Abstract
High ethanol tolerance is an exquisite characteristic of the yeast Saccharomyces cerevisiae, which enables this microorganism to dominate in natural and industrial fermentations. Up to now, ethanol tolerance has only been analyzed in laboratory yeast strains with moderate ethanol tolerance. The genetic basis of the much higher ethanol tolerance in natural and industrial yeast strains is unknown. We have applied pooled-segregant whole-genome sequence analysis to map all quantitative trait loci (QTL) determining high ethanol tolerance. We crossed a highly ethanol-tolerant segregant of a Brazilian bioethanol production strain with a laboratory strain with moderate ethanol tolerance. Out of 5974 segregants, we pooled 136 segregants tolerant to at least 16% ethanol and 31 segregants tolerant to at least 17%. Scoring of SNPs using whole-genome sequence analysis of DNA from the two pools and parents revealed three major loci and additional minor loci. The latter were more pronounced or only present in the 17% pool compared to the 16% pool. In the locus with the strongest linkage, we identified three closely located genes affecting ethanol tolerance: MKT1, SWS2, and APJ1, with SWS2 being a negative allele located in between two positive alleles. SWS2 and APJ1 probably contained significant polymorphisms only outside the ORF, and lower expression of APJ1 may be linked to higher ethanol tolerance. This work has identified the first causative genes involved in high ethanol tolerance of yeast. It also reveals the strong potential of pooled-segregant sequence analysis using relatively small numbers of selected segregants for identifying QTL on a genome-wide scale.
Genetic analysis of polygenic traits remains an important challenge (Swinnen et al. 2012). It requires, in the first instance, reliable scoring of many genetic markers covering the whole genome. In yeast, the first successful approaches to simultaneously mapping multiple genetic loci that were either independent (Winzeler et al. 1998) or involved in a quantitative trait (QTL) (Steinmetz et al. 2002) made use of SNP markers that were scored by hybridization of genomic DNA from individual segregants to gene expression microarrays. Subsequently, a similar approach was used to map QTL involved in traits such as sporulation efficiency (Deutschbauer and Davis 2005), gene expression (Brem et al. 2002), acetic acid production (Marullo et al. 2007), cell morphology (Nogami et al. 2007), and resistance to small-molecule drugs (Perlstein et al. 2007).
The advent of high-throughput sequencing technologies provides a new way to score large numbers of SNPs as genetic markers. Application to individual segregants remains cumbersome because of the high costs involved. On the other hand, whole-genome sequence analysis of pooled segregants has recently been used to identify multiple QTL throughout the genome (Ehrenreich et al. 2010; Parts et al. 2011). In these studies, very large pools of segregants were used with or without selection to enrich for beneficial alleles. Validation of this methodology through identification of all causative genes in the QTL remains a challenge. Parts et al. (2011) were able to reduce the size of the mapped intervals by inbreeding and subsequent selection of large segregant pools for the trait of interest. For genetic analysis of industrially important traits, enrichment of segregants is mostly impossible. The use of very large pools of segregants is also cumbersome because the precise phenotyping of such traits usually requires elaborate experimental procedures and is, therefore, difficult to apply to large numbers of segregants. The use of small numbers of segregants is particularly important in higher eukaryotic organisms, where phenotyping of commercially important traits is a major bottleneck in genetic analysis.
We have now applied pooled-segregant whole-genome sequence analysis for the mapping of QTL involved in tolerance to high ethanol levels (16% and 17%) in an industrial yeast strain. High ethanol tolerance is an exquisite characteristic of the yeast Saccharomyces cerevisiae. It is of prime importance for survival in its natural sugar-rich niche environments, since yeast produces high levels of ethanol to inhibit competing microorganisms. High ethanol tolerance is also crucial for the use of yeast in the fermentation industries (production of bioethanol, beer, wine, and other alcoholic beverages), since it strongly influences the rate and completion of fermentation. Until now, ethanol tolerance in yeast has been studied mostly in laboratory yeast strains and always with low to moderately high ethanol concentrations (5%–12%). These studies have revealed that properties like membrane lipid composition, chaperone protein expression, and trehalose content are important determinants of ethanol tolerance (D'Amore and Stewart 1987; Ding et al. 2009). Genome-wide transcriptomics and screening of deletion mutants have revealed many genes required for tolerance to low or moderate ethanol concentrations (Fujita et al. 2006; van Voorst et al. 2006; Lewis et al. 2010). In contrast, nothing is known about the genetic loci or gene polymorphisms that are responsible for the much higher ethanol tolerance in natural and industrial yeast strains compared to laboratory strains.
In this work, we show that pooled-segregant whole-genome sequence analysis can be used for straightforward mapping of QTL responsible for a typical polygenic trait of industrial importance in yeast. We demonstrate that this can be successfully accomplished using relatively small populations of segregants and without any enrichment procedures. We have identified and validated three genetic loci in a Brazilian bioethanol production strain that are responsible for tolerance to high ethanol levels. In addition, we have dissected the locus with the strongest linkage and identified two novel genes with a previously unrecognized, positive function in ethanol tolerance. The locus also contained a mutant allele with a negative contribution to high ethanol tolerance, which was located in between the two genes with a positive contribution. Application of whole-genome sequence analysis to two pools of segregants tolerant to 16% or 17% ethanol showed that more stringent phenotypic screening reveals additional minor QTL.
Results
Characterization of parent strains with high and low ethanol tolerance
VR1 is a former bioethanol production strain originally isolated as a wild yeast strain from a fermentor in a Brazilian plant. We isolated a segregant, called VR1-5B, that displayed similarly high ethanol tolerance as the VR1 parent strain. Ethanol tolerance was, thereby, defined as growth on solid YP plates with ethanol as the sole carbon source. Because high ethanol tolerance is only relevant toward the end of yeast fermentation when the sugar level has dropped to low values, we determined ethanol tolerance in the absence of any other sugar or carbon source. The VR1 parent strain could grow in medium containing up to 16% ethanol, while the VR1-5B segregant showed growth in medium containing up to 18% ethanol (Fig. 1). Both strains were clearly more ethanol-tolerant than the control haploid BY4741 and diploid BY laboratory strains, which could grow only slightly in medium with 14% ethanol (Fig. 1). The diploid VR1-5B/BY4741 strain displayed similarly high ethanol tolerance at 14% and 16% ethanol compared to the VR1-5B parent strain (and the original VR1 strain), indicating that under these conditions the high ethanol tolerance in this strain is largely a dominant property (Fig. 1).
Figure 1.
Ethanol tolerance of the Brazilian bioethanol production strain VR1 and its segregant VR1-5B. The ethanol tolerance of VR1 (diploid) and VR1-5B (haploid) was determined by scoring growth of tenfold dilutions on YP plates with different concentrations of ethanol. Both strains, as well as the heterozygous VR1-5B/BY4741 strain (diploid) showed a clearly higher ethanol tolerance than the control laboratory strains BY4741 (haploid) and BY (diploid), the latter of which was obtained by crossing BY4741 with BY4742.
Pooled-segregant whole-genome sequence analysis
From the cross between VR1-5B and BY4741, we obtained 5974 segregants that were phenotyped for ethanol tolerance by scoring growth on YP with different concentrations of ethanol. The segregants with extreme phenotypes were subsequently classified in two pools. The first pool contained 136 segregants with a tolerance to at least 16% ethanol (“16% pool”) and the second pool contained 31 segregants from the first pool with a tolerance to at least 17% ethanol (“17% pool”). All segregants were individually grown up to saturation, after which equal amounts of cells based on dry weight were combined to obtain the 16% and 17% pools. The genomic DNA from both pools and the parent strains was extracted and submitted to custom sequence analysis using Illumina HiSeq 2000 technology (GATC Biotech AG). The sequencing was performed at 40 times or greater coverage and generated paired-end short reads of ∼100 bp, allowing a highly precise alignment of the reads. The VR1-5B and BY4741 sequences were aligned to the reference S288c genome sequence (Cherry et al. 1997), and SNPs between VR1-5B and BY4741 with a coverage of more than 20 times and a ratio of at least 80% were selected. The ratio of at least 80% was chosen based upon the plots of the SNPs between the two parent strains VR1-5B and BY4741 (see Supplemental Fig. S1 for an example of chromosome XIV). There are two distinct groups of SNPs present, one at the top and one at the bottom. We consider the cloud at the top to contain reliable SNPs. Its lower border is ∼80%. We, therefore, assume that all SNPs with a frequency above 80% can be considered reliable if they have been sequenced at least 20 times. One might increase the cut-off value to, e.g., 90% or 95% but must take into account that removing extra SNPs will negatively influence the smoothness of the estimated curve. The minimal coverage of 20 is motivated by the finding of Dohm et al. (2008) that a 20-fold sequencing coverage is sufficient to compensate for errors by the number of correct reads. Subsequently, the sequence of each pool was aligned to the BY4741 sequence, and the nucleotide frequency of each selected SNP was plotted against its chromosomal position.
The SNP nucleotide frequency curve obtained by whole-genome sequencing of DNA extracted from the 16% pool fluctuated around 50% in most areas in the genome (Fig. 2). On the other hand, three loci showed a strong deviation from 50% inheritance. They were located on chromosomes V, X, and XIV. The significance of the deviation in SNP nucleotide frequency could be confirmed by scoring a single SNP from the center of each locus in at least 96 individual highly ethanol-tolerant segregants by PCR. The QTL on chromosome V (QTL1) and chromosome XIV (QTL3) showed the strongest link, with, respectively, 92.6% and 94.1% of the highly ethanol-tolerant segregants harboring the nucleotide from VR1-5B. The locus on chromosome X (QTL2) showed a much weaker link, with only 72.9% of the segregants showing VR1-5B inheritance. Scoring the same SNPs in an unselected pool of at least 80 segregants resulted in an association percentage of 50.0%, which is consistent with random segregation (data not shown). We next examined the joint effect of the three QTL on high ethanol tolerance by determining the appearance of each of the eight combinations in 85 highly ethanol-tolerant segregants (Table 1). The combination between the VR1-5B alleles from QTL1 and QTL3 was most prevalent in the segregants. Taken together, 88.2% of the highly ethanol-tolerant segregants carried the VR1-5B alleles from QTL1 and QTL3, indicating that inheriting both alleles is strongly advantageous for high ethanol tolerance. These results revealed that the VR1-5B alleles from QTL1 and QTL3 are the major contributors to the high- ethanol-tolerance phenotype and that QTL2 is less important.
Figure 2.
Genetic mapping of QTL involved in high ethanol tolerance by whole-genome sequence analysis. QTL were mapped by whole-genome sequence analysis of DNA extracted from a pool of 136 segregants tolerant to at least 16% ethanol (16% pool; green line) and from a pool of 31 segregants tolerant to at least 17% ethanol (17% pool; red line). The genomic DNA of the parents, VR1-5B and BY4741, and of the two pools, was sequenced and aligned to identify SNPs. The nucleotide frequency of quality-selected SNPs in the sequence of each pool was plotted against the chromosomal position. Significant deviations from the average of 0.5 indicate candidate QTL linked to high ethanol tolerance. Upward deviations indicate linkage to QTL in the ethanol-tolerant parent VR1-5B. The three major QTL on chromosomes V, X, and XIV are not significantly different between the two pools. However, in several instances, e.g., on chromosomes II, XII, and XV, minor loci can be identified, showing a significant difference between the two pools. These candidate QTL are more distinctive in the 17% pool compared to the 16% pool. The difference in SNP frequency between the two pools is certainly significant when the simultaneous confidence bands do not overlap.
Table 1.
Appearance of each QTL combination in highly ethanol-tolerant segregants
The three identified QTL were also found after whole-genome sequence analysis of DNA extracted from the 17% pool (Fig. 2), which is consistent with the requirement for the same causative genes in tolerance to 16% and 17% ethanol. However, these data also revealed significant upward deviations from 50% inheritance at several other loci, which appear to represent minor loci determining high ethanol tolerance (Fig. 2). For example, there are regions on chromosomes II and XV that did not show a significant deviation from random segregation in the pool of segregants tolerant to 16% ethanol, whereas a clear deviation was observed in the pool of segregants tolerant to 17% ethanol (Fig. 2). Interestingly, at the position of ∼200,000 bp on chromosome XIV, there appears to be a significant downward deviation in the 17% pool, which is absent in the 16% pool. This may indicate a genetic element in the BY4741 strain that can contribute to tolerance to high ethanol levels in spite of the poor overall ethanol tolerance of that strain.
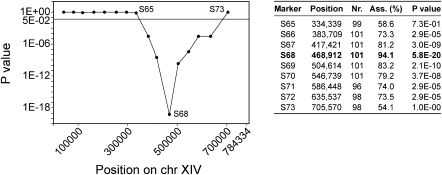
The boundaries of QTL3, the locus with the strongest linkage identified in both pools by pooled-segregant whole-genome sequence analysis, were determined by scoring selected SNP markers in chromosome XIV for at least 96 individual segregants that composed the 16% pool by PCR. We calculated the P-value for each SNP using an exact binomial test with a confidence level of 95% and correction for multiple testing by a false discovery rate (FDR) control according to Benjamini and Yekutieli (2005). The P-values were plotted over the length of chromosome XIV (Fig. 3).
Figure 3.
Detailed linkage statistics of QTL3, the locus with the strongest linkage to high ethanol tolerance. The table shows for each marker in the mapped QTL3 on chromosome XIV the position of the marker, the number of segregants in which the marker was scored, the association percentage, and the P-value. The association percentage represents the percentage of segregants with VR1-5B inheritance, i.e., the nucleotide from VR1-5B. The marker with the strongest linkage is shown in bold.
Genetic dissection of QTL3 reveals two positive and one negative genetic element
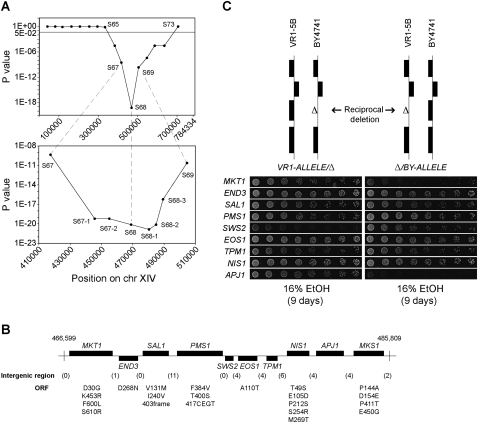
Since QTL3 showed the strongest linkage with high ethanol tolerance, we submitted this locus to detailed analysis to identify the causative gene(s). The 370-kb QTL3 was fine-mapped using selected SNPs to reduce the size of the interval to a practical number of candidate genes for further functional analysis. The P-values for eight SNP markers (S67, S67-1, S67-2, S68, S68-1, S68-2, S68-3, S69) defined a smaller locus of 16 kb between markers S68 and S68-2, which had the strongest linkage (Fig. 4A). The locus contained 10 annotated genes (Fig. 4B). Sanger sequence analysis of this region was performed to detect all nucleotide polymorphisms between VR1-5B and BY4741 (Fig. 4B). We observed that VR1-5B and BY4741 were highly divergent with a polymorphism, on average, every 167 bp. All genes except TPM1 had at least one polymorphism in their ORF, being silent mutations for the genes APJ1 and SWS2, and missense mutations in the other seven genes. In addition, all genes had at least one polymorphism in their putative promoter and/or terminator. Given the difficulty of predicting the effect of both coding and noncoding polymorphisms on phenotypes (Tabor et al. 2002), we, therefore, could not use the sequence data to exclude genes from further functional analysis.
Figure 4.
Fine-mapping and identification of the causative genes in QTL3. (A) The 87-kb locus defined by SNP markers S67, S68, and S69 in QTL3 showed the lowest probability of random segregation in 101 highly ethanol-tolerant segregants. Further fine-mapping was achieved by scoring five additional markers within the 87-kb interval in the same segregants. Calculation of the P-values revealed the strongest linkage for a 16-kb locus defined by markers S68, S68-1, and S68-2. (B) The name and location of each ORF in the fine-mapped locus is shown as annotated in SGD (Cherry et al. 1997). The interval from nucleotide 466,599 to 485,809 was sequenced in VR1-5B and BY4741, which revealed 115 polymorphisms, of which part were in intergenic regions (numbers in parentheses). For the ORFs, only polymorphisms that change the amino acid sequence are indicated (amino acid in BY4741, followed by position in the protein and amino acid in VR1-5B). SAL1 has a frame shift mutation in BY4741 resulting in an earlier stop codon and truncation of the protein, which is assumed to be a loss-of-function gene product (Dimitrov et al. 2009). PMS1 has an insertion of four amino acids at position 417 in VR1-5B. The sequence of BY4741 in this interval is the same as that of S288c (Cherry et al. 1997), except for one nucleotide in SAL1 that causes an amino acid change at position 131 (valine in BY4741 and methionine in S288c and VR1-5B). (C) Reciprocal hemizygosity analysis. For the nine genes in the fine-mapped locus, two diploid strains were constructed in the VR1-5B/BY4741 hybrid background that carried either the VR1-5B (left) or BY4741 (right) allele from the gene. The rest of the genome was identical between the two hybrids. The reciprocal deletions were engineered in the haploid strains, after which the proper haploids were crossed to obtain the diploid hybrids. The ethanol tolerance of the diploid hybrids was determined by scoring the growth of twofold dilutions on 16% ethanol after 9 d. This revealed different contributions of the parental alleles from MKT1, SWS2, and APJ1 to high ethanol tolerance. The strain pairs were always spotted on the same plate. The results were assembled from different plates, thus slight differences in growth may be present between hybrid pairs that otherwise do not show differences in ethanol tolerance. Hence, only growth differences between strains within a hybrid pair are relevant. The growth of the wild-type diploid hybrid was similar to that of the hybrid pairs whose ethanol tolerance was unaltered.
We have applied reciprocal hemizygosity analysis (RHA) to identify the causative genes in the locus. RHA allows analyzing whether the two parental alleles have a different contribution to the phenotype in an otherwise uniform genetic background (Steinmetz et al. 2002). For nine genes, two heterozygous strains were constructed in the VR1-5B/BY4741 hybrid background that only differed genetically in the candidate gene, i.e., they carried either one copy of the VR1-5B or the BY4741 allele while the other copy was deleted (Fig. 4C). Comparing the ethanol tolerance of each pair of heterozygous strains revealed a difference in the phenotypic contribution between the parental alleles from MKT1, SWS2, and APJ1 (Fig. 4C). The presence of the VR1-5B allele from the MKT1 and APJ1 gene resulted in higher ethanol tolerance compared to the BY4741 allele. Surprisingly, for SWS2 the opposite was true, as the BY4741 allele was advantageous over the VR1-5B allele. Hence, although SWS2 clearly affects ethanol tolerance, it cannot be one of the causative genes responsible for the high ethanol tolerance of VR1-5B. These experiments were carried out with two independently constructed sets of strains, and all strains were spotted twice on different plates, which gave consistent results.
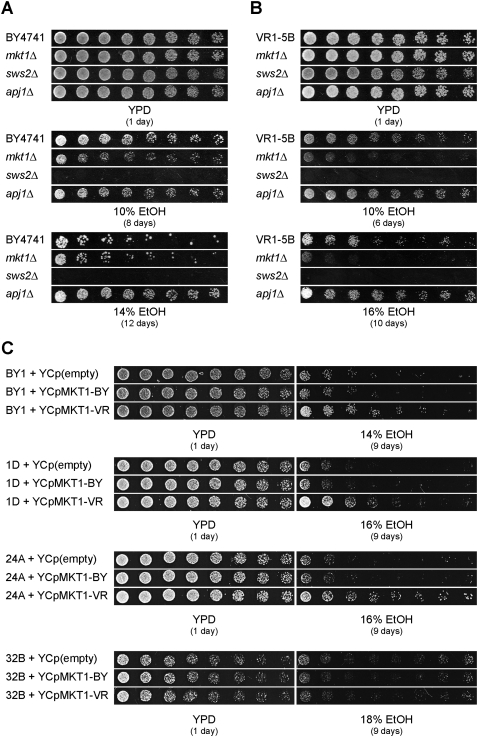
One potential complication with RHA is that the hybrid diploid background used in the assay is different from the haploid segregants background used in the QTL mapping experiment. For this reason, we determined the deletion phenotypes of MKT1, SWS2, and APJ1 in the BY4741 and VR1-5B haploid strains. For the BY4741 background, which has a moderate ethanol tolerance, we tested 10%, 12%, 14%, 15%, and 16% ethanol. For the VR1-5B background, which has a high ethanol tolerance, we tested 10%, 12%, 14%, 15%, 16%, 17%, 18%, and 19% ethanol. In the BY4741 background, the mkt1Δ strain showed only a minor growth reduction for all ethanol concentrations tested (Fig. 5A and data not shown). In the VR1-5B background, deletion of MKT1 caused a strong reduction in growth for all ethanol concentrations tested (Fig. 5B and data not shown).
Figure 5.
Effect of MKT1, SWS2, and APJ1 on ethanol tolerance. (A) The ethanol tolerance of BY4741 (inferior wild type) and the mkt1Δ, sws2Δ, and apj1Δ mutants thereof was determined by scoring growth of twofold dilutions on 10% ethanol after 8 d and 14% ethanol after 12 d. (B) The ethanol tolerance of VR1-5B (superior wild type) and the mkt1Δ, sws2Δ, and apj1Δ mutants thereof was determined by scoring growth of twofold dilutions on 10% ethanol after 6 d and 16% ethanol after 10 d. (C) The VR1-5B allele of MKT1 is beneficial for high ethanol tolerance. MKT1-BY and MKT1-VR, including 534 bp upstream and 344 bp downstream regions of the ORF, were cloned into the low-copy-number plasmid YCplac111 and expressed in BY4741 (BY1) and three segregants from VR1-5B/BY4741 that inherited the MKT1-BY allele (1D, 24A, and 32B). The ethanol tolerance was determined in twofold dilutions on different concentrations of ethanol.
In the BY4741 background, the apj1Δ strain grew equally well as the wild-type strain on 10% ethanol but grew clearly better on all higher ethanol concentrations tested (Fig. 5A and data not shown). In the VR1-5B background, deletion of APJ1 caused a small improvement in ethanol tolerance at all ethanol concentrations tested (Fig. 5B and data not shown). The improvement of ethanol tolerance by deletion of APJ1 is consistent with the APJ1 gene product acting negatively on ethanol tolerance, at least under our test conditions. When this is combined with the result from RHA and the absence of nonsynonymous mutations in the open reading frame, it suggests that the beneficial effect on ethanol tolerance of the APJ1 VR1-5B allele may be due to lower expression compared to that of the BY4741 allele. Determination of APJ1 expression by real-time PCR in the BY4741 and VR1-5B strains during fermentation confirmed a higher expression level in the BY4741 strain (normalized to 1.0 ± 0.19) compared to the VR1-5B strain (0.43 ± 0.12), especially in the beginning of the fermentation. Although the expression in VR1-5B was lower, it was clearly detectable, consistent with deletion of APJ1 causing further enhanced ethanol tolerance in VR1-5B.
Deletion of SWS2 resulted in complete loss of growth on all ethanol concentrations tested and in both genetic backgrounds (Fig. 5A,B and data not shown). The results obtained for the three genes are in agreement with those of the screening of the BY deletion strain collection, in which an ethanol-sensitive growth phenotype was only observed for the sws2Δ strain (Teixeira et al. 2009; Yoshikawa et al. 2009).
The relevance of MKT1 for high ethanol tolerance was confirmed by expressing both parental alleles in BY4741 and in segregants from VR1-5B/BY4741 that have the BY4741 allele from MKT1. Expression of the VR1-5B allele from MKT1, in contrast to the BY4741 allele, resulted in higher ethanol tolerance in BY4741 and two out of the three segregants (Fig. 5C). This confirmed the result from RHA, suggesting that the VR1-5B allele is advantageous for high ethanol tolerance. On the other hand, as we did not observe an effect in all segregants, it seems that MKT1 alone is not sufficient to enhance ethanol tolerance. Comparing ethanol tolerance in the strains BY4741 and BY4741mkt1Δ revealed that the BY4741 allele from MKT1 behaves as a near loss-of-function allele under our conditions, since no difference in ethanol tolerance was observed at 14% and only a slight difference at 10% (Fig. 5A). In contrast, deletion of MKT1 in VR1-5B caused a pronounced drop in ethanol tolerance (Fig. 5B), which confirms that a loss-of-function mutation in MKT1 decreases ethanol tolerance.
Discussion
Genetic analysis of complex traits remains a challenging endeavor. Because multiple genetic elements are required for expression of the trait of interest, relatively large numbers of segregants have to be phenotyped to identify enough segregants displaying the trait of interest. In addition, the latter segregants must all be scored for genetic markers throughout the genome. Bulked segregant analysis provides a convenient way to reduce the workload and expenses involved in the scoring of markers (Michelmore et al. 1991). This has been demonstrated for scoring of SNPs by microarray detection (Segrè et al. 2006) and whole-genome sequence analysis using very large numbers of segregants with or without enrichment procedures (Ehrenreich et al. 2010; Parts et al. 2011). We have now shown that genome-wide detection of SNPs by whole-genome sequence analysis can also be applied successfully to pools with relatively small numbers of selected segregants. Although we originally phenotyped 5974 segregants, the results obtained with the pool of 31 segregants tolerant to 17% ethanol indicate that, for traits in which 4 to 5 QTL are involved, phenotyping of 500–1000 segregants would be enough to map all QTL. This is of major importance for the genetic analysis of industrially important traits, which often require elaborate procedures for precise phenotyping.
The results obtained with pooled-segregant whole-genome sequence analysis of the 17% pool indicate that application of a more stringent phenotypic selection can enhance the sensitivity of QTL detection with this methodology. In this case, more segregants have to be phenotyped in order to obtain enough segregants with the phenotype of interest for the pool. A higher number of segregants (136 in the 16% pool versus 31 in the 17% pool) does not seem to increase the sensitivity of the method, at least as judged from the major QTL. The more stringent selection in the 17% pool appears to have more effect than the higher number of segregants in the 16% pool.
In principle, all major genetic loci acting in an interdependent way (i.e., additively or synergistically) can be identified with this methodology. It must also be emphasized that causative genetic elements unique to the VR1-5B strain (or to the other parent) (e.g., insertion of a new DNA sequence) can be mapped based on their linkage with the SNPs in regions adjacent to the unique genetic element and common to the two parent strains. Additional sequences are not unlikely since whole-genome sequence analysis has revealed unique sequences in many yeast strains, including the Brazilian bioethanol production strain PE-2 (Argueso et al. 2009). Hence, once the QTL have been mapped, it is important to check the precise sequence in the center of all QTL in the superior parent strain by Sanger sequence analysis and compare it with the corresponding region in the control parent strain, in our case, BY4741. At present, it is unclear whether the current methodology could also detect QTL in very large insertions or in chromosomal rearrangements. Inhibitory loci in the superior parent strain should also be visible as a deviation of the SNP frequency curve below the 50% mean value. Two or more loci that can provide independently of each other the same, nonadditive contribution to the phenotype (e.g., due to two or more duplicates of the same gene in unlinked positions in the genome) will be difficult to detect, as a segregant can have either one of the loci and still show the same phenotype. As a result, the SNP nucleotide frequency will only be 66.7% for each locus in the case of two such independent loci/genes and will further decrease to 50% if more independent loci/genes are involved. To identify such independent polymorphisms contributing to the phenotype of interest, several backcrosses with the control parent strain can be performed to separate the independent polymorphisms from one another. Pools of selected segregants from different backcrosses can then be submitted to whole-genome sequence analysis to identify the different independent polymorphisms contributing to the phenotype of interest.
Detailed analysis of QTL3, which exhibited the strongest linkage to high ethanol tolerance, led to the identification of three genes affecting high ethanol tolerance, MKT1, SWS2, and APJ1. SWS2, however, had an inferior allele in VR1-5B and thus cannot be one of the causative genes responsible for its high ethanol tolerance. In spite of this, the identification of MKT1 and APJ1 as causative genes confirms the importance of QTL3 for high ethanol tolerance and validates the genetic mapping result obtained by pooled-segregant whole-genome sequence analysis. To our knowledge, this is the first time that MKT1 has been conclusively associated with ethanol tolerance in S. cerevisiae. MKT1 seems to be important for diverse aspects of cellular function under stressful conditions since previous QTL mapping experiments have identified MKT1 as a quantitative trait gene determining high temperature growth (Steinmetz et al. 2002; Sinha et al. 2006), sporulation efficiency (Deutschbauer and Davis 2005), induction of petite mutants (Dimitrov et al. 2009), and drug resistance (Demogines et al. 2008; Kim and Fay 2009; Ehrenreich et al. 2010). In all of the above studies, the mapping was performed against the BY/S288c background. The pleiotropic effect of MKT1 on cellular function can most likely be attributed to its recently established regulatory role in global gene expression (Zhu et al. 2008). Lee et al. (2009) have later established that the D30G and K453R polymorphisms are responsible for deficiency of the Mkt1 protein in the BY strain. The study of Tadauchi et al. (2004) investigating the regulation of HO expression suggests that Mkt1 may control gene expression at a post-transcriptional step. It was also suggested that Mkt1 is recruited to the polysomes through the activity of Pbp1 (Tadauchi et al. 2004). As Mkt1 physically interacts with Pbp1, we investigated whether PBP1 itself is a quantitative trait gene determining high ethanol tolerance. We, therefore, performed RHA for PBP1 in the VR1-5B/BY4741 hybrid background but did not observe allele-specific contributions of the gene to high ethanol tolerance (data not shown).
SWS2 has previously been associated with ethanol tolerance in two studies in which the BY deletion strain collection was screened for ethanol-sensitive mutants: One study investigated growth of the deletion strains on solid plates containing 8% ethanol (Teixeira et al. 2009), and the other study determined the specific growth rate of the deletion strains in liquid medium containing 8% ethanol (Yoshikawa et al. 2009). It was surprising to find in our work that the SWS2 allele from the low ethanol-tolerant parent strain was beneficial for high ethanol tolerance. On the other hand, this result may explain the higher ethanol tolerance of the VR1-5B/BY4741 diploid strain in comparison to the original VR1 strain, assuming that the negative mutation in SWS2 is homozygous in VR1. SWS2 encodes a mitochondrial ribosomal protein that is essential for respiratory growth (Merz and Westermann 2009). Moreover, SWS2 has been identified as a quantitative trait gene for sporulation efficiency in a cross between the high efficiency strain SK1 and the low efficiency strain S288c (Ben-Ari et al. 2006). It was shown that the SK1 allele from SWS2 was advantageous for high-sporulation efficiency. The SK1 and S288c alleles from SWS2 did not contain nonsynonymous mutations in the ORF but were polymorphic in the putative promoter and terminator. These polymorphisms resulted in a higher SWS2 mRNA and protein level in S288c compared to SK1. The VR1-5B and BY4741 alleles from SWS2 contained one synonymous polymorphism in the ORF and several polymorphisms in the putative promoter. This suggests that the different contribution of both alleles to high ethanol tolerance may also result from a difference in SWS2 mRNA and protein level rather than a change in the amino acid sequence of the Sws2 protein.
Until the present study, the APJ1 gene has not been directly associated with ethanol tolerance. It was one of many genes found to be induced at least threefold in a study investigating the transcriptional response of S288c to a short-term ethanol shock (7% ethanol for 30 min) (Alexandre et al. 2001). However, the relevance of these genes for ethanol tolerance was not investigated. APJ1 encodes a putative chaperone protein of the Hsp40 family that is known to stimulate the activity of members of the Hsp70 chaperone family (Qiu et al. 2006). Like Sws2, Apj1 is localized in the mitochondria, and its effect may have to do with a requirement for efficient mitochondrial respiration during growth on high levels of ethanol as a sole carbon source. The VR1-5B and BY4741 alleles from APJ1 differ by five synonymous polymorphisms in the ORF and several polymorphisms in the putative promoter and terminator regions. This suggested that a difference in the expression level of APJ1 in VR1-5B and BY4741 may account for the difference in their ethanol tolerance. Surprisingly, deletion of APJ1 enhanced ethanol tolerance, consistent with the APJ1 gene product having a negative effect on ethanol tolerance and suggesting that the superior character of the VR1-5B allele from APJ1 may be due to its lower expression compared to the BY4741 allele. Real-time PCR analysis showed that APJ1 expression, especially in the beginning of fermentation, was about 2.5-fold higher in the BY4741 strain than in the VR1-5B strain. Although this would be consistent with the previous interpretation, the conditions of the expression analysis and the growth experiments on plates are necessarily different, and thus, it is not possible to make a definite conclusion yet. Alternative possibilities, such as interaction with one of the other identified causative genes or with elements in the genetic background of the strain, cannot be fully ruled out. On the other hand, our interpretation agrees with the observation that deletion of APJ1 in the VR1-5B background caused less improvement in ethanol tolerance than in the BY4741 background, although this also may have another explanation such as the inherently higher ethanol tolerance of that strain and/or interference with other beneficial polymorphisms. Interestingly, APJ1 seems to be a gene specifically involved in tolerance to high ethanol levels. Its deletion had no effect in the BY4741 strain for growth on 10% ethanol, and it was also never identified in any screen for ethanol tolerance in laboratory strains in which relatively low ethanol levels have been used (5%–12%). This indicates that it is unlikely that we can obtain a full understanding of tolerance to high ethanol levels with studies only in laboratory yeast strains.
Further analysis of QTL1 identified the URA3 gene as the sole causative gene in that locus. The BY4741 strain used as a control strain for low ethanol tolerance is ura3Δ and has several other auxotrophic mutations. This has led to a detailed study of the effect of auxotrophic mutations on tolerance to low and high ethanol levels and to other stress conditions (S Swinnen, A Goovaerts, K Schaerlaekens, F Dumortier, P Verdyck, K Souvereyns, M. Foulquié-Moreno, and JM Thevelein, in prep.).
In conclusion, we have shown that pooled-segregant whole-genome sequencing using relatively low numbers of selected segregants is a powerful and convenient approach to identify major genetic loci involved in complex, quantitative traits of industrial importance, such as high ethanol tolerance. We have validated the approach by identifying new causative alleles, MKT1 and APJ1, with a clear effect on the phenotype under study, i.e., high ethanol tolerance. More stringent phenotypic screening as well as future improvements of the whole-genome sequencing technology may improve the detection of minor genetic elements contributing to the phenotype of interest.
Methods
Strains and growth conditions
Yeast cells were grown at 30°C in YPD medium containing 1% (w/v) yeast extract, 2% (w/v) Bacto peptone, and 2% (w/v) glucose. Selection of transformants was done with 100 μg/mL geneticin. Selection for amino acid prototrophy was performed in minimal media containing a complete supplement mixture without the amino acid under study, 0.17% (w/v) yeast nitrogen base without amino acids and ammonium sulfate, 0.5% (w/v) ammonium sulfate, and 2% (w/v) glucose (pH 5.5). For solid plates, 1.5% (w/v) Bacto agar was added, and the pH was adjusted to 6.5.
Escherichia coli cells (TOP10; genotype F- mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (StrR) endA1 nupG) were grown at 37°C in Luria Broth (LB) medium containing 0.5% (w/v) yeast extract, 1% (w/v) Bacto tryptone, and 1% (w/v) sodium chloride (pH 7.5). For solid plates, 1.5% (w/v) Bacto agar was added, and the pH was adjusted to 6.5. Selection of transformants was done with 100 μg/mL ampicillin.
The yeast strains used in this study are listed in Supplemental Table S1 online.
General molecular biology methods
Genomic DNA was extracted from yeast according to Hoffman and Winston (1987). When required, additional purification was performed by ethanol precipitation. Polymerase chain reaction (PCR) was performed with Accuprime (Invitrogen) for cloning and sequencing purposes and with ExTaq (TAKARA) for diagnostic purposes. Yeast was transformed with the LiAc/PEG method (Gietz et al. 1995). Cloning was performed by standard techniques. Dephosphorylation was performed with rAPid Alkaline Phosphatase (Roche) and ligation with T4 DNA ligase (Roche). E. coli was transformed with the CaCl2 method (Sambrook et al. 1989) and plasmid DNA isolated according to Del Sal et al. (1988). The plasmids used in this study are listed in Supplemental Table S2 online.
Mating, sporulation, and tetrad analysis
Mating, sporulation, and tetrad analysis were performed by standard procedures (Sherman and Hicks 1991). The mating type of the segregants was determined by diagnostic PCR for the MAT locus (Huxley et al. 1990).
Ethanol tolerance assay
Strains were inoculated in YPD and grown at 30°C for 3 d until stationary phase. Cultures were diluted to an OD600 of 0.5, and 5 μl of a twofold (10° to 8.10−3) or tenfold (10° to 10−3) dilution range was spotted on YPD and YP with different concentrations of ethanol. Growth was scored after 1 d for control YPD plates and 9–11 d for plates with ethanol. All spot tests were repeated at least twice, starting from independent cultures.
Genotyping of SNP markers by PCR
For each SNP marker, two primers were constructed that differed only at their 3′-terminal end nucleotide. In particular, one primer contained the VR1-5B nucleotide, while the other primer contained the BY4741 nucleotide. Both primers were always applied in separate PCR reactions with a common indirect primer. The two primer pairs were investigated for their ability to specifically amplify the VR1-5B or BY4741 sequence by performing four PCR reactions at different hybridization temperatures that differed in the combination of DNA and primer pairs. The combinations were: (1) DNA from BY4741 with primer pair for BY4741, (2) DNA from BY4741 with primer pair for VR1-5B, (3) DNA from VR1-5B with primer pair for BY4741, and (4) DNA from VR1-5B with primer pair for VR1-5B. Reactions were performed in 20-μl mixtures containing 10 ng template DNA, 10 pmol of forward and reverse primers, and appropriate amounts of dNTPs, ExTaq polymerase, and ExTaq buffer, according to the manufacturer's guidelines (TAKARA). The following cycling parameters were used: 4-min initial template denaturation at 94°C, and 32 cycles comprising a 15-sec denaturation step at 94°C, followed by annealing for 30 sec, and a 1-min elongation step at 72°C. The reactions were performed at annealing temperatures ranging from 58°C to 66°C (in 2°C-increments). The annealing temperature at which the VR1-5B and BY4741 sequences were found to be specifically amplified (see Supplemental Table S3 online) was subsequently applied to genotype the SNP marker in individual highly ethanol-tolerant segregants. Each SNP marker check included VR1-5B and BY4741 as controls.
Preparation of DNA samples and whole-genome sequence analysis
The two parent strains VR1-5B and BY4741 and all segregants with high ethanol tolerance were grown individually to saturation in 50 mL YPD at 30°C for 3 d. Exactly 10 mL of each culture was filtered, after which the cells were dried in the microwave and weighed to establish the relationship between optical density and dry weight. The remaining culture volumes were stored at −80°C. The two pools of segregants were constructed by combining equal amounts of cells from the stored cultures based on dry weight. The genomic DNA from the parent strains and the pools was extracted according to Johnston (1994). At least 3 μg of each DNA sample was provided to GATC Biotech AG for sequence analysis using Illumina HiSeq 2000 technology. Paired-end short reads of ∼100 bp were generated for the three samples (11.1 M, 11.9 M, and 11.7 M reads for VR1-5B, the 16% pool, and the 17% pool, respectively). The assembled sequences had an average coverage of 76, 58, and 55, respectively.
Reciprocal hemizygosity analysis
All deletions for reciprocal hemizygosity analysis were made in the haploid backgrounds. The BY4741 deletion strains were obtained from the deletion strain collection (Giaever et al. 2002). The deletions in the VR1-5B background were made using the same primers and strategy as the International Deletion Consortium (Winzeler et al. 1999; Giaever et al. 2002). The transformants were selected on geneticin plates and verified by PCR with several combinations of internal and external primers. The haploid strains were subsequently crossed to construct the hybrid diploid strains. The presence of both the wild-type and deletion allele from the gene in the diploid hybrids was verified by PCR. The reciprocal hemizygosity analysis was performed twice starting from independent PCR amplifications and transformations.
Real-time PCR
For measurement of APJ1 expression, samples were taken from early exponential-phase grown cells of BY4741 and VR1-5B. Pellets were frozen in liquid nitrogen and stored at −80°C. RNA extraction was performed using the phenol chloroform method. cDNA was prepared following the instructions of the GoScript Reverse Transcription System kit (Promega). Relative quantification of APJ1 and 18S was performed using a StepOnePlus Real-time PCR system (Applied Biosystems), primers: Fw APJ1 (TGATGGGCACGGTGGTCTA), Rv APJ1 (TTGAATACCTTGCCCTTTGCA), Fw 18S (CACTTCTTAGAGGGACTATCGGTTTC) and Rv 18S (CAGAACGTCTAAGGGCATCACA).
Statistical analysis
For each chromosome, the quantified frequencies of the detected SNPs were considered to be binomially distributed. The underlying structure in the SNP scatterplot of a given chromosome (Fig. 2) was identified by fitting smoothing splines in the generalized linear mixed model framework (Ruppert et al. 2003). The number of knots of the spline was chosen such that they were spaced at ∼40-kb intervals. Simultaneous confidence bands (Ruppert et al. 2003) for the fitted smoother were constructed and allowed identification of regions that are significantly different from a baseline, i.e., a SNP frequency of 50%.
For chromosome II and XV, the data from both pools of segregants (16% and 17% ethanol) were simultaneously modeled with generalized additive mixed models with a smoother for the mean trend (Fig. 2B) and for the difference between both pools (data not shown). For graphical representation, we have chosen to represent the resulting fit for each pool and their simultaneous confidence bands. The difference in SNP frequency between the two pools is certainly significant when the simultaneous confidence bands do not overlap.
Data access
All sequence data have been submitted to the NCBI Sequence Read Archive (SRA) (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession number SRA049724.
Acknowledgments
We thank Henrique Amorim and Mario Lucio Lopes (Fermentec, Piracicaba, Brazil) for the gift of the VR1 strain and for stimulating discussions. We also acknowledge Ben Souffriau, Elke Nevoigt, Huyen Nguyen, Tomasz Burzykowski, and Ziv Shkedy for helpful discussions. We thank Catherina Coun, Martine De Jonge, Kjell Lenaers, Ilse Palmans, Paul Vandecruys, and Evy Vanderheyden for technical help with the experiments, and Nico Vangoethem for preparation of the figures. This work has been supported by a predoctoral fellowship to S.S. and a post-doctoral fellowship to K.S. from the Agency for Innovation by Science and Technology (IWT-Flanders), SBO grants (IWT 50148 and IWT 90043) from IWT-Flanders, the EC 7th Framework program (NEMO project), IOF-Knowledge platform (IKP/10/002 ZKC 1836), and BOF-Program financing (project NATAR) to J.M.T.
Authors' contributions: S.S. and A.G. performed the experiments; S.S., K.S., T.P., G.H., Y.Y., M.D., M.F., F.D., and J.T. analyzed the data; J.C. and L.C. provided expert statistical analysis; and S.S., M.F., and J.T. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.131698.111.
References
- Alexandre H, Ansanay-Galeote V, Dequin S, Blondin B 2001. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett 498: 98–103 [DOI] [PubMed] [Google Scholar]
- Argueso JL, Carazzolle MF, Mieczkowski PA, Duarte FM, Netto OV, Missawa SK, Galzerani F, Costa GG, Vidal RO, Noronha MF, et al. 2009. Genome structure of a Saccharomyces cerevisiae strain widely used in bioethanol production. Genome Res 19: 2258–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari G, Zenvirth D, Sherman A, David L, Klutstein M, Lavi U, Hillel J, Simchen G 2006. Four linked genes participate in controlling sporulation efficiency in budding yeast. PLoS Genet 2: e195 doi: 10.1371/journal.pgen.0020195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D 2005. Quantitative trait loci analysis using the false discovery rate. Genetics 171: 783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755 [DOI] [PubMed] [Google Scholar]
- Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, Adler C, Dunn B, Dwight S, Riles L, Mortimer RK, et al. 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature 387: 67–73 [PMC free article] [PubMed] [Google Scholar]
- D'Amore T, Stewart GG 1987. Ethanol tolerance of yeast. Enzyme Microb Technol 9: 322–330 [Google Scholar]
- Del Sal G, Manfioletti G, Schneider C 1988. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res 16: 9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demogines A, Smith E, Kruglyak L, Alani E 2008. Identification and dissection of a complex DNA repair sensitivity phenotype in baker's yeast. PLoS Genet 4: e1000123 doi: 10.1371/journal.pgen.1000123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer AM, Davis RW 2005. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat Genet 37: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Dimitrov LN, Brem RB, Kruglyak L, Gottschling DE 2009. Polymorphisms in multiple genes contribute to the spontaneous mitochondrial genome instability of Saccharomyces cerevisiae S288C strains. Genetics 183: 365–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Huang X, Zhang L, Zhao N, Yang D, Zhang K 2009. Tolerance and stress response to ethanol in the yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol 85: 253–263 [DOI] [PubMed] [Google Scholar]
- Dohm JC, Lottaz C, Borodina T, Himmelbauer H 2008. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res 36: e105 doi: 10.1093/nar/gkn425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich IM, Torabi N, Jia Y, Kent J, Martis S, Shapiro JA, Gresham D, Caudy AA, Kruglyak L 2010. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464: 1039–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K, Matsuyama A, Kobayashi Y, Iwahashi H 2006. The genome-wide screening of yeast deletion mutants to identify the genes required for tolerance to ethanol and other alcohols. FEMS Yeast Res 6: 744–750 [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360 [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272 [DOI] [PubMed] [Google Scholar]
- Huxley C, Green ED, Dunham I 1990. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet 6: 236. [DOI] [PubMed] [Google Scholar]
- Johnston JR. 1994 Molecular genetics of yeast: A practical approach. Oxford University Press, New York. [Google Scholar]
- Kim HS, Fay JC 2009. A combined-cross analysis reveals genes with drug-specific and background-dependent effects on drug sensitivity in Saccharomyces cerevisiae. Genetics 183: 1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SI, Dudley AM, Drubin D, Silver PA, Krogan NJ, Pe'er D, Koller D 2009. Learning a prior on regulatory potential from eQTL data. PLoS Genet 5: e1000358 doi: 10.1371/journal.pgen.1000358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Elkon IM, McGee MA, Higbee AJ, Gasch AP 2010. Exploiting natural variation in Saccharomyces cerevisiae to identify genes for increased ethanol resistance. Genetics 186: 1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marullo P, Aigle M, Bely M, Masneuf-Pomarede I, Durrens P, Dubourdieu D, Yvert G 2007. Single QTL mapping and nucleotide-level resolution of a physiologic trait in wine Saccharomyces cerevisiae strains. FEMS Yeast Res 7: 941–952 [DOI] [PubMed] [Google Scholar]
- Merz S, Westermann B 2009. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance, and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol 10: R95 doi: 10.1186/gb-2009-10-9-r95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci 88: 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogami S, Ohya Y, Yvert G 2007. Genetic complexity and quantitative trait loci mapping of yeast morphological traits. PLoS Genet 3: e31 doi: 10.1371/journal.pgen.0030031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parts L, Cubillos FA, Warringer J, Jain K, Salinas F, Bumpstead SJ, Molin M, Zia A, Simpson JT, Quail MA, et al. 2011. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res 21: 1131–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlstein EO, Ruderfer DM, Roberts DC, Schreiber SL, Kruglyak L 2007. Genetic basis of individual differences in the response to small-molecule drugs in yeast. Nat Genet 39: 496–502 [DOI] [PubMed] [Google Scholar]
- Qiu XB, Shao YM, Miao S, Wang L 2006. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci 63: 2560–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert D, Wand MP, Carroll RJ 2003. Semiparametric regression. Cambridge University Press, New York [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T 1989. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Segrè AV, Murray AW, Leu JY 2006. High-resolution mutation mapping reveals parallel experimental evolution in yeast. PLoS Biol 4: e256 doi: 10.1371/journal.pbio.0040256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Hicks J 1991. Micromanipulation and dissection of asci. Methods Enzymol 194: 21–37 [DOI] [PubMed] [Google Scholar]
- Sinha H, Nicholson BP, Steinmetz LM, McCusker JH 2006. Complex genetic interactions in a quantitative trait locus. PLoS Genet 2: e13 doi: 10.1371/journal.pgen.0020013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, McCusker JH, Davis RW 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330 [DOI] [PubMed] [Google Scholar]
- Swinnen S, Thevelein JM, Nevoigt E 2012. Genetic mapping of quantitative phenotypic traits in Saccharomyces cerevisiae. FEMS Yeast Res 12: 215–227 [DOI] [PubMed] [Google Scholar]
- Tabor HK, Risch NJ, Myers RM 2002. Candidate-gene approaches for studying complex genetic traits: Practical considerations. Nat Rev Genet 3: 391–397 [DOI] [PubMed] [Google Scholar]
- Tadauchi T, Inada T, Matsumoto K, Irie K 2004. Posttranscriptional regulation of HO expression by the Mkt1-Pbp1 complex. Mol Cell Biol 24: 3670–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MC, Raposo LR, Mira NP, Lourenco AB, Sa-Correia I 2009. Genome-wide identification of Saccharomyces cerevisiae genes required for maximal tolerance to ethanol. Appl Environ Microbiol 75: 5761–5772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Voorst F, Houghton-Larsen J, Jonson L, Kielland-Brandt MC, Brandt A 2006. Genome-wide identification of genes required for growth of Saccharomyces cerevisiae under ethanol stress. Yeast 23: 351–359 [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Richards DR, Conway AR, Goldstein AL, Kalman S, McCullough MJ, McCusker JH, Stevens DA, Wodicka L, Lockhart DJ, et al. 1998. Direct allelic variation scanning of the yeast genome. Science 281: 1194–1197 [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Yoshikawa K, Tanaka T, Furusawa C, Nagahisa K, Hirasawa T, Shimizu H 2009. Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res 9: 32–44 [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang B, Smith EN, Drees B, Brem RB, Kruglyak L, Bumgarner RE, Schadt EE 2008. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet 40: 854–861 [DOI] [PMC free article] [PubMed] [Google Scholar]