Abstract
Improvements in technology have reduced the cost of DNA sequencing to the point that the limiting factor for many experiments is the time and reagent cost of sample preparation. We present an approach in which 192 sequencing libraries can be produced in a single day of technician time at a cost of about $15 per sample. These libraries are effective not only for low-pass whole-genome sequencing, but also for simultaneously enriching them in pools of approximately 100 individually barcoded samples for a subset of the genome without substantial loss in efficiency of target capture. We illustrate the power and effectiveness of this approach on about 2000 samples from a prostate cancer study.
Improvements in technology have reduced the sequencing cost per base by more than a 100,000-fold in the last decade (Lander 2011). The amount of sequence data that is needed per sample, for example, for studying small target regions or low-coverage sequencing of whole genomes is often less than the commercial cost of “library” preparation, so that library preparation is now often the limiting cost for many projects. To reduce library preparation costs, researchers can purchase kits and produce libraries in their own laboratories or use published library preparation protocols (Mamanova et al. 2010; Meyer and Kircher 2010; Fisher et al. 2011). However, this approach has two limitations. First, available kits have limited throughput so that scaling to thousands of samples is difficult without automation. Second, an important application of next-generation sequencing technology is to enrich sample libraries for a targeted subsection of the genome (like all the exons) (Albert et al. 2007; Hodges et al. 2007; Gnirke et al. 2009), and then to sequence this enriched pool of DNA, but such experiments are expensive because of the high costs of target capture reagents. One way to save funds is to pool samples prior to target enrichment (after barcoding to allow them to be distinguished after the data are gathered). Although the recently introduced Nextera DNA Sample Prep Kit (Illumina) together with “dual indexing” (12 × 8 indices and two index reads) allows higher sample throughput for library preparation (Adey et al. 2010) and pooling of up to 96 libraries, the long indexed adapter may interfere during pooled hybrid selection (see below).
We report a method for barcoded library preparation that allows highly multiplexed pooled target selection (hybrid selection or hybrid capture). We demonstrate its usefulness by generating libraries for more than 2000 samples from a prostate cancer study that we have enriched for a 2.2-Mb subset of the genome of interest for prostate cancer. We also demonstrate the effectiveness of libraries produced with this strategy for whole-genome sequencing, both by generating 40 human libraries and sequencing them to fivefold coverage, and by generating 12 microbial libraries and sequencing them to 150-fold coverage. Our method was engineered for high-throughput sample preparation and low cost, and thus we implemented fewer quality-control steps and were willing to accept a higher rate of duplicated reads compared with methods that have been optimized to maximize library complexity and quality (Meyer and Kircher 2010; Fisher et al. 2011). Because of this, our method is not ideal for deep sequencing of large genomes (e.g., human genome at 30×), where sequencing costs are high enough that it makes sense to use a library that has as low a duplication rate as possible. However, our method is advantageous for projects in which a modest amount of sequencing is needed per sample, so that the savings in sample preparation outweigh costs due to sequencing duplicated molecules or failed libraries. Projects that fall into this category include low-pass sequencing of human genomes, microbial sequencing, and target capture of human exomes and smaller genomic targets.
Our method reduces costs and increases throughput by parallelizing the library preparation in 96-well plates, reducing enzyme volumes at a cost-intensive step, using inexpensive paramagnetic beads for size selection and buffer exchange steps (DeAngelis et al. 1995; Lennon et al. 2010; Meyer and Kircher 2010), and automation (Farias-Hesson et al. 2010; Lennon et al. 2010; Lundin et al. 2010; Fisher et al. 2011). To permit highly multiplexed sample pooling prior to target enrichment or sequencing, we attach “internal” barcodes directly to sheared DNA from a sample that is being sequenced, and flank the barcoded DNA fragments by partial sequencing adapters that are short enough that they do not strongly interfere during enrichment (the adapters are then extended after the enrichment step). By combining these individual libraries in pools and enriching them for a subset of the genome, we show that we obtain data that are effective for polymorphism discovery, without substantial loss in capture efficiency.
Outline
Our method is based on a blunt-end-ligation method originally developed for the 454 Life Sciences (Roche) platform (Stiller et al. 2009), which we have extensively modified for the Illumina platform to reduce costs and increase sample processing speed, by parallelizing the procedure in 96-well format and automating the labor-intensive cleanup steps (Figs. 1, 2A; Methods; Supplemental Notes). Some of the modifications adapt ideas from the literature, such as DNA fragmentation on the Covaris E210 instrument in 96-well PCR plates (Lennon et al. 2010) or replacing the gel-based size selection by a bead-based, automatable, size selection (Lennon et al. 2010; Borgstrom et al. 2011). Another change is to replace a commonly used commercial kit (AMPure XP kit) for SPRI-cleanup steps with a homemade mix. An important feature of our libraries compared with almost all other Illumina library preparation methods (Cummings et al. 2010; Mamanova et al. 2010; Meyer and Kircher 2010; Teer et al. 2010) is that we add a 6-bp “internal” barcoded adapter to each fragment (Craig et al. 2008). These adapters are ligated directly to the DNA fragments, leading to “truncated” libraries with 34- and 33-bp overhanging adapters at the end of each DNA fragment. Adapters at this stage in our library preparation are sufficiently short that they interfere with each other minimally during hybrid capture, compared with what we have found when long adapters are used (64 and 61 bp on either side, including the 6-bp internal barcode). The truncated adapter sites are then extended to full length after hybrid capture allowing the libraries to be sequenced (Fig. 2A). To assess how this strategy works in different-sized pools (between 14 and 95), we applied it to 2.2 Mb of the genome of interest for prostate cancer, where it reduces the capture reagent that is required by two orders of magnitude while still producing highly useful data. Sequencing these libraries shows that we can perform pooled target capture on at least 95 barcoded samples simultaneously without substantial reduction in capture efficiency.
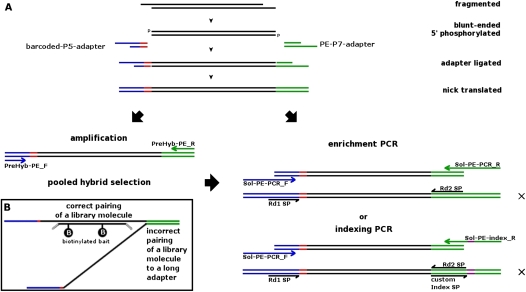
Figure 1.
Experimental workflow of the library preparation protocol for 95 samples for pooled hybrid capture.
Figure 2.
(A) Schematic overview of the library preparation procedure using the Illumina PE adapter (internal barcode in red). After a cascade of enzymatic reactions and cleanup steps, enrichment PCR can be performed to complete the adapter sites for Illumina PE sequencing (Rd1 SP, Rd2 SP are PE sequencing primers). Alternatively, libraries can be pooled for hybrid selection (if desired), and then enrichment PCR can be performed after hybrid selection. To achieve an even higher magnitude of pooling for sequencing, “indexing PCR” can be performed instead of “enrichment PCR,” whereby unique indices (in purple) are introduced to the adapter, and a custom index sequencing primer (index-PE-sequencing-Primer) is used to read out the index in a separate read. Finished libraries that have all the adapters necessary to allow sequencing are marked with an X. (B) Schematic figure of “daisy-chaining” during pooled solution hybrid capture, which may explain why a large proportion of molecules are empirically observed to be off-target when using long adapters. Library molecules exhibiting the target sequences hybridize to biotinylated baits, but unwanted library molecules can also hybridize to the universal adapter sites. The adapters of our “truncated” libraries (including barcode: 34 and 33 bp) are about half the length of regular “long” adapters (64 and 61 bases), and thus may be less prone to binding DNA fragments that do not belong to the target region.
The fact that we are using internal strategy in which barcoded oligonucleotides are ligated directly to fragmented DNA is a nonstandard strategy, which deserves further discussion. First, when combined with indexing (introducing a second barcode via PCR after pooling) (Fig. 2A; Meyer and Kircher 2010), an almost unlimited number of samples can be pooled and sequenced in one lane. We are currently using this strategy in our prostate cancer study to test library quality and to assess the number of sequence-able molecules per library prior to equimolar pooling for hybrid capture. Second, a potential concern of our strategy of directly ligating barcodes is that differences in ligation efficiency for different barcodes in principle could cause some barcodes to perform less efficiently than others. However, to date, we have used each of 138 barcodes at least 15 times and have not found evidence of particular barcodes performing worse than others as measured by the number of sequenced molecules per library. Third, the blunt-end ligation used in our protocol results in a loss of 50% of the input DNA because two different adapters have to be attached to either side. This is not a concern for low-coverage and small-target studies using input DNA amounts of 500 ng or higher but is not an ideal strategy for samples with less input material. Fourth, chimeras of blunted DNA molecules can be created during blunt-end ligation. In our protocol, the formation of chimeras is reduced by using adapter oligonucleotides in such vast excess to the sample DNA that the chance of ligating barcodes to the DNA is much higher than ligating two sample molecules (while the adapters can form dimers, these are removed during bead cleanup). Fifth, when using our internal barcodes, it is important to pool samples in each lane in such a way that the base composition of the barcodes is balanced, because the Illumina base-calling software assumes balanced nucleotide composition especially during the first few cycles. This is of particular importance when only a few barcoded samples are being pooled. To prevent base-calling problems in such unbalanced pools, a PhiX library can be spiked into the library to increase diversity.
We performed a rough calculation breaking the cost for our method down into (a) reagents, (b) technician time, and (c) capital equipment (Table 1). The reagents and consumables cost is about $9 per sample without taking into account discounts that would be available for a project that produced large numbers of libraries. The cost for technician time is $3 per sample, assuming that an individual makes 480 libraries on five plates per week. Capital costs are difficult to compute (because some laboratories may already have the necessary equipment), but if one computes the cost of a Covaris LE220 instrument, a PCR machine, and an Agilent Bravo liquid handling platform, and divides by the cost of 100,000 libraries produced over the 2–3-yr lifetime of these instruments, this would add about $3 more to the cost per sample. This accounting does not include administrative overhead, space rental, process management, quality control on the preparation of reagents, bioinformatic support, data analysis, and research and development, all of which could add significantly to cost.
Table 1.
Cost and time assumptions for library preparation
Application 1: Enrichment of more than 2000 human samples by solution hybrid capture
For many applications, it is of interest to enrich a DNA sample for a subset of the genome; for example, in medical genetics, a candidate region for disease risk, or all exons. The target-enriched (captured) sample can then be sequenced. To perform studies with statistical power to detect subtle genetic effects with genome-wide significance, however, it is often necessary to study thousands of samples (Kryukov et al. 2009; Lango Allen et al. 2010), which can be prohibitively expensive given current sample preparation and target enrichment costs. We designed our protocol with the aim of allowing barcoded and pooled samples to be captured simultaneously. Specifically, our libraries have internal barcodes that are tailored to pooled hybrid capture, whereas most other libraries have external barcodes in the long adapters. It has been hypothesized that hybridization experiments using libraries that already have long adapters do not work efficiently in pooled hybridizations because a proportion of library molecules not only hybridize to the “baits” but also catch unwanted off-target molecules with the long adapter (“daisy-chaining”) (Mamanova et al. 2010; Nijman et al. 2010), thus reducing capture efficiency (Fig. 2B). In the Supplemental Notes (“Influence of Adapter Length in Pooled Hybrid Capture”), we present experiments showing that the number of reads mapping to the target region increased from 29% to 73% when we shortened the adapters (Supplemental Table S1), providing evidence for the hypothesis that interference between barcoded adapters is lowered by short adapters. Our results show empirically that short adapters improve hybridization efficiency.
To investigate the empirical performance of our libraries in the context of target capture, we produced libraries for 189 human samples starting from 0.2–4.8 μg of DNA (98% <1 μg for fragmentation), prepared in two 96-well plates as in Supplemental Figure S1. We combined the samples into differently sized pools of libraries (14, 28, 52, and 95) and then enriched the pooled libraries using a custom Agilent SureSelect Target Enrichment Kit in the volume recommended for a single sample (the target was a 2.2-Mb subset of the genome containing loci relevant to prostate cancer). We sequenced the three smaller pools together on one lane of the Genome Analyzer II instrument (36 bp, single reads) and the 95-sample pool on one lane of a HiSeq2000 instrument (50 bp, paired-end reads). We aligned the reads to the human genome using BWA (Li and Durbin 2009), after removing the first six bases of the first read that we used to identify the sample. We removed PCR duplicates using Picard's (http://picard.sourceforge.net) MarkDuplicates and computed hybrid selection statistics with Picard's CalculateHsMetrics. For the 95-sample pool (unnormalized before hybrid capture), f2 = 93% of samples had a mean target coverage of within a factor of 2 of the median, f1.5 = 67% within a factor of 1.5 of the median, and the coefficient of variation (standard deviation divided by mean coverage) was CV = 0.40. For the three smaller pools where normalization was performed, coverage was in general more uniform: For the pool of 14, f2 = 93%, f1.5 = 86%, CV = 0.66; for the pool of 28, f2 = 100%, f1.5 = 96%, CV = 0.19; and for the pool of 52, f2 = 100%, f1.5 = 94%, CV = 0.19 (Supplemental Table S2). In the 95-sample experiment, the percentage of selected bases, defined as “on bait” or within 250 bp of either side of the baits, was 70%–79% across samples (Table 2; Supplemental Table S2), comparable to the literature for single-sample selections (Supplemental Table S3). Results on the 95-sample pool are as good as the 14-, 28-, and 52-sample pools.
Table 2.
Sequencing results
To demonstrate that pooled target capture using our libraries is amenable to an experiment on the scale that is relevant to medical genetic association studies, we used the library preparation method to prepare 2152 DNA samples from one population (African-Americans) in the space of 2 mo. We normalized these samples to the lowest concentrated sample in each pool, combined them into 15 pools of between 138 and 144 samples, and enriched these 15 pools for the 2.2-Mb target. We sequenced the captured products on a HiSeq 2000 instrument using 75-bp paired-end reads to an average coverage of 4.1 in nonduplicated reads (data not shown). The duplication rate of the reads was on average 72%, an elevation above the levels reported in Table 2 and Supplemental Table S2 that we hypothesize is due to dilution to the lowest-complexity library within the pools. We were able to solve this problem by replacing the dilution with a cherry-picking approach that combines samples of similar complexity. We tested this approach by pooling 81 prostate cancer libraries with similar complexity (allowing no more than a 5× difference in molecule count per library), resulting in a duplication rate of 24% on average at 7× coverage (Supplemental Table S2e).
The experiment was highly sensitive for detecting polymorphisms in the targeted regions. After restricting to sites with at least one-fourth of the average coverage, we discovered 35,211 polymorphisms at high confidence (10,000:1 probability of being real based on their quality score from BWA). This is more than double the 16,457 sites discovered by the 1000 Genomes Project in 167 African ancestry samples over the same nucleotides (February 2011 data release) (The 1000 Genomes Project Consortium 2010). Exploring this in more detail, we found that we rediscovered 99.7% of sites in the 1000 Genomes Project with minor allele frequency >5% and 83% of 1000 Genomes Project sites with lower frequency in the African samples. As a second measure of the quality of our data, we compared n = 1642 African-American samples that had previously been genotyped on an Illumina 1M array at 1367 SNPs that overlapped between that array and the 2.2-Mb target region of the capture experiments. We found that 99.77% of the mapped de-duplicated reads are consistent with the “gold standard” results from genotyping. As a third measure of data quality, we checked for a potential reference bias by counting the reads matching the reference and variant allele at the 1367 SNPs where we knew the true genotypes. As shown in Supplemental Figure S2, there is a slight bias (Nref/Ntot = 1,289,080/2,537,488 = 50.8%) for the reference allele, which is sufficiently small that we do not expect it to cause a major problem for most applications such as identification of heterozygous sites.
Application 2: Whole-genome sequencing of 40 human libraries to 5× coverage
Whole-genome shotgun sequencing (WGS) of mammalian genomes to high coverage (e.g., 30×) is still a process that is dominated by sequencing costs. However, lighter sequencing is of interest for some applications. For example, Genomewide Association Studies (GWAS), which have discovered more than 1300 associations to human phenotypes (Manolio 2010), cost hundreds of dollars per sample on SNP arrays, which is less than commercial costs of library preparation, and hence sequence-based GWAS are not economical. However, the situation would change if library production costs were lower. If libraries were inexpensive, sequencing the genome to light coverage followed by imputing missing data using a reference panel of more deeply sequenced or genotyped samples, in theory would allow more cost-effective GWAS (Li et al. 2011). With sufficiently low library production costs, sequencing may begin to compete seriously with SNP array–based analysis for medical genetic association studies, as is already occurring in studies of gene expression analysis, where RNA-seq is in the process of replacing array-based methods (Majewski and Pastinen 2010).
To test if our method can produce libraries appropriate for whole-genome sequencing, we prepared 40 libraries using an earlier version of our protocol that used microTUBES for shearing instead of plates and a slightly different enrichment PCR procedure (Supplemental Fig. S3). (A more up-to-date protocol, which involves shearing in plates and which we used to produce libraries for the prostate cancer study, further reduces costs by about $5 per sample.) Table 2 and Supplemental Table S4 show the results of sequencing these libraries to an average of 5.4× coverage using 100-bp paired-end reads on 58 lanes on Illumina HiSeq 2000 instruments. A high proportion (95%) of the reads align to the human reference genome (hg19) using BWA (Li and Durbin 2009), and duplicates were removed. We found that 99.86% of the mapped reads are concordant with the “gold standard” SNP array data previously collected on these samples (Li et al. 2008) (sequences with quality ≥30 for the 40 libraries were compared at 585,481 SNPs). Thus, we have demonstrated that our protocol can produce libraries that are useful for low-pass whole-genome human sequencing.
Application 3: Sequencing of 12 Escherichia coli strains to 150× coverage
An important application of high-throughput sequencing is the study of microbial genomes, for example, in an epidemiological context where it is valuable to study strains from many patients to study the spread of an epidemic, or in the same individual to study the evolution of an infection. Microbial genomes are small so that the required amount of sequencing per sample can be small, and thus the limiting cost is often sample preparation. To explore the utility of our library preparation protocol for microbial sequencing, we produced libraries for 12 E. coli strains for a project led by M. Lajoie, F. Isaacs, and G. Church (whom we thank for allowing us to report the data) (Isaacs et al. 2011). We produced these libraries as a single row on a 96-well plate with an input DNA amount of 1 μg together with human libraries that we were producing for another study following the protocol in Supplemental Figure S4. Table 2 and Supplemental Table S5 report the results of the sequencing of these 12 libraries on a single lane of a HiSeq 2000 (50-bp paired-end reads). We analyzed the data after separating the libraries by sample using internal barcodes and mapping to the E. coli reference (strain K12 substrain MG1655, Refseq NC_000913) using BWA (Li and Durbin 2009). Overall, 97% of reads mapped, with an average of 147-fold coverage and 1% duplicated reads.
Discussion
We have reported a high-throughput library preparation method for next-generation sequencing, which has been designed to allow an academic laboratory to generate thousands of barcoded libraries at a cost that is one to two orders of magnitude less than the commercial cost of library preparation. These libraries are appropriate for whole-genome sequencing of large and small genomes. A particularly important feature of these libraries is that they are effective for pooling approximately a hundred samples together and enriching them for a subset of the genome of interest. We have proven that the method is practical at a scale that is relevant to medical genetics by generating more than 2000 libraries for a prostate cancer study, enriching them for more than 2 Mb of interest, and obtaining sequencing data that are concordant with previously reported genotype calls.
From an engineering point of view, our method was designed with a different set of goals than have driven most previous library preparation methods. In most methods, the emphasis has been on producing libraries with maximal complexity (as measured by the number of unique molecules) and length uniformity (as measured by the tightness of the distribution of insert sizes) given the large amount of sequencing that was planned for each library. Our goal is different: to increase throughput and decrease reagent cost, while building libraries that are appropriate for pooled target capture. In this study, we empirically show that the human libraries produced by our method are complex enough that when shotgun-sequenced to a coverage of around 5×, they give duplication rates of 9%–20%. This duplication rate is somewhat higher than some published protocols, and the problem of duplication becomes greater as coverage increases, so that for deep-sequencing studies (e.g., whole-genome sequencing at 30×) in which thousands of dollars are invested per sample, it may be more economical to use a more expensive library preparation protocol that minimizes duplication rates. One reason for an increased duplication rate in our libraries is our distribution of fragment insert sizes. Because size selection with beads is not as tight as gel-based size selection, fragment insert sizes of the libraries produced with our protocol are variable. Longer fragments are more prone to duplicated reads (“optical duplicates”), in which the Illumina software identifies one cluster as two adjacent clusters. Another reason for an increased duplication rate is the low input DNA amount per ligation reaction (0.75 μg for each of the four ligation reactions per sample), much less than the recommended 3–5 μg for standard whole-genome sequencing library protocols; we also lose complexity because 50% of molecules are lost during blunt-end ligation due to wrong adapter combinations. Coverages of 10-fold or less, a level where our libraries have reasonable duplication rates, have been shown to be highly effective for SNP discovery and genotype imputation (The 1000 Genomes Project Consortium 2010), and thus our libraries are valuable for most medical genetic applications. The high duplication rate for our prostate cancer target capture enrichment study (72% at about 4× coverage) arose from the normalization strategy of diluting to the lowest complex library within each pool. We were able to lower the duplication rate to 24% at about 7× coverage when we pooled similarly complex libraries and hope to be able to lower this even further in the future.
The method we have presented is tailored to paired-end sequencing using Illumina technology but is easy to adapt to multiplexing (we recently switched to the Multiplexing-P7 adapter) and to other technologies, for example, 454 Life Sciences (Roche), Applied Biosystems SOLiD (Life Technologies), and Ion Torrent (Life Technologies). While these technologies are different at the detection stage, they are similar in sample preparation, in that technology-specific adapters are attached to DNA fragments, and the fragments are subjected to enrichment PCR to complete the adapter sites, allowing clonal amplification of the libraries and subsequent sequencing-by-synthesis. Thus, a method for one technology can be modified for use with the others. Although we only used the Agilent SureSelect platform for hybrid selections, we expect that similar hybridization-based target enrichment systems, such as the Illumina TruSeq Enrichment kits (Clark et al. 2011), the Roche/NimbleGen SeqCap EZ Hybridization kits, and array-based hybridization (Hodges et al. 2007), would enrich multiplexed samples as efficiently as the Agilent system if the libraries are prepared with short adapters.
There are several potential improvements to our method, which should make it possible to produce libraries at even higher throughput, and to further improve library quality. A bottleneck at present is the machine time required for sample shearing. On the Covaris E210 instrument, 21 h are required to shear to a mean insert size of 200–300 bp for a plate of 96 samples (although this takes negligible technician time), and thus two instruments would be required to produce enough sheared samples for a full-time technician. However, this bottleneck could be eliminated by a recently released instrument, the Covaris LE220, which is able to shear eight samples simultaneously. The number of samples that can be pooled per lane is 159 with our 6-mer 5′-barcodes, but may not be enough if, for example, the target size is small and the desired coverage is low. When combining the barcoding strategy with indexing via PCR, a much greater number of samples can be pooled. Another way to increase the number of samples that can be pooled is to either extend the number of barcode nucleotides or to ligate two different adapters on either side of the molecule. Further improvements to the protocol and quality-control steps are important directions, which should improve the usefulness of these libraries even further.
Methods
We discuss each of the steps of the protocol in turn, highlighting modifications that achieve substantial savings in terms of reagents or technician time compared with previously published protocols (Fig. 1 presents the workflow of the library preparation for the hybrid selection application). A detailed protocol for all three applications (pooled hybrid selection, human shotgun sequencing, microbial sequencing) is given in the Supplemental Notes.
DNA fragmentation
We used the Covaris E210 AFA instrument (Woburn, MA) (Quail et al. 2008, 2009; Fisher et al. 2011) to shear the DNA into fragments of a desired length. A previous study on automated 454 library preparation (Lennon et al. 2010) showed that it is possible to use 96-well PCR plates on the E210, and we adapted the method to another plate type and a shorter fragment size (Supplemental Notes, “DNA Fragmentation”). This reduces consumable costs for the shearing step by ∼90-fold compared with the microTUBE plates provided by Covaris.
Reaction cleanup
Library preparation involves a cascade of enzymatic reactions as well as intermediate cleanup steps for buffer-exchange. To make the cleanup steps efficient, our method heavily uses paramagnetic carboxyl-coated beads in a PEG/NaCl buffer, known as SPRI technology (DeAngelis et al. 1995), as a replacement for column-based cleanup. The beads have three advantages. First, they allow parallelization of the procedure in a way that is impossible using column-based methods (Farias-Hesson et al. 2010; Lennon et al. 2010; Lundin et al. 2010; Meyer and Kircher 2010; Fisher et al. 2011). Second, they permit size selection, which is important for removing PCR primer or adapter dimers (Quail et al. 2008). Third, they permit a “dual size selection” to reduce not only small DNA molecules but also long molecules (Lennon et al. 2010; Borgstrom et al. 2011) (see below, “Fragment Size Selection”). The commercially available kits are expensive. Thus, we used a homemade mix by combining Carboxyl-modified Sera-Mag Magnetic Speed-beads (Fisher Scientific, cat. #65152105050250) in a PEG/NaCl buffer (MagNA; see Supplemental Notes, “Reaction Clean-Up”). We have found empirically that this combination of reagents attains performance that is comparable to the commercial kit with respect to yield and retained fragment sizes for our application (Supplemental Fig. S5). Using commercial bead kits instead of a homemade mix would raise reagent costs for our protocol from $8.80 to about $16.80 (as the commercial cost is about $8 per sample).
Fragment size selection
Gel-based fragment size selections are not amenable to high-throughput sample preparation even using fully automated systems such as the Pippin-Prep (SageScience) or the LabChip XT Chip Prep (Caliper). Dual SPRI size selection is faster and can be automated more easily (Lennon et al. 2010; Borgstrom et al. 2011). Because SPRI purification is already part of our protocol at all cleanup and concentration steps, we used the beads to perform size selection for whole-genome shotgun sequencing applications. We aimed for a mean insert of 300 bp, and thus we attempted to remove fragments larger than 400 bp and smaller than 200 bp (Supplemental Notes, “Dual Fragment Size Selection”). Size selection can be performed at any stage of the protocol, although in the examples reported here we performed it after fragmentation.
Sample barcoding
We are using a blunt-end ligation procedure to add barcoded, truncated adapter to the fragmented end-repaired DNA (Stiller et al. 2009). Specifically, one of the two truncated partially double-stranded adapters includes a 6-mer molecular barcode that is directly ligated to the blunted and 5′-phosphorylated DNA fragments and is therefore detected in the first six cycles of the first sequencing read. Because the adapters are not 5′-phosphorylated (to prevent adapter dimer formation and to reduce cost), a nick fill-in-step has to be performed before enrichment PCR, which then completes the truncated adapter sites so that the libraries can be sequenced (Fig. 2A). This PCR finishes the library preparation for the two WGS applications, but not for hybrid selection. For hybrid selection, we have modified the protocol so that the enrichment PCR (to complete the adapter to full length) is performed after hybrid capture, since we have found that the long adapters interfere with hybrid capture (see Supplemental Notes, “Influence of Adapter Length in Pooled Hybrid Capture”). No indexing read is needed to read out the internal barcode, but because cluster identification is performed in these cycles in the Illumina technology, care has to be taken to equally balance the four nucleotides at any of the six positions within the barcodes that will be sequenced together. Reaction conditions and overviews about the procedure can be found in Figures 1 and 2A (Supplemental Notes, “Sample Barcoding”) and Supplemental Figures S1, S3, and S4.
Automation
To achieve high-throughput library production, it is crucial to use automated liquid handling robots (although a multichannel pipettor can be used for eight or 12 samples at once). The simple liquid handler that we used for the libraries we produced for microbial whole-genome shotgun sequencing and hybrid capture has a 96-tip head and is thus appropriate for the cleanup and transfer steps. Over time, we slightly modified the protocol (in particular, for elution volumes), so that it is now (as for the microbial libraries) even more automated. It would be possible to further automate the protocol if the robot were capable of moving plates between positions and had heating and cooling elements. We anticipate that with a robot with all of these capabilities, the technician time for library production would be ∼1.5 h per plate; in particular, a single dedicated technician could produce 384 libraries in a workday simply by replacing tip boxes and providing plates with master mixes and buffer solutions on two robots in parallel.
Normalization and pooling
To achieve an even read coverage across samples that are being sequenced simultaneously, it is necessary to measure the number of sequence-able fragments per library before pooling. For the WGS of microbial samples and the smaller sample pools for pooled hybrid selection (14, 28, and 52 samples per pool), a quantitative real-time PCR assay was used, and for the whole-genome shotgun sequencing of human samples, one lane of Illumina sequencing was performed to determine the number of sequence-able molecules per library. Libraries were subsequently pooled in equimolar ratios per application and sequenced for the WGS experiments or enriched prior to sequencing for the pooled hybrid capture experiments. No normalization was carried out for the 95-sample pool for hybrid selection, but for the prostate cancer project, we used sequencing to determine the copy number per truncated library before pooling for hybrid capture. Because we are reusing the barcoded adapters (159 total) (Supplemental Table S6), copy number determination via sequencing for a total of 2152 libraries was achieved by sequencing these samples all together on just one lane (pooling 138–144 libraries each and using indexing PCR to introduce a unique index to one of the adapters to each of these pools) (Fig. 2A), a cost of approximately $0.67 per library. Normalization was then performed. Detailed experimental conditions are given in the Supplemental Notes (“Copy Number Determination for Equimolar Library Pooling”).
Pooled hybrid selection
A custom Agilent SureSelect Target Enrichment Kit with a target size of 2.2 Mb was developed for a medical genetics study of prostate cancer risk loci. Here, we focus on results for 189 barcoded libraries that we pooled into four pools with 14–95 libraries and performed a single hybrid selection per pool. Experimental conditions for the hybridization were as given in the instructions with one modification. Since our libraries only exhibit truncated adapter sites, the blocking oligonucleotides (Block #3) from the kit were replaced by Univ_Block (see Supplemental Notes, Methods, and Supplemental Table S6). We performed 15 cycles of enrichment PCR after hybridization to complete the adapter sites for sequencing.
Data access
Raw sequence data from the human and microbial whole-genome shotgun data as well as from the pooled hybrid selection experiment (Influence of Adapter Length) have been submitted to the NCBI Sequence Read Archive (SRA) (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession number SRA047577. The dbGaP (http://www.ncbi.nlm.nih.gov/dbgap) accession number for the prostate cancer sequence data is phs000306.v3.p1.
Competing interest statement
Harvard University has filed a patent on the techniques discussed in the manuscript. N.R. and D.R. are named as coinventors.
Acknowledgments
We are grateful to Swapan Mallick, Heng Li, and Andrew Kernytsky for assistance with data analysis. We thank Matthias Meyer, Brendan Blumenstiel, and Daniel Herman for technical advice; David Altshuler, George Church, Sheila Fisher, Eric Lander, Erica Mazaika, Steve McCarroll, Matthias Meyer, Bogdan Pasaniuc, Alkes Price, Mark DePristo, Robert Steen, and James Wilson for critical comments; Marc Lajoie, Farren Isaacs, and George Church for allowing us to report on the bacterial data; Jacob Kitzman and Jay Shendure for allowing us to report on the whole-genome sequencing data; and Christopher Haiman and Brian Henderson for allowing us to report on the prostate cancer target capture data. We are grateful to the Biopolymers Facility at Harvard Medical School for sequencing services, to Christine and Jonathan Seidman for access to the Covaris E210, and to Stacy Gabriel and Christine Stevens for support. This research was supported by NIH grants CA129435, HL084107, CA63464, and HG004726 and NSF HOMINID grant no. 1032255.
Authors' contributions: N.R. and D.R. conceived the experiments and wrote the manuscript. N.R. performed the experiments.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.128124.111.
References
- The 1000 Genomes Project Consortium 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adey A, Morrison HG, Asan, Xun X, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, et al. 2010. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol 11: R119 doi: 10.1186/gb-2010-11-12-r119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. 2007. Direct selection of human genomic loci by microarray hybridization. Nat Methods 4: 903–905 [DOI] [PubMed] [Google Scholar]
- Borgstrom E, Lundin S, Lundeberg J 2011. Large scale library generation for high throughput sequencing. PLoS ONE 6: e19119 doi: 10.1371/journal.pone.0019119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MJ, Chen R, Lam HY, Karczewski KJ, Euskirchen G, Butte AJ, Snyder M 2011. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol 29: 908–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, Corneveaux JJ, Pawlowski TL, Laub T, Nunn G, Stephan DA, et al. 2008. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods 5: 887–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings N, King R, Rickers A, Kaspi A, Lunke S, Haviv I, Jowett JB 2010. Combining target enrichment with barcode multiplexing for high throughput SNP discovery. BMC Genomics 11: 641 doi: 10.1186/1471-2164-11-641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis MM, Wang DG, Hawkins TL 1995. Solid-phase reversible immobilization for the isolation of PCR products. Nucleic Acids Res 23: 4742–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias-Hesson E, Erikson J, Atkins A, Shen P, Davis RW, Scharfe C, Pourmand N 2010. Semi-automated library preparation for high-throughput DNA sequencing platforms. J Biomed Biotechnol 2010: 617469 doi: 10.1155/2010/617469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, Young G, Fennell TJ, Allen A, Ambrogio L, et al. 2011. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol 12: R1 doi: 10.1186/gb-2011-12-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. 2009. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27: 182–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, et al. 2007. Genome-wide in situ exon capture for selective resequencing. Nat Genet 39: 1522–1527 [DOI] [PubMed] [Google Scholar]
- Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, et al. 2011. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science 333: 348–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Shpunt A, Stamatoyannopoulos JA, Sunyaev SR 2009. Power of deep, all-exon resequencing for discovery of human trait genes. Proc Natl Acad Sci 106: 3871–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES 2011. Initial impact of the sequencing of the human genome. Nature 470: 187–197 [DOI] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, et al. 2010. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467: 832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon NJ, Lintner RE, Anderson S, Alvarez P, Barry A, Brockman W, Daza R, Erlich RL, Giannoukos G, Green L, et al. 2010. A scalable, fully automated process for construction of sequence-ready barcoded libraries for 454. Genome Biol 11: R15 doi: 10.1186/gb-2010-11-2-r15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319: 1100–1104 [DOI] [PubMed] [Google Scholar]
- Li Y, Sidore C, Kang HM, Boehnke M, Abecasis G 2011. Low coverage sequencing: Implications for the design of complex trait association studies. Genome Res 21: 940–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin S, Stranneheim H, Pettersson E, Klevebring D, Lundeberg J 2010. Increased throughput by parallelization of library preparation for massive sequencing. PLoS ONE 5: e10029 doi: 10.1371/journal.pone.0010029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Pastinen T 2010. The study of eQTL variations by RNA-seq: From SNPs to phenotypes. Trends Genet 27: 72–79 [DOI] [PubMed] [Google Scholar]
- Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ 2010. Target-enrichment strategies for next-generation sequencing. Nat Methods 7: 111–118 [DOI] [PubMed] [Google Scholar]
- Manolio TA 2010. Genomewide association studies and assessment of the risk of disease. N Engl J Med 363: 166–176 [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc doi: 10.1101/pdb.prot5448 [DOI] [PubMed] [Google Scholar]
- Nijman IJ, Mokry M, van Boxtel R, Toonen P, de Bruijn E, Cuppen E 2010. Mutation discovery by targeted genomic enrichment of multiplexed barcoded samples. Nat Methods 7: 913–915 [DOI] [PubMed] [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ 2008. A large genome center's improvements to the Illumina sequencing system. Nat Methods 5: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail MA, Swerdlow H, Turner DJ 2009. Improved protocols for the illumina genome analyzer sequencing system. Curr Protoc Hum Genet 62: 18.2.1–18.2.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller M, Knapp M, Stenzel U, Hofreiter M, Meyer M 2009. Direct multiplex sequencing (DMPS)—a novel method for targeted high-throughput sequencing of ancient and highly degraded DNA. Genome Res 19: 1843–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer JK, Bonnycastle LL, Chines PS, Hansen NF, Aoyama N, Swift AJ, Abaan HO, Albert TJ, Margulies EH, Green ED, et al. 2010. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res 20: 1420–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]