soc Trop Med Hyg 78: 339–341 [DOI] [PubMed] [Google Scholar]
- Campbell TL, De Silva EK, Olszewski KL, Elemento O, Llinas M 2010. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathog 6: e1001165 doi: 10.1371/journal.ppat.1001165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chookajorn T, Dzikowski R, Frank M, Li F, Jiwani AZ, Hartl DL, Deitsch KW 2007. Epigenetic memory at malaria virulence genes. Proc Natl Acad Sci 104: 899–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeaux CA, Coleman BI, Bei AK, Whitehurst N, Duraisingh MT 2011. Functional analysis of epigenetic regulation of tandem RhopH1/clag genes reveals a role in Plasmodium falciparum growth. Mol Microbiol 80: 378–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés A 2005. A chimeric Plasmodium falciparum Pfnbp2b/Pfnbp2a gene originated during asexual growth. Int J Parasitol 35: 125–130 [DOI] [PubMed] [Google Scholar]
- Cortés A 2008. Switching Plasmodium falciparum genes on and off for erythrocyte invasion. Trends Parasitol 24: 517–524 [DOI] [PubMed] [Google Scholar]
- Cortés A, Benet A, Cooke BM, Barnwell JW, Reeder JC 2004. Ability of Plasmodium falciparum to invade Southeast Asian ovalocytes varies between parasite lines. Blood 104: 2961–2966 [DOI] [PubMed] [Google Scholar]
- Cortés A, Carret C, Kaneko O, Yim Lim BY, Ivens A, Holder AA 2007. Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PLoS Pathog 3: e107 doi: 10.1371/journal.ppat.0030107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson RM, Hall N, Ouzounis CA 2004. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome Res 14: 1548–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley VM, Rovira-Graells N, de Pouplana LR, Cortés A 2011. Heterochromatin formation in bistable chromatin domains controls the epigenetic repression of clonally variant Plasmodium falciparum genes linked to erythrocyte invasion. Mol Microbiol 80: 391–406 [DOI] [PubMed] [Google Scholar]
- Daily JP, Scanfeld D, Pochet N, Le Roch K, Plouffe D, Kamal M, Sarr O, Mboup S, Ndir O, Wypij D, et al. 2007. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature 450: 1091–1095 [DOI] [PubMed] [Google Scholar]
- Dodd IB, Micheelsen MA, Sneppen K, Thon G 2007. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell 129: 813–822 [DOI] [PubMed] [Google Scholar]
- Eksi S, Haile Y, Furuya T, Ma L, Su X, Williamson KC 2005. Identification of a subtelomeric gene family expressed during the asexual–sexual stage transition in Plasmodium falciparum. Mol Biochem Parasitol 143: 90–99 [DOI] [PubMed] [Google Scholar]
- Flueck C, Bartfai R, Volz J, Niederwieser I, Salcedo-Amaya AM, Alako BT, Ehlgen F, Ralph SA, Cowman AF, Bozdech Z, et al. 2009. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog 5: e1000569 doi: 10.1371/journal.ppat.1000569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Functional Genomics Workshop Group. 2007. Mechanisms of gene regulation in Plasmodium. Am J Trop Med Hyg 77: 201–208 [PubMed] [Google Scholar]
- Ganesan K, Ponmee N, Jiang L, Fowble JW, White J, Kamchonwongpaisan S, Yuthavong Y, Wilairat P, Rathod PK 2008. A genetically hard-wired metabolic transcriptome in Plasmodium falciparum fails to mount protective responses to lethal antifolates. PLoS Pathog 4: e1000214 doi: 10.1371/journal.ppat.1000214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Escobar N, Amambua-Ngwa A, Walther M, Okebe J, Ebonyi A, Conway DJ 2010. Erythrocyte invasion and merozoite ligand gene expression in severe and mild Plasmodium falciparum malaria. J Infect Dis 201: 444–452 [DOI] [PubMed] [Google Scholar]
- Gonzales JM, Patel JJ, Ponmee N, Jiang L, Tan A, Maher SP, Wuchty S, Rathod PK, Ferdig MT 2008. Regulatory hotspots in the malaria parasite genome dictate transcriptional variation. PLoS Biol 6: e238 doi: 10.1371/journal.pbio.0060238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S 2007. Heterochromatin revisited. Nat Rev Genet 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Gunasekera AM, Myrick A, Le Roch K, Winzeler E, Wirth DF 2007. Plasmodium falciparum: Genome wide perturbations in transcript profiles among mixed stage cultures after chloroquine treatment. Exp Parasitol 117: 87–92 [DOI] [PubMed] [Google Scholar]
- Hayton K, Gaur D, Liu A, Takahashi J, Henschen B, Singh S, Lambert L, Furuya T, Bouttenot R, Doll M, et al. 2008. Erythrocyte binding protein PfRH5 polymorphisms determine species-specific pathways of Plasmodium falciparum invasion. Cell Host Microbe 4: 40–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Llinas M, Li J, Preiser PR, Bozdech Z 2007. Selection of long oligonucleotides for gene expression microarrays using weighted rank-sum strategy. BMC Bioinformatics 8: 350 doi: 10.1186/1471-2105-8-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Cabrera A, Kono M, Mok S, Chaal BK, Haase S, Engelberg K, Cheemadan S, Spielmann T, Preiser PR, et al. 2010. Transcriptional profiling of growth perturbations of the human malaria parasite Plasmodium falciparum. Nat Biotechnol 28: 91–98 [DOI] [PubMed] [Google Scholar]
- Jeffares DC, Pain A, Berry A, Cox AV, Stalker J, Ingle CE, Thomas A, Quail MA, Siebenthall K, Uhlemann AC, et al. 2007. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet 39: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Lopez-Barragan MJ, Jiang H, Mu J, Gaur D, Zhao K, Felsenfeld G, Miller LH 2010. Epigenetic control of the variable expression of a Plasmodium falciparum receptor protein for erythrocyte invasion. Proc Natl Acad Sci 107: 2224–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussell E, Leibler S 2005. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309: 2075–2078 [DOI] [PubMed] [Google Scholar]
- Lavazec C, Sanyal S, Templeton TJ 2007. Expression switching in the stevor and Pfmc-2TM superfamilies in Plasmodium falciparum. Mol Microbiol 64: 1621–1634 [DOI] [PubMed] [Google Scholar]
- Lemieux JE, Gomez-Escobar N, Feller A, Carret C, Amambua-Ngwa A, Pinches R, Day F, Kyes SA, Conway DJ, Holmes CC, et al. 2009. Statistical estimation of cell-cycle progression and lineage commitment in Plasmodium falciparum reveals a homogeneous pattern of transcription in ex vivo culture. Proc Natl Acad Sci 106: 7559–7564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roch KG, Johnson JR, Ahiboh H, Chung DW, Prudhomme J, Plouffe D, Henson K, Zhou Y, Witola W, Yates JR, et al. 2008. A systematic approach to understand the mechanism of action of the bisthiazolium compound T4 on the human malaria parasite, Plasmodium falciparum. BMC Genomics 9: 513 doi: 10.1186/1471-2164-9-513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinas M, Bozdech Z, Wong ED, Adai AT, DeRisi JL 2006. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res 34: 1166–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez Rivas R, Scherf A 2007. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol 66: 1296–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rubio JJ, Mancio-Silva L, Scherf A 2009. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe 5: 179–190 [DOI] [PubMed] [Google Scholar]
- Mackinnon MJ, Marsh K 2010. The selection landscape of malaria parasites. Science 328: 866–871 [DOI] [PubMed] [Google Scholar]
- Mackinnon MJ, Li J, Mok S, Kortok MM, Marsh K, Preiser PR, Bozdech Z 2009. Comparative transcriptional and genomic analysis of Plasmodium falciparum field isolates. PLoS Pathog 5: e1000644 doi: 10.1371/journal.ppat.1000644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick CJ, Duraisingh MT 2010. Epigenetics in Plasmodium: what do we really know? Eukaryot Cell 9: 1150–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok BW, Ribacke U, Winter G, Yip BH, Tan CS, Fernandez V, Chen Q, Nilsson P, Wahlgren M 2007. Comparative transcriptomal analysis of isogenic Plasmodium falciparum clones of distinct antigenic and adhesive phenotypes. Mol Biochem Parasitol 151: 184–192 [DOI] [PubMed] [Google Scholar]
- Mu J, Awadalla P, Duan J, McGee KM, Keebler J, Seydel K, McVean GA, Su XZ 2007. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet 39: 126–130 [DOI] [PubMed] [Google Scholar]
- Mu J, Myers RA, Jiang H, Liu S, Ricklefs S, Waisberg M, Chotivanich K, Wilairatana P, Krudsood S, White NJ, et al. 2010. Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat Genet 42: 268–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natalang O, Bischoff E, Deplaine G, Proux C, Dillies MA, Sismeiro O, Guigon G, Bonnefoy S, Patarapotikul J, Mercereau-Puijalon O, et al. 2008. Dynamic RNA profiling in Plasmodium falciparum synchronized blood stages exposed to lethal doses of artesunate. BMC Genomics 9: 388 doi: 10.1186/1471-2164-9-388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndonwi M, Burlingame OO, Miller AS, Tollefsen DM, Broze GJ Jr, Goldberg DE 2011. Inhibition of antithrombin by Plasmodium falciparum histidine-rich protein II. Blood 117: 6347–6354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguitragool W, Bokhari AA, Pillai AD, Rayavara K, Sharma P, Turpin B, Aravind L, Desai SA 2011. Malaria parasite clag3 genes determine channel-mediated nutrient uptake by infected red blood cells. Cell 145: 665–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes MC, Goldring JP, Doerig C, Scherf A 2007. A novel protein kinase family in Plasmodium falciparum is differentially transcribed and secreted to various cellular compartments of the host cell. Mol Microbiol 63: 391–403 [DOI] [PubMed] [Google Scholar]
- Oakley MS, Kumar S, Anantharaman V, Zheng H, Mahajan B, Haynes JD, Moch JK, Fairhurst R, McCutchan TF, Aravind L 2007. Molecular factors and biochemical pathways induced by febrile temperature in intraerythrocytic Plasmodium falciparum parasites. Infect Immun 75: 2012–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochola LI, Tetteh KK, Stewart LB, Riitho V, Marsh K, Conway DJ 2010. Allele frequency-based and polymorphism-versus-divergence indices of balancing selection in a new filtered set of polymorphic genes in Plasmodium falciparum. Mol Biol Evol 27: 2344–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petter M, Haeggstrom M, Khattab A, Fernandez V, Klinkert MQ, Wahlgren M 2007. Variant proteins of the Plasmodium falciparum RIFIN family show distinct subcellular localization and developmental expression patterns. Mol Biochem Parasitol 156: 51–61 [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J 2006. TM4 microarray software suite. Methods Enzymol 411: 134–193 [DOI] [PubMed] [Google Scholar]
- Salcedo-Amaya AM, van Driel MA, Alako BT, Trelle MB, van den Elzen AM, Cohen AM, Janssen-Megens EM, van de Vegte-Bolmer M, Selzer RR, Iniguez AL, et al. 2009. Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum. Proc Natl Acad Sci 106: 9655–9660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargeant TJ, Marti M, Caler E, Carlton JM, Simpson K, Speed TP, Cowman AF 2006. Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol 7: R12 doi: 10.1186/gb-2006-7-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A, Lopez-Rubio JJ, Riviere L 2008. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol 62: 445–470 [DOI] [PubMed] [Google Scholar]
- Spielmann T, Fergusen DJ, Beck HP 2003. etramps, a new Plasmodium falciparum gene family coding for developmentally regulated and highly charged membrane proteins located at the parasite–host cell interface. Mol Biol Cell 14: 1529–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamez PA, Bhattacharjee S, van Ooij C, Hiller NL, Llinas M, Balu B, Adams JH, Haldar K 2008. An erythrocyte vesicle protein exported by the malaria parasite promotes tubovesicular lipid import from the host cell surface. PLoS Pathog 4: e1000118 doi: 10.1371/journal.ppat.1000118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez M, Matesanz F, Alcina A 2003. The C-terminal domain of the Plasmodium falciparum acyl-CoA synthetases PfACS1 and PfACS3 functions as ligand for ankyrin. Mol Biochem Parasitol 129: 191–198 [DOI] [PubMed] [Google Scholar]
- Templeton TJ 2009. The varieties of gene amplification, diversification and hypervariability in the human malaria parasite, Plasmodium falciparum. Mol Biochem Parasitol 166: 109–116 [DOI] [PubMed] [Google Scholar]
- Templeton TJ, Kaslow DC 1999. Identification of additional members define a Plasmodium falciparum gene superfamily which includes Pfs48/45 and Pfs230. Mol Biochem Parasitol 101: 223–227 [DOI] [PubMed] [Google Scholar]
- Trojer P, Reinberg D 2007. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell 28: 1–13 [DOI] [PubMed] [Google Scholar]
- Van Tyne D, Park DJ, Schaffner SF, Neafsey DE, Angelino E, Cortese JF, Barnes KG, Rosen DM, Lukens AK, Daniels RF, et al. 2011. Identification and functional validation of the novel antimalarial resistance locus PF10_0355 in Plasmodium falciparum. PLoS Genet 7: e1001383 doi: 10.1371/journal.pgen.1001383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Smits WK, Kuipers OP 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62: 193–210 [DOI] [PubMed] [Google Scholar]
- Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, Milner DA Jr, Daily JP, Sarr O, Ndiaye D, Ndir O, et al. 2007. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet 39: 113–119 [DOI] [PubMed] [Google Scholar]
- Walliker D, Quakyi IA, Wellems TE, McCutchan TF, Szarfman A, London WT, Corcoran LM, Burkot TR, Carter R 1987. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science 236: 1661–1666 [DOI] [PubMed] [Google Scholar]
- Winter G, Kawai S, Haeggstrom M, Kaneko O, von Euler A, Kawazu S, Palm D, Fernandez V, Wahlgren M 2005. SURFIN is a polymorphic antigen expressed on Plasmodium falciparum merozoites and infected erythrocytes. J Exp Med 201: 1853–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Johnson JR, Benner C, Yan SF, Chen K, Le Roch KG, Zhou Y, Winzeler EA 2008. In silico discovery of transcription regulatory elements in Plasmodium falciparum. BMC Genomics 9: 70 doi: 10.1186/1471-2164-9-70 [DOI] [PMC free article] [PubMed] [Google Scholar]