Abstract
Meiotic DNA double-stranded breaks (DSBs) initiate genetic recombination in discrete areas of the genome called recombination hotspots. DSBs can be directly mapped using chromatin immunoprecipitation followed by sequencing (ChIP-seq). Nevertheless, the genome-wide mapping of recombination hotspots in mammals is still a challenge due to the low frequency of recombination, high heterogeneity of the germ cell population, and the relatively low efficiency of ChIP. To overcome these limitations we have developed a novel method—single-stranded DNA (ssDNA) sequencing (SSDS)—that specifically detects protein-bound single-stranded DNA at DSB ends. SSDS comprises a computational framework for the specific detection of ssDNA-derived reads in a sequencing library and a new library preparation procedure for the enrichment of fragments originating from ssDNA. The use of our technique reduces the nonspecific double-stranded DNA (dsDNA) background >10-fold. Our method can be extended to other systems where the identification of ssDNA or DSBs is desired.
Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) is a powerful and widely used method for the identification of protein-bound DNA sequences. An intrinsic problem of ChIP-seq is the limited efficiency of the immunoprecipitation step (Park 2009). Even if high-affinity antibodies are available, the identification of target DNA sequences can be difficult if the specific protein–DNA complexes are found only in a small fraction of cells. Here we develop a procedure we call SSDS (ssDNA sequencing) that significantly improves the efficiency of ChIP-seq when the target is single-stranded DNA.
One system where the identification of protein-bound DNA targets is particularly challenging is at meiotic hotspots of recombination in mammals. Meiosis is a specialized cell division program in sexually reproducing organisms that leads to the generation of haploid gametes. An intrinsic part of meiosis is genetic recombination. In addition to its role in generating genetic diversity homologous recombination is essential for the faithful segregation of homologous chromosomes during meiosis. Mutations that substantially reduce or abolish recombination lead to infertility or aneuploidy-associated birth defects, which include multiple developmental disabilities and mental retardation. Meiotic recombination is initiated through DNA cleavage by the SPO11 protein (Petes 2001; Neale and Keeney 2006). Following DSB formation by SPO11 and 5′-DSB termini resection, 3′-ssDNA overhangs are covered by the strand exchange proteins DMC1 and RAD51 (Neale and Keeney 2006). In Saccharomyces cerevisiae, a model organism where meiotic hotspots are studied in great detail, DSBs have been mapped by the purification of covalent Spo11–DNA complexes (Gerton et al. 2000; Mieczkowski et al. 2007; Pan et al. 2011), enrichment of ssDNAs at resected ends (Blitzblau et al. 2007; Buhler et al. 2007), or ChIP of DNA–protein complexes (Borde et al. 2009). Two factors make direct meiotic DSB identification in the mouse much more difficult compared with yeast. First, although the number of DSBs introduced per meiosis is similar in mice and yeast (Buhler et al. 2007; Moens et al. 2007; Mancera et al. 2008), the mouse genome is 200 times larger. Second, unlike yeast, where cells can be induced to synchronously enter sporulation and initiate meiosis (Govin and Berger 2009), meiotic progression is asynchronous in mammals and at any given moment <2% of the cells in the mouse gonads contain DSBs (Bellve et al. 1977; Meistrich 1977). Unfortunately, sperm genotyping (Li et al. 1988; Cui et al. 1989; Hogstrand and Bohme 1994)—the method of choice for high-resolution recombination hotspot mapping in mammals—does not scale to genome-wide applications (for reviews, see Arnheim et al. 2007; Kauppi et al. 2009; Paigen and Petkov 2010).
Recently, we used ChIP-seq for DMC1 and RAD51 proteins to map meiotic recombination initiation hotspots in male mice (Smagulova et al. 2011). Though anti-DMC1 ChIP-seq yielded a high quality DSB hotspot map, this method required the use of Psmc3ip (also known as Hop2) knockout mice (Psmc3ip−/−) to enrich for cells containing unrepaired DSBs. Meiotic progression is blocked at the pachytene-like stage and DSBs are not repaired in Psmc3ip−/− mice (Petukhova et al. 2003). The need to introduce a Psmc3ip mutation and the considerable amount of sequencing necessary to obtain a representative data set limits the applicability of this method to other strains and species.
To further distinguish hotspots from the bulk of genomic DNA we incorporated ssDNA detection and enrichment steps into the ChIP-seq protocol. Here we describe a computational framework for the differential detection of ssDNA and double-stranded DNA derived fragments in a sequencing library. We also report a modified library preparation protocol for the removal of virtually all dsDNA background, leaving only ssDNA-derived fragments. We applied SSDS to map DSB hotspots in male mice and observe a much improved sensitivity and specificity of hotspot detection. Although we utilized our method only to map meiotic DSB hotspots, it can be used for many other applications where the detection of ssDNA is desired.
Results
Detection of ssDNA-originated fragments in sequencing libraries
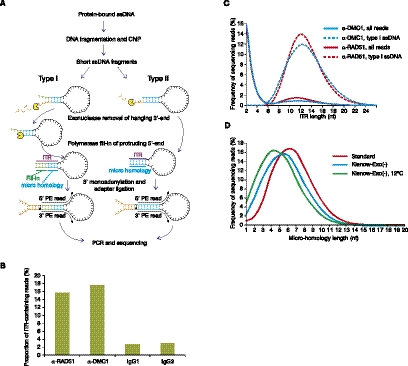
A distinctive property of ssDNA is the ability to anneal intra-molecularly at short micro-homologies and form hairpins (Fig. 1A). We used this potential for hairpin formation to devise a strategy for sequencing library preparation from ssDNA (Supplemental Fig. 1). As in the standard Illumina library preparation protocol, ssDNA hairpins are blunt-ended and then 3′-monoadenylated before ligation of the sequencing adapters.
Figure 1.
Detection of ssDNA-derived fragments in sequencing libraries. (A) Strategy for sequencing library preparation from single-stranded DNA fragments. (B) The proportion of ITR-containing reads (ITR length 6–20 nt) in α-DMC1, α-RAD51, and two control IgG samples. (C) The distribution of ITR lengths in α-DMC1 and α-RAD51 libraries. Frequency of sequencing read pairs having ITRs of a given length for all fragments (all reads) and type I ssDNA fragments (type I ssDNA) is plotted separately. (D) The length of micro-homology depends on the temperature of the reaction and on the end-repair enzyme.
Most hairpin-loop structures formed via micro-homology annealing will have both 5′ and 3′ overhangs as it is unlikely that both microhomologies will be at the very ends of the ssDNA (Fig. 1A; Supplemental Fig. 2). If both 5′ and 3′ overhangs are present, the end-repair enzyme mix of Klenow and T4 polymerases will fill in the protruding 5′ end after the exonucleolytic digestion of the 3′ overhang (type I hairpins; Fig. 1A; Supplemental Fig. 2). If the micro-homology is at the very 5′ end, the reaction will stop after the digestion of the 3′ overhang (type II hairpins; Fig. 1A; Supplemental Fig. 2). For the minority of ssDNA hairpins without overhangs, no end-repair will be performed as the ssDNA hairpin will already be blunt-ended. In all cases, if the resulting duplex stem is sufficiently stable it may serve as an efficient substrate for 3′ monoadenylation and subsequent adapter ligation.
To check whether we can detect such hairpins derived from ssDNA fragments present at DSBs, we prepared sequencing libraries from mouse meiotic cells. We first immunoprecipitated chromatin from Psmc3ip−/− mouse testes with α-DMC1 or α-RAD51 antibodies and then constructed and sequenced a paired-end library. Because meiotic DSBs contain ∼1-kb-long ssDNA nucleoprotein filaments covered with DMC1 and RAD51 (Neale and Keeney 2006), such samples should contain ssDNA after the immunoprecipitation step. Indeed, 15.7% and 17.6% of sequenced fragments from α-RAD51 and α-DMC1 libraries, respectively (Fig. 1B) contain inverted terminal repeats (ITRs) of 6–20 nt in length (Fig. 1C) compared with 2.7%–3.0% in the IgG control samples (Fig. 1B). Thus, a large number of sequencing reads containing long ITRs can be found in the sequencing libraries prepared from meiotic cells. These reads were derived from DNA fragments that preferentially interact with DMC1 and RAD51, both known to bind ssDNA (Sung 1994; Li et al. 1997). These data are thus consistent with ssDNA recovery via a hairpin-mediated mechanism.
Although both type I and type II hairpins may be ligated to sequencing adapters following repair, there is an important difference between them. While type II hairpins are completely collinear with genomic DNA, in type I hairpins the 3′ end of the fragment is copied from the 5′ end. This specific structure of the 3′ end is not found in the genome and is created only when the hairpin is extended and stabilized by the polymerase. Thus, the presence of a short sequence complementary to that at the opposite end of the DNA fragment and distinct from the target genomic DNA creates a unique signature that indicates that the fragment was single-stranded rather than double-stranded at the stage of library preparation (Fig. 1A; Supplemental Figs. 2, 3). Although type II hairpins can be derived from ssDNA as well, we cannot differentiate them from dsDNA fragments having inverted repeats at the ends. Because simply detecting ITRs at the ends of sequencing read pairs is not sufficient for specific detection of ssDNA, we devised a computational strategy to parse all reads into type I hairpins, type II hairpins, and dsDNA (Supplemental Figs. 2, 3). This strategy requires that for each fragment both reads map to the genome on the same chromosome, are on opposite strands, and within 400–500 nt of each other (see Methods for details). For both type I and II hairpins, we require an ITR >5 nt, and for type I hairpins, we also require that the fill-in ITR part (see Supplemental Figs. 2, 3) is >2 nt. Finally, we defined dsDNAs as read pairs with an ITR <3 nt. Using these criteria, 88%–91% of DNA fragments with ITRs >5 nt can be assigned to either type I or type II hairpins, with the majority being of type I (69% for both samples). Consistent with the requirement for having a fill-in part of the ITR, ITRs are longer in type I hairpins compared with type II hairpins (not shown). We subsequently exclusively used the more specific type I hairpins to detect ssDNA. However, type II hairpins that account for a substantial fraction of the true ssDNA-derived reads can be useful for more accurate strength estimates in the hotspots that have been already unambiguously identified by the presence of type I hairpins (data not shown).
Genomic coverage of putative ssDNA hairpins
According to our model (Fig. 1A), a sequence at the 3′ end of a ssDNA fragment must anneal to a complementary target near the 5′ end in order to form a hairpin. Consequently, only those fragments that have sufficiently stable micro-homology regions close to the 3′ and 5′ ends of the fragment will be recovered. This selection for micro-homologies might reduce the sensitivity of our approach and bias the detection of ssDNAs, because if the requirement for micro-homology is quite stringent, only a small subset of the genome would have properly positioned inverted repeats.
To evaluate the extent of biases introduced by the selection for micro-homology, we first estimated the lengths of these micro-homologies in our sequencing libraries (Fig. 1D). Under standard conditions, the average micro-homology is 6–7 nt long and, importantly, ∼10% of sequences have micro-homology of 4 nt or less (Fig. 1D). Next, we computed the co-occurrence of all short oligonucleotides found in an inverted orientation in the mouse genome within 50–200 nt of each other (the size range of ssDNA fragments in our libraries) (Supplemental Fig. 4). Such short DNA fragments with closely positioned inverted repeats represent potential ssDNAs that can be recovered by our method. We subsequently calculated the proportion of 1-kb windows, the approximate size of ssDNA-nucleoprotein filaments at DSBs, containing at least one such inverted repeat and found that 100% of 1-kb windows have 6- and 7-nt putative micro-homology targets within 100 nt of each other (Supplemental Fig. 5). In fact, even 9-nt targets are found in 43% of 1-kb windows (Supplemental Fig. 5). Genomic coverage of closely spaced inverted repeats is also quite deep and uniform (Supplemental Fig. 6). We calculate that every 1-kb window contains, on average, 56 six-nucleotide-long inverted repeats <100 bp apart (Supplemental Fig. 6A,B). For 95% of 1-kb windows, the variation in coverage by inverted repeats is only threefold (32 to 101 putative targets per window), while <0.1% of 1-kb windows have <20 targets. Because in many cases micro-homologies anneal with mismatches (see, for example, Supplemental Fig. 3), our estimate based on perfect palindrome occurrence is conservative. We conclude that the requirement for micro-homology is relatively mild and does not significantly impede the sensitivity or quantitative accuracy of our method when detecting kilobase-sized hotspots.
While the detection of moderately sized genomic targets such as hotspots is efficient under our standard conditions, more dense coverage by micro-homologies may be required for smaller DNA targets. We thus assessed two approaches to facilitate the selection of less stable micro-homologies. One way to enforce such selection is to prevent exonucleolytic digestion of the free 3′ end of ssDNA. Under these conditions, a blunt-ended substrate may only form if the micro-homology is at the very 3′ end of the ssDNA molecule. Thus, more stable micro-homologies which may be present away from the ssDNA 3′ end cannot be exploited. Indeed, in samples where the end repair was performed using only an exonuclease-deficient version of the Klenow fragment of Polymerase I, micro-homologies are shorter (Fig. 1D). To shorten the micro-homologies even further we lowered the reaction temperature. Micro-homologies must anneal to each other to be extended by the polymerase; therefore, lowering the reaction temperature of the ITR stabilization step should reduce average micro-homology length. In agreement with our model, lowering the reaction temperature shifts the micro-homology distribution farther to the left and, in a sample prepared at 12°C, >30% of sequences have micro-homologies <4 nt (Fig. 1D).
Kinetic enrichment of ssDNA-derived products
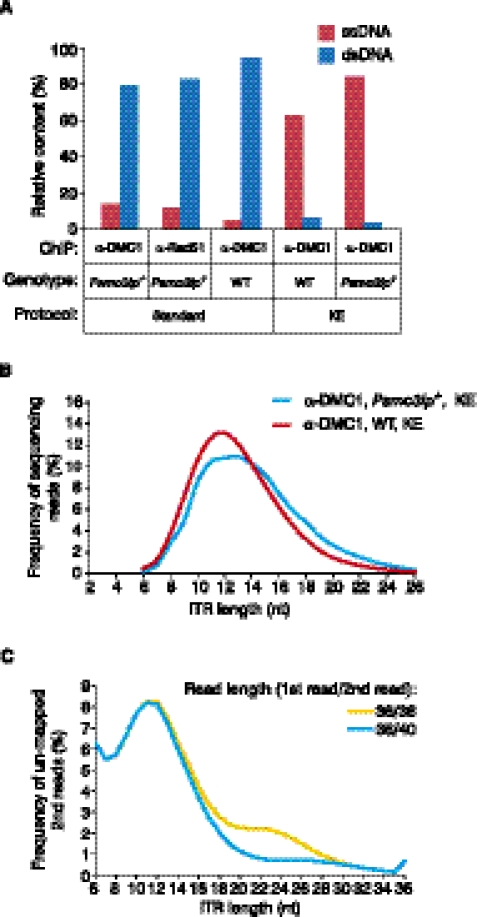
Although we can effectively identify ssDNA using the type I hairpin signature, the yield of ssDNA in ChIP-seq samples is frequently low and varies from sample to sample. Out of all the fragments that we can classify, the ssDNA-derived fragments represent 14.1% of all reads in the Psmc3ip−/− α-DMC1 sample and 12.1% in the Psmc3ip−/− α-RAD51 sample (Fig. 2A). In the α-DMC1 sample from wild-type (wt) mice, which are not enriched for meiotic cells, the ssDNA content is only 4.7%, and across a wider range of samples the ssDNA yield can be as low as 1% (Fig. 2A and not shown). To improve the yield of ssDNA-derived library fragments we introduced a kinetic enrichment (KE) step to the standard ChIP-seq procedure (Fig. 3).
Figure 2.
Enrichment of ssDNA-derived fragments in sequencing libraries. (A) Kinetic enrichment increases the proportion of ssDNA-derived reads in the sequencing libraries. The percentage of type I ssDNA- (ssDNA) and dsDNA-derived (dsDNA) reads in sequencing libraries prepared with (KE) or without (standard) kinetic enrichment. Antibody used in ChIP (α-DMC1 or α-RAD51) and genotypes (Psmc3ip−/− or wt) of the mice used are indicated on the graph. (B) The distribution of ITR lengths in type I ssDNA fragments from two KE libraries. (C) Asymmetric sequencing largely corrects bias against longer ITRs. ITR length distribution for read pairs where the first read is uniquely mapped and the second read is not mapped using either 36 nt (36/36) or 40 nt (36/40) long second reads is plotted on the graph.
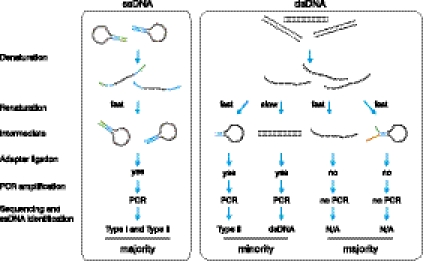
Figure 3.
Schematic representation of the kinetic enrichment approach. During the quick renaturation step the ssDNA-derived hairpins re-form almost immediately, generating ends suitable for adapter ligation (left). Complete renaturation of the dsDNA does not occur within this time frame, and the majority of the partially annealed products do not have ends suitable for ligation (right).
KE is based on the much faster annealing time of ssDNA hairpins compared with that of dsDNAs. For example, at a concentration of 1 μg/μL, the reannealing time for single-copy dsDNA fragments from a genomic library could be on the order of 10 d, while the renaturation of hairpins is virtually instantaneous (Britten and Kohne 1968). Thus, if we denature and then quickly renature a library consisting of both ssDNA hairpins and dsDNA fragments, hairpins will be reconstituted while duplex DNAs will not have enough time to reanneal and will remain single-stranded or will form hairpins with non-blunt ends (Fig. 3). We then ligate sequencing adapters to the reconstituted dsDNA ends of hairpins and PCR-amplify adapter-ligated fragments. Since we do not perform end-repair after reannealing, the majority of dsDNA-derived products will not be ligated to the adapters and PCR-amplified. The consequence is the virtually complete removal of dsDNA-derived fragments from the sequencing library and enhancement of the ssDNA-derived signal (Fig. 2A). The exception will be those few dsDNA fragments that contain sufficiently long inverted repeats right at the DNA ends. They will be recovered as type II hairpins. Assuming a minimal ITR length of 6 nt we would estimate that only 1 out of 46 dsDNA fragments or ∼0.025% will be recovered as type II hairpins. Following KE, the type I ssDNA content in a wt α-DMC1 sample and in a Psmc3ip−/− α-DMC1 sample is 62.6% and 83.8%, respectively. The distribution of ITR lengths after KE (Fig. 2B) is similar to the distribution of ITR lengths observed in samples prepared without KE (Fig. 1C; Supplemental Fig. 7). Thus, KE can be used to improve ssDNA yield without introducing any apparent distortion in the distribution of fragments.
To ensure a more equal representation of ITRs of different lengths we introduced another modification, asymmetric sequencing. The rationale for the introduction of asymmetric sequencing comes from the bias against longer ITR structures in the detected ssDNA (Fig. 2C). The fill-in part of the ITR is not collinear with the genome but copied from the other end of ssDNA fragment. Thus, the longer the fill-in part of the ITR becomes, the shorter the remaining part of the read collinear to the genome will be. This will reduce the number of reads with long fill-in parts of the ITR (and consequently, long ITRs) that can be mapped uniquely to the genome. Sequencing an extra 4 nt from the second end (36 nt for the first read/40 nt for the second read) of each fragment largely corrects this bias (Fig. 2C). The same effect can be achieved by using longer symmetrical paired-end sequencing (≥40 nt from both ends), but 36 nt first read/40 nt second read sequencing is sufficient to largely correct this bias against longer ITRs.
Identification of ssDNA in ChIP-seq libraries improves detection of meiotic DSB hotspots
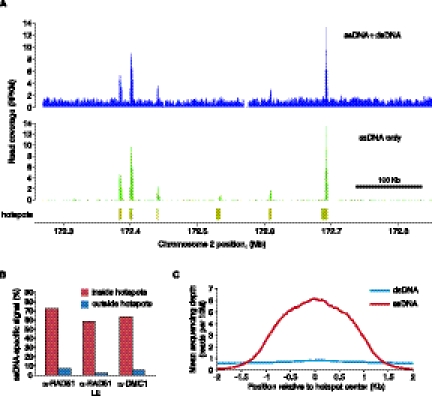
Above we have shown that ssDNAs can be detected and enriched in sequencing libraries. To evaluate the effect of ssDNA identification on the detection of DSB hotspots we have further analyzed our Psmc3ip−/− α-RAD51 and α-DMC1 sequencing libraries. A visual examination of ssDNA and ssDNA + dsDNA coverage profiles clearly shows that background is drastically reduced and hotspots are better defined in the ssDNA-specific subset of the sequencing reads (Fig. 4A). To quantify the proportion of hotspots-specific signal that is derived from ssDNA, we calculated the ssDNA-specific read density both inside and outside hotspots. We find that, inside hotspots, 60%–70% of the signal is ssDNA-specific, while the background signal comes primarily from dsDNA (Fig. 4B). In fact, using only ssDNA fragments reduces the background 30-fold in the α-RAD51 sample with an apparent low enrichment obtained in the immunoprecipitation step (α-RAD51 LE) (Fig. 4B). Background reduction is more modest for more representative higher quality α-RAD51 or α-DMC1 libraries but is still >10-fold (Fig. 4B). The proportion of ssDNA-specific signal inside hotspots increases to 80%–85% if we consider both type I and type II hairpins, but the background signal also increases (not shown). Next, we compared the sample specificity (the proportion of sequencing reads inside peaks) for all reads and for only those reads derived from ssDNA. While only 4.6% of all reads reside inside hotspots, over half (54%) of the ssDNA reads are hotspot derived in the α-RAD51 LE sample. Thus, the signal specificity increases >10-fold using ssDNA data. Next, we plotted mean coverage profiles across all previously identified hotspots (Smagulova et al. 2011) using both ssDNA and dsDNA reads (Fig. 4C). In agreement with earlier analyses (Fig. 4A,B) we see a clear accumulation of ssDNA within hotspots while the coverage profile of dsDNA across the same regions is nearly flat. Taken together, our data suggest that almost all hotspot-specific DNA fragments are single-stranded.
Figure 4.
Most of the DSB hotspot signal is ssDNA-specific. (A) ssDNA identification strongly reduces background in low-enrichment ChIP α-RAD51 Psmc3ip−/−LE library. ssDNA + dsDNA or type I ssDNA only coverage profiles (reads per 1 kb, per million reads, RPKM) are plotted for a region on mouse chromosome 2. Coverage profiles were normalized to total number of ssDNA + dsDNA reads. α-RAD51 Psmc3ip−/− LE library was prepared without using kinetic enrichment and ssDNA was computationally identified in the sequencing data. (B) The proportion of ssDNA-specific signal inside or outside all hotspots. While most of the signal inside hotspots is retained after ssDNA identification, outside of the hotspots, the background is reduced 10-fold or more. Sample specificity is plotted for α-DMC1 Psmc3ip−/− (α-DMC1), α-RAD51 Psmc3ip−/− (α-RAD51), and α-RAD51 Psmc3ip−/− LE (α-RAD51 LE) samples. (C) Most of the signal inside hotspots originates from ssDNA. Mean depth of ssDNA and dsDNA read coverage (reads per 10 million reads) across all 9874 published mouse hotspots are plotted in 5-bp increments.
The proportion of sequencing reads originating from hotspots is the main determinant of peak detection sensitivity. Previously we have used Psmc3ip−/− mice for hotspot mapping, because these mice are enriched for meiotic cells with unrepaired DSBs. To evaluate if KE of ssDNAs permits efficient hotspot mapping in mice without such enrichment, we compared the KE sample prepared from wt mice with the samples prepared without KE (Fig. 5). Similarly to the substantial reduction of background achieved by the use of only ssDNA-derived reads (Fig. 4A), physical enrichment of ssDNA-derived fragments by KE results in the much lower background in the wt KE sample compared with that in the wt non-KE sample and even when compared with the reference sample prepared from Psmc3ip−/− mice without KE (Fig. 5A; Smagulova et al. 2011). Therefore, KE does not dramatically increase background. To estimate the quality of the wt KE sample more accurately we calculated its specificity. Although the 35% specificity of the wt KE sample is lower than the specificity of the ssDNA-specific subset of our Psmc3ip−/− libraries (54% for the low ChIP enrichment α-RAD51 LE sample or up to 80% for high ChIP enrichment α-DMC1 samples), it is still nearly four times higher than the 9.4% specificity of the reference Psmc3ip−/− data set generated without ssDNA detection or KE, and 10 times higher than the specificity of the wt non-KE sample. Similarly, the outside-of-hotspots background is >10 times lower in the wt KE sample (Fig. 5A) compared with the reference Psmc3ip−/− data set. KE also does not appear to preferentially enrich hotspot-specific ssDNAs. The calculated specificities of the wt KE sample and of ssDNA-derived reads in the wt sample prepared using regular ChIP are similar (not shown). Thus, KE increases the yield of ssDNA-derived fragments but does not change their distribution.
Figure 5.
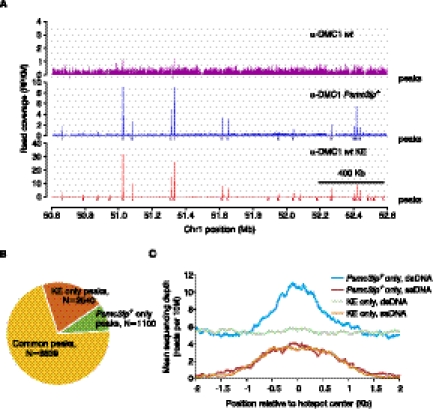
Mapping meiotic DSB hotspots in wt mice using SSDS. (A) Meiotic DSB maps obtained using wt mice and regular ChIP-seq (α-DMC1 wt), Psmc3ip−/− mice, and regular ChIP-seq (α-DMC1 Psmc3ip−/−, reference data set; Smagulova et al. 2011) or using wt mice and SSDS (α-DMC1 wt KE) in a 2 Mb region of chromosome 1. We plot read coverage for all reads without ssDNA identification for regular ChIP-seq samples. Although peaks are located at the same places, hotspot detection is more sensitive in the KE library because of the lower background. Peaks are virtually undetectable in the wt sample prepared without kinetic enrichment. (B) Most hotspots are shared between the reference Psmc3ip−/− data set and the wt KE samples. We detected hotspots in the α-DMC1 wt KE sample and compared their positions with the published hotspots (Smagulova et al. 2011). The number of peaks shared by and unique to these two data sets are plotted. (C) Hotspot detection in the wt KE sample is more specific. Mean depth of ssDNA and dsDNA read coverage (reads per 10 million reads) of Psmc3ip−/− only (reference set) and KE only hotspots.
Previously (Supplemental Figs. 5, 6) we estimated that the putative targets of our hairpin-based method, closely positioned short inverted repeats, are densely distributed in the genome and allow potential identification of nearly all hotspots. However, inverted repeat content could potentially affect the accuracy of quantification of the hotspot strength. We therefore asked whether the number of sequencing reads in the hotspots (estimated hotspot strength) is dependent on the number of putative hairpins within the hotspot. We found very little correlation between the number of inverted repeats per hotspot and the number of hotspot-derived reads (Supplemental Fig. 8). We also did not detect substantial biases toward longer ITRs/microhomologies or ITRs with higher GC content in stronger hotspots (Supplemental Fig. 9). We therefore conclude that hotspots strength quantification is not strongly affected by either the nonuniform distribution of inverted repeats in the genome or by their composition.
Next, we identified DSB hotspots in a subset of wt KE data corresponding to a single lane of an Illumina GAII flowcell (Fig. 5A,B). We restricted our analysis to a subset of data to evaluate the sensitivity of SSDS when applied to smaller data sets and to give a practical reference point for the amount of sequencing required for SSDS. Reassuringly, most of the hotspots identified with the subset of wt KE sample coincide with previously published DSB hotspots including all of the 1000 hottest previously identified hotspots (Fig. 5B; Smagulova et al. 2011). This reinforces the conclusion that there is little effect of KE on the genomic distribution of library fragments. Even though the number of reads for the wt KE sample (∼6 million) and for the reference Psmc3ip−/− data set (107 million) are vastly different, we detect 15% more hotspots in the KE sample (11,376 hotspots versus 9874) (Fig. 5B). In fact, if we use all 18 million available ssDNA reads for peak detection, we detect >2000 additional hotspots for a total of 13,937 hotspots. As for common hotspots, the majority of these newly defined KE-only hotspots overlap H3K4me3 peaks and the hotspot motif (Supplemental Fig. 10). This increased hotspot detection sensitivity in the wt KE sample is likely explained by the high content of hotspot-specific reads. Consistent with the specific ssDNA content being the primary determinant of hotspot detection sensitivity regardless of the sample identity, we can detect ∼5100 hotspots in a 6 million read subset of the reference Psmc3ip−/− data set, but <2000 hotspots in a 6 million read subset of the wt non-KE sample. Thus, while a single sequencing lane is sufficient to generate a representative DSB hotspot map using SSDS from wt mice, one needs to have >10 times more sequencing reads to define a hotspot map of similar quality using standard ChIP-seq, even when utilizing mice with the Psmc3ip gene disruption.
In addition to the 2540 hotspots found in the wt KE sample (KE only), there are 1100 hotspots found only in the reference Psmc3ip−/− sample (Fig. 4B). While the ssDNA coverage profiles across hotspots unique to each sample are similar, dsDNA coverage is denser exclusively in the hotspots detected only in the reference Psmc3ip−/− data set (Fig. 5C). Furthermore, these dsDNAs appear to be confined to the weakest hotspots (Supplemental Fig. 11). Thus, it appears that a subset of weak Psmc3ip−/− hotspots was identified due to nonspecific accumulation of dsDNA. Based on the comparison of read count distribution in the Psmc3ip−/− only and common hotspots we define a hotspot as dsDNA-derived if less than six ssDNA fragments were found within the hotspot (Supplemental Fig. 12). By this definition, 482 (44%) Psmc3ip−/− only hotspots were dsDNA-derived compared with 33 (0.4%) common hotspots (Supplemental Fig. 12), which in aggregate is in good agreement with the previously estimated false discovery rate (Smagulova et al. 2011). Thus, the hotspot detection in the KE sample is not only more sensitive, but also more specific.
Discussion
ChIP followed by paired-end sequencing with KE and computational ssDNA identification (SSDS) is a powerful and simple method for mapping meiotic DSB hotspots in complex genomes. Broadly speaking our technique is most similar to genome-wide ssDNA mapping approaches (Blitzblau et al. 2007; Buhler et al. 2007; Borde et al. 2009) that were used successfully to map DSB hotspots in yeast. To achieve the high enrichment levels that are necessary in mammals, our method combines specific computational ssDNA detection with physical enrichment of ssDNA-derived products by KE and ChIP. An important advantage of our method is its simplicity and efficiency. KE represents basically a two-minute addition to the standard Illumina sample preparation protocol and requires no specialized reagents. Although a hairpin-mediated ssDNA recovery mechanism has been proposed for library preparation for directional RNA sequencing (Croucher et al. 2009), we are the first to identify and use a specific hairpin signature (type I hairpins) for detection of ssDNA.
In this manuscript we focused mostly on the identification of ssDNA. An important advantage of SSDS is the ability to detect both ss- and dsDNA simultaneously. To our knowledge, SSDS is the first method that allows simultaneous detection of ssDNA and dsDNA by sequencing. We utilized this ability of SSDS to demonstrate that ∼500 weak hotspots are likely explained by the nonspecific accumulation of dsDNA. One can imagine further applications where the ability of our approach to distinguish single-stranded and double-stranded substrates would be beneficial. It may, for example, allow us to differentiate ssDNA and dsDNA targets of proteins that can bind both ssDNA and dsDNA, such as RAD51, DMC1, or Smc5 (Sung 1994; Li et al. 1997; Roy et al. 2011).
SSDS was originally designed and used to map meiotic DSB hotspots in the mouse. In mice, the use of SSDS alleviates the need to enrich for cells containing meiotic recombination intermediates (e.g., the Psmc3ip gene disruption) and, therefore, permits the use of adult wt mice for hotspot mapping. Both the high yield of SSDS and the high quality of the data make the analysis of multiple biological samples both feasible and affordable. Our method can be immediately applied to construct DSB hotspot maps in other mammalian species including humans. Importantly, SSDS is not confined to detection of only meiotic DSBs. We can also detect DSBs of any origin provided that the DSB ends are processed to form ssDNA overhangs.
In addition to mapping meiotic DSB hotspots, our method can be adapted to study biological processes where single-stranded intermediates are formed either normally or in pathological states and for detection of exogenous ssDNA. For example, a straightforward extension of our method would be the sensitive detection of ssDNA viruses and bacteriophages. Although as presented our method is well suited for relatively large, several hundred nucleotide-long targets, the use of less stringent conditions for end repair and for hairpin formation should allow the detection of 100-nt-long or even shorter products. This will allow one to extend this method to the detection of smaller targets as found in other fundamental processes including DNA replication, where ssDNA intermediates are normally formed near replication forks and in cancer, where such intermediates could be associated with genomic rearrangements.
Methods
Mouse strains and antibodies
For consistency and to allow comparison with our previously generated hotspot map we have used 9R/13R F1 mice in this study. 9R (alternative name C57Bl/10.S, Jackson Labs stock number 001650) and 13R (C57Bl/10.F, Jackson Labs stock number 001818) have been received from Dr. N. Arnheim, University of Southern California. All experiments were performed using adult (2–6 mo old) mice on a 9R/13R F1 background. All animal procedures have been approved by the USUHS Animal Care and Use Committee or were performed according to NIH Guide for the Care and Use of Laboratory Animals.
The following antibodies were used: anti-DMC1, Santa Cruz (C-20, sc-8973), anti-RAD51, Santa Cruz (H92, sc-8349), normal goat or rabbit IgGs, Santa Cruz (sc-2028 and sc-2027, respectively).
Chromatin immunoprecipitation and sequencing
Chromatin immunoprecipitation and high-throughput sequencing were performed as previously described with minor modifications (Smagulova et al. 2011). Briefly, testes were fixed for 10 min in 1% formaldehyde and the tissue was homogenized and washed in the following buffers: (1) PBS (twice); (2) 0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM Tris-HCl, pH 8.0; (3) 0.2 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM Tris-HCl, pH 8.0. Cells were lysed in 1.5 mL of the lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.0 plus complete protein inhibitor cocktail [Roche]) and the chromatin was sheared to ∼1000 bp by sonication. Chromatin was dialyzed against ChIP buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.0, 167 mM NaCl), pre-cleared with Protein G beads (Sigma) and incubated with appropriate antibodies overnight at 4°C followed by a 2-h incubation with Protein G beads. Beads were washed in the following buffers: (1) 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl; (2) 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 500 mM NaCl; (3) 0.25 M LiCl, 1% Igepal, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0, 1% Deoxycholic acid; (4) TE (twice). The chromatin was eluted by 1% SDS, 0.1 M NaHCO3, pH 9.0 at 65°C and cross-linking was reversed at 65°C overnight. Standard sequencing library construction was done using enzymes from New England Biolabs according to the protocol provided by Illumina, with the exception that DNA size fractionation was done after the amplification step. “Klenow exo(−)” sample was prepared by substituting Klenow fragment and T4 DNA polymerase from the end repair step of the standard protocol with Klenow exo(−) enzyme and performing end repair at 37°C for 1 h. “Klenow exo(−), 12 °C” sample was prepared the same way followed by incubation at 20°C for 2 h and at 12°C overnight.
Sequencing was performed on Illumina GA, GAII, or HiSeq 2000 instruments in the NIDDK Genomics core.
Kinetic enrichment and asymmetric sequencing
The standard protocol was followed until the adapter ligation step. Before ligation of adapters the DNA was denatured at 95°C for 2 min and cooled to room temperature within 2 min. Moderate increases of the length of the denaturation and annealing steps for up to 30 min did not critically affect success of the procedure. Then the adapter mix and T4 DNA ligase were added and the mixture was incubated at 20°C for 30 min. The rest of the procedure was as described above. We used 36/40 nt asymmetric sequencing in conjunction with 36-cycle sequencing kits and the Illumina GAII. On that platform, two 36-cycle kits routinely allowed sequencing for 76 cycles, thus resulting in no cost increase compared with symmetrical 2 × 36 PE sequencing. On other sequencing platforms, such as Illumina HiSeq 2000, different combinations of cycles might be more cost effective.
Sample list and brief description
α-DMC1 Psmc3ip−/−: 9R/13R F1, Psmc3ip−/−, standard protocol, GSM851661.
α-RAD51 Psmc3ip−/−: 9R/13R F1, Psmc3ip−/−, standard protocol, GSM851662.
α-RAD51 Psmc3ip−/− LE: 9R/13R F1, Psmc3ip−/−, standard protocol, GSM851663.
IgG1, IgG2: 9R/13R Psmc3ip−/− F1, standard protocol, GSM851664, GSM851665.
α-DMC1 Psmc3ip−/− Klenow (exo-) 12C: 9R/13R F1, Psmc3ip−/−, modified protocol, GSM851667.
α-DMC1 Psmc3ip−/−: Klenow (exo-): 9R/13R F1, Psmc3ip−/−, modified protocol, GSM851666.
α-DMC1 wt: 9R/13R F1, standard protocol, GSM851668.
α-DMC1 Psmc3ip−/− KE: 9R/13R F1, Psmc3ip−/−, KE protocol, GSM851669.
α-DMC1 wt KE: 9R/13R F1, KE protocol, GSM851670.
wt KE IgG: 9R/13R F1, KE protocol, GSM851671.
The full paired-end data set is submitted to the GEO under accession GSE34592. Reference Psmc3ip−/− data set (Smagulova et al. 2011) is a pooled α-DMC1 single-end sample, GSM602194. First read data for α-DMC1 Psmc3ip−/−, α-RAD51 Psmc3ip−/−, IgG1, and IgG2 samples prepared with the standard protocol were used as a single-end data in our previous work (Smagulova et al. 2011).
Computational ssDNA detection in the sequencing libraries
To identify type I and type II ssDNA-derived reads in paired-end sequencing data we have developed a computational pipeline. First, base calling and data processing were performed using the standard Illumina pipeline. We then retained only quality-filtered reads and aligned them to the genome using BWA (Li and Durbin 2009). While the 5′ read is fully complementary to the genome, the 3′ read of type I hairpins contains a fill-in ITR part at the very 5′ end of the read (Fig. 1; Supplemental Fig. 1). To align such reads with mismatches at the very 5′ end we have modified the BWA program to search for the longest mappable suffix in the query. This modification, BWA-RA (“right-align”) progressively removes 5′ nucleotides from each read until it can be mapped to the genome.
After initial alignment to the genome, reads were paired using the “sampe” command of BWA (Li and Durbin 2009). Before determining if reads are properly paired, BWA calculates average insert size and shape parameters of the distribution of the fragment lengths in the library. The maximum fragment cutoff size is then set to the mean of this distribution plus seven standard deviations. For our libraries with mean fragment size of ∼100 nt, this maximum fragment size was generally between 400 and 500 nt. We then post-process the output SAM/BAM files using SAMtools (Li et al. 2009) and a custom script to generate type I, type II ssDNA- and dsDNA-specific subsets.
To be defined as a type I hairpin, we require that a sequenced fragment have an ITR >5 nt, a fill-in part of the ITR >2 nt, and that all of the remaining sequence (except for the fill-in ITR part) must map to the genome close to the 5′ read but to the opposite strand (“properly paired” as defined by BWA). The fill-in part of the ITR must be complementary to the 5′-end of the first read but not homologous to the target genomic DNA. Type II hairpins were defined identically to type I except that the fill-in part should be <3 nt. The requirement for 3 nt derived from the other end of the fragment effectively guarantees that this short fill-in part is not due to PCR and/or sequencing mutations. Taking into account that per-base error-rates are currently <1%, the probability of finding such an exact match by chance is negligibly low, P < 10−6. The distribution of ITR lengths in our libraries shows a steep drop-off between 4 and 6 nt. Therefore, the cutoff for minimal ITR length in ssDNA was set to 6 nt.
dsDNA-derived fragments were defined as fragments with both ends properly mapped and having an ITR <3 nt. Three-nucleotide-long ITRs are unlikely to be sufficiently stable for hairpin formation and therefore unlikely to be derived from ssDNAs. This is an arbitrary cutoff and some dsDNA-derived fragments can have longer ITRs. We cannot, however, distinguish such dsDNAs from type II hairpins. ITRs were computed allowing for one mismatch.
Although we primarily based our work on BWA-generated genome alignments, any other aligner can be used provided that they will find the longest 3′ end subsequence for the second read. Our ssDNA parsing script, ITR-id, takes as an input standard SAM/BAM files. We also used ELAND for genome alignment for some of the data sets. Our programs for parsing PE sequencing data, BWA-RA and ITR-id, are available upon request.
Hotspot identification and computational data analysis
Hotspots in the wt α-DMC1 KE sample were identified using MACS v 1.3.7., P < 0.0001. For all of the analyses (e.g., calculation of the ssDNA/dsDNA coverage profiles, sample specificity calculations) with the exception of analyses presented on Figure 5 and Supplemental Figure 11 we used the 9874 hotspots defined previously (Smagulova et al. 2011). These hotspots were recentered relative to the center of mass of all nonduplicate reads in the 5-kb region around the center of MACS-defined peaks. We did not recenter peaks identified in the KE data using MACS when calculating hotspot overlaps. The set of 11,376 hotspots detected in α-DMC1 wt KE is provided as Supplemental Data set 1. For the hotspot-associated motif search we used previously determined position-weight matrix (Smagulova et al. 2011). Motif locations were detected in the mouse genome using FIMO (Grant et al. 2011) and a minimal log-likelihood score cutoff of 10. Positions of testis-specific H3K4me3 peaks were defined previously (Smagulova et al. 2011).
Data access
Sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE34592.
Acknowledgments
We thank Peggy Hsieh and Michael Lichten for helpful discussion and Shaila Sharmeen for help with high-throughput sequencing. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov). This research was supported by the NIDDK Intramural Research Program; by Basil O'Connor Starter Scholar Research Award Grant No. 5-FY07-667 from the March of Dimes Foundation (G.V.P.); NIH grant 1R01GM084104-01A1 from NIGMS (G.V.P.); New Investigator Start-up Grants FS71HU, R071HU, and CS71HU from USUHS (G.V.P.).
Authors' contributions: P.P.K. conceived the method. F.S. performed all experiments. P.P.K. and K.M.B. performed computational data analyses and wrote the analysis code. All authors contributed to experimental design. G.V.P. and R.D.C.-O. designed and supervised the study. P.P.K. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.130583.111.
References
- Arnheim N, Calabrese P, Tiemann-Boege I 2007. Mammalian meiotic recombination hot spots. Annu Rev Genet 41: 369–399 [DOI] [PubMed] [Google Scholar]
- Bellve AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M 1977. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol 74: 68–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A 2007. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr Biol 17: 2003–2012 [DOI] [PubMed] [Google Scholar]
- Borde V, Robine N, Lin W, Bonfils S, Geli V, Nicolas A 2009. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J 28: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Kohne DE 1968. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science 161: 529–540 [DOI] [PubMed] [Google Scholar]
- Buhler C, Borde V, Lichten M 2007. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol 5: e324 doi: 10.1371/journal.pbio.0050324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Fookes MC, Perkins TT, Turner DJ, Marguerat SB, Keane T, Quail MA, He M, Assefa S, Bahler J, et al. 2009. A simple method for directional transcriptome sequencing using Illumina technology. Nucleic Acids Res 37: e148 doi: 10.1093/nar/gkp811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XF, Li HH, Goradia TM, Lange K, Kazazian HH Jr, Galas D, Arnheim N 1989. Single-sperm typing: Determination of genetic distance between the Gγ-globin and parathyroid hormone loci by using the polymerase chain reaction and allele-specific oligomers. Proc Natl Acad Sci 86: 9389–9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD 2000. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci 97: 11383–11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J, Berger SL 2009. Genome reprogramming during sporulation. Int J Dev Biol 53: 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS 2011. FIMO: Scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogstrand K, Bohme J 1994. A determination of the frequency of gene conversion in unmanipulated mouse sperm. Proc Natl Acad Sci 91: 9921–9925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, May CA, Jeffreys AJ 2009. Analysis of meiotic recombination products from human sperm. Methods Mol Biol 557: 323–355 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Gyllensten UB, Cui XF, Saiki RK, Erlich HA, Arnheim N 1988. Amplification and analysis of DNA sequences in single human sperm and diploid cells. Nature 335: 414–417 [DOI] [PubMed] [Google Scholar]
- Li Z, Golub EI, Gupta R, Radding CM 1997. Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc Natl Acad Sci 94: 11221–11226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich ML 1977. Separation of spermatogenic cells and nuclei from rodent testes. Methods Cell Biol 15: 15–54 [DOI] [PubMed] [Google Scholar]
- Mieczkowski PA, Dominska M, Buck MJ, Lieb JD, Petes TD 2007. Loss of a histone deacetylase dramatically alters the genomic distribution of Spo11p-catalyzed DNA breaks in Saccharomyces cerevisiae. Proc Natl Acad Sci 104: 3955–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Marcon E, Shore JS, Kochakpour N, Spyropoulos B 2007. Initiation and resolution of interhomolog connections: Crossover and non-crossover sites along mouse synaptonemal complexes. J Cell Sci 120: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Neale MJ, Keeney S 2006. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Petkov P 2010. Mammalian recombination hot spots: Properties, control and evolution. Nat Rev Genet 11: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu XA, Neale MJ, Jasin M, Socci ND, et al. 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PJ 2009. ChIP-seq: Advantages and challenges of a maturing technology. Nat Rev Genet 10: 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD 2001. Meiotic recombination hot spots and cold spots. Nat Rev Genet 2: 360–369 [DOI] [PubMed] [Google Scholar]
- Petukhova GV, Romanienko PJ, Camerini-Otero RD 2003. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev Cell 5: 927–936 [DOI] [PubMed] [Google Scholar]
- Roy MA, Siddiqui N, D'Amours D 2011. Dynamic and selective DNA-binding activity of Smc5, a core component of the Smc5-Smc6 complex. Cell Cycle 10: 690–700 [DOI] [PubMed] [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265: 1241–1243 [DOI] [PubMed] [Google Scholar]