Abstract
The global spread of drug-resistant parasites is a serious problem for the treatment of malaria. Although identifying drug-resistance genes is crucial for the efforts against resistant parasites, an effective approach has not yet been developed. Here, we report a robust method for identifying resistance genes from parasites by using a Plasmodium artificial chromosome (PAC). Large genomic DNA fragments (10–50 kb) from the drug-resistant rodent malaria parasite Plasmodium berghei were ligated into the PAC and directly introduced into the drug-sensitive (i.e., wild-type) parasite by electroporation, resulting in a PAC library that encompassed the whole genomic sequence of the parasite. Subsequently, the transformed parasites that acquired resistance were selected by screening with the drug, and the resistance gene in the PAC was successfully identified. Furthermore, the drug-resistance gene was identified from a PAC library that was made from the pyrimethamine-resistant parasite Plasmodium chabaudi, further demonstrating the utility of our method. This method will promote the identification of resistance genes and contribute to the global fight against drug-resistant parasites.
The emergence and spread of drug-resistant parasites are major problems that prevent the control of malaria. Parasite resistance to most anti-malarial drugs, such as chloroquine, sulphadoxine-pyrimethamine, quinine, and mefloquine, has increased globally, rendering these drugs useless in most malaria-endemic areas (Wongsrichanalai et al. 2002). The World Health Organization currently recommends artemisinin-based combination therapy (ACT) as the first-line treatment for malaria; however, a decline in the effectiveness of ACT was recently reported in the border region between Thailand and Cambodia, indicating the emergence of an artemisinin-resistant parasite (Dondorp et al. 2009; World Health Organization 2010). If such drug-resistant parasites become widely distributed, global malaria control efforts will be significantly hampered.
The identification of genes responsible for drug resistance is crucial for combating drug-resistant parasites because the mutations found in these genes are useful molecular markers for the surveillance of the emergence and spread of drug-resistant parasites. In the past, the identification of resistance genes was carried out by restriction fragment length polymorphism (RFLP) analysis of the progeny generated by genetic crosses between drug-resistant and drug-sensitive parasites using primates and mosquitoes. The chloroquine-resistance gene (pfcrt, MAL7P1.27) of Plasmodium falciparum was identified by this approach and facilitated the elucidation of the mechanisms of chloroquine resistance and the global surveillance of this resistant parasite (Wellems et al. 1991; Su et al. 1997; Djimde et al. 2001a,b). However, because this approach requires a large amount of time and effort, resistance genes for other anti-malarial drugs, such as mefloquine and quinine, have not yet been identified. Moreover, unknown resistance genes are thought to be present even in the chloroquine-resistant parasite (Valderramos et al. 2010). Now, whole-genome sequencing and microarray technologies are used for the identification of single nucleotide polymorphisms (SNPs) involved in the drug resistance of parasites (Hunt et al. 2010; Rottmann et al. 2011). However, it may be difficult to identify the drug-resistance genes from field-isolated parasites using these two technologies because these parasites have more than 10,000 SNPs (Dharia et al. 2010). Therefore, a more effective method for the identification of drug-resistance genes in parasites is required.
The artificial chromosome—which consists of three essential elements, the centromere, the telomere, and the replication origin—is an attractive genetic tool for molecular biology–based studies in eukaryotes. The first eukaryotic artificial chromosome was developed in the budding yeast Saccharomyces cerevisiae and is known as the yeast artificial chromosome (YAC) (Murray and Szostak 1983). The YAC can be stably maintained throughout mitosis and meiosis and has been widely used for cloning large DNA fragments. The development of artificial chromosomes in other eukaryotes, including humans, has been attempted by combining these three essential elements but has not yet been successful. Human artificial chromosomes are integrated at the centromere or the telomere of the original chromosome during cell division and are therefore not episomally maintained (Ikeno et al. 1998).
In a previous study, we made a Plasmodium artificial chromosome (PAC) from the small centromere (1189 bp) and the short telomeric fragment (<250 bp) of the rodent malaria parasite Plasmodium berghei, which is widely used as a model for human malaria (Iwanaga et al. 2010). This PAC is the second successful development of a functional eukaryotic artificial chromosome after the YAC. Although it is small (<10 kb), the PAC behaves as an actual parasite chromosome and segregates into daughter cells with >99.9% efficiency during multiple rounds of cell divisions. After cell division, it is stably maintained as a single copy without integration throughout the parasite life cycle. Notably, the transfection efficiency of the PAC is significantly higher than that of a normal plasmid. These unique features of the PAC overcome the current technical limitations of parasite genetic modification technology and could be applied in various parasite research fields. In the present study, we demonstrate that the PAC can be utilized for the identification of drug-resistance genes from parasites. Finally, we discuss the advantages of the method for identifying drug-resistance genes.
Results
The transfection efficiency of the PAC is extremely high
Based on the unique features of the PAC, we hypothesized that it could be utilized for identifying drug-resistance genes from parasites as follows (Fig. 1): In step 1, the genomic library of a drug-resistant parasite is constructed in a drug-sensitive parasite with the PAC; in step 2, transformed parasites that acquired resistance are selected in rats by drug screening; and, in step 3, the drug-resistance genes incorporated into the PAC are identified from the selected parasites. To achieve this, the PAC library needs to encompass the entire genomic sequence of the parasite; thus, the PAC ligated with DNA fragments has to be introduced into the parasites with high efficiency. Moreover, the PAC library needs to be generated from large genomic DNA fragments because short fragments would not completely cover the regulatory regions, exons, and introns of the genes. According to the average gene density, which has been estimated to be 4.5 kb in Plasmodium spp. (Carlton et al. 2008), DNA fragments that are longer than 10 kb must be inserted into the PAC.
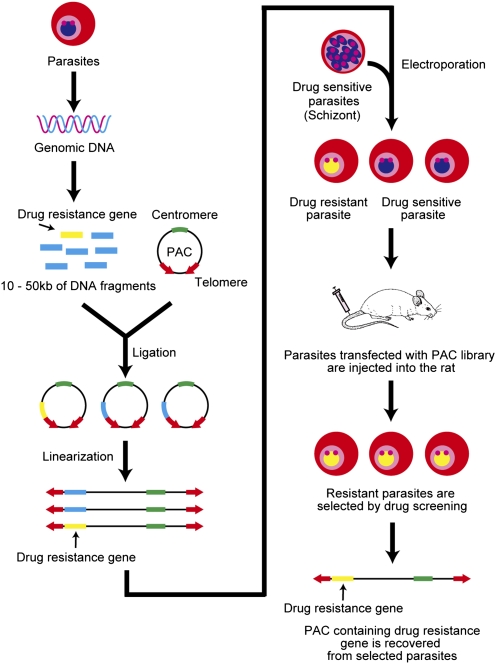
Figure 1.
A schematic representation of the method for the identification of drug-resistance genes from the PAC library. The genomic DNA isolated from drug-resistant parasites is partially digested with a restriction enzyme, and the DNA fragments ranging from 10–50 kb are then incorporated into the PAC. Subsequently, the PAC that includes the DNA fragments is linearized by digestion with PmeI, and the constructed PAC library is directly introduced into drug-sensitive parasites by electroporation. Parasites that acquire resistance via the PAC (including the drug-resistance gene) are selected by drug screening in vivo. Finally, the drug-resistance genes are identified by analyzing the DNA fragments in the PAC from the selected parasites.
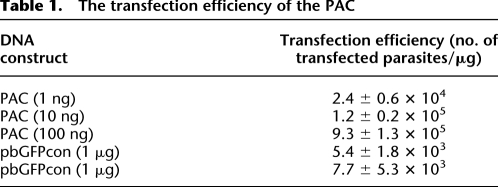
First, to examine whether the transfection efficiency of the PAC was sufficient to generate a library that covered the entire genome sequence of the parasite, we transfected parasites with 1–100 ng of the linearized PAC (Fig. 2) and determined the transfection efficiency as the initial number of transfected parasites per microgram of DNA, as described in the Methods. The transfection efficiency of the PAC was calculated to be 9.3 × 105 to 2.4 × 104 in each experiment and was ∼100-fold higher than that of a control plasmid, which contained neither centromere nor telomere sequences (Table 1). If the average size of the inserted DNA fragments was 10 kb, as discussed above, 2.5 × 103 independent clones would cover the entire parasite genome, which has been estimated to be ∼25 Mb (Janse et al. 1994). Therefore, based on the transfection efficiency of the PAC, we concluded that a genomic library that covered approximately 10 to hundreds of genome equivalents would theoretically be generated by the transfection of 1 μg of the PAC.
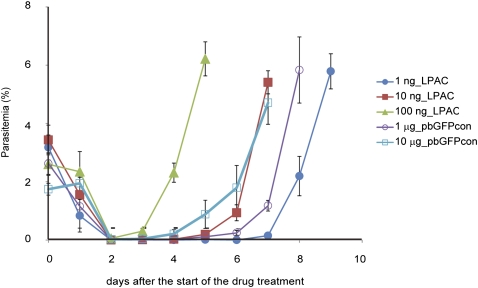
Figure 2.
The transfection efficiency of the PAC. The parasitemia of parasites transfected with various amounts of the linearized PAC are shown. To determine the transfection efficiency of the PAC, the parasites were transfected with 1 ng (closed circle), 10 ng (closed square), or 100 ng (closed triangle) of the PAC. In addition, parasite transfection with 1 μg (open circle) or 10 μg (open square) of pbGFPcon was performed as controls. The drug treatment of the parasites was initiated 24 h after the transfection, as indicated by the arrow.
Table 1.
The transfection efficiency of the PAC
A high-coverage PAC library can be made from large DNA fragments
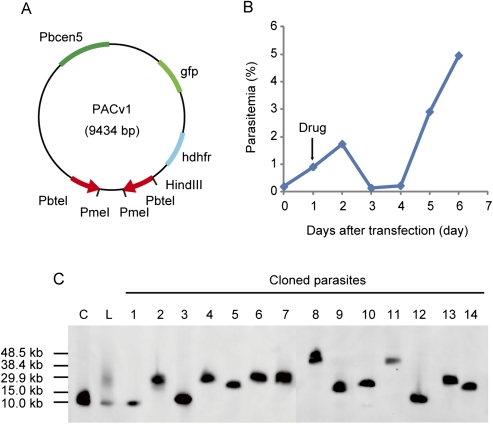
Next, we examined whether the PAC library could be made from DNA fragments longer than 10 kb. Briefly, the genomic DNA of the wild-type P. berghei ANKA strain was partially digested with HindIII. Then, 3 μg of the DNA fragments, ranging from 10–50 kb, was purified and ligated to the same quantity of PACv1, which contained the centromere and the telomere sequences and the gfp and human dihydrofolate reductase (hdhfr) genes as fluorescent and drug-selectable marker genes, respectively (Fig. 3A, 9434 bp). After linearizing PACv1 by digestion with PmeI, the reaction mixture was directly introduced into mature schizonts of wild-type parasites by electroporation, and the transfected parasites were immediately injected into rats, which were then treated with drug. GFP-expressing parasites, which contained PACv1, were microscopically observed immediately before drug screening (24 h after transfection), and all parasites were GFP-positive 4 d after transfection. The percentage of parasitemia was 0.2% at this point (Fig. 3B), and thus, the initial number of transfected parasites was calculated to be 5.6 × 104, suggesting that the PACv1 library with large DNA fragments was introduced into the parasites with high efficiency.
Figure 3.
The construction of a PAC library from the genomic DNA of the wild-type parasite. (A) A schematic drawing of PACv1. (B) The PACv1 library was made from the genomic DNA of wild-type parasites and directly introduced into these parasites by electroporation. The parasitemia was monitored every 24 h after transfection. The arrow indicates the initiation of the drug treatment of the parasites. (C) Southern blot analysis of PACv1 (including DNA fragments) was performed using the hdhfr gene as the probe. (Lanes C and L) Linearized PACv1 without an inserted DNA fragment and the PAC library of the total population of transfected parasites, respectively. (Lane 1) Parasites with self-ligated PACv1. The remaining 13 lanes show parasites with PACv1 (including DNA fragments).
To further examine if large DNA fragments could indeed be introduced into the parasites, the PACv1 isolates that contained the DNA fragments from the total parasite population were separated by contour-clamped homogeneous electrical field (CHEF) electrophoresis and analyzed by Southern blotting using the hdhfr gene as the probe (Fig. 3C). In this analysis, a strong signal at ∼10 kb and a broad signal ranging from 15.0–38.4 kb were detected. The size of the strong signal was comparable to that of the linearized form of the original PACv1, indicating that it was derived from the self-ligation of PACv1. In contrast, the broad signal was derived from the PACv1 clones that contained various lengths of DNA fragments. Given the size of PACv1, this result showed that DNA fragments ranging from 5–30 kb could be successfully introduced into the parasites. Subsequently, each transfected parasite was cloned by limiting dilution. Southern blot analysis of the resulting parasites showed that 13 of the 25 parasite clones harbored PACv1 that contained DNA fragments, and the remaining 12 parasite clones contained the self-ligated PACv1 (Fig. 3C). The PACv1 clones that contained DNA fragments were found to have sizes ranging from 15.0–40.0 kb, and the average size of the inserted fragments was ∼16.6 kb. The number of independent parasite clones with PACv1 that contained DNA fragments was calculated to be 2.9 × 104 based on the initial number of transfected parasites and the ratio of the number of parasites with PACv1 that contained DNA fragments to the total parasite population (i.e., 5.6 × 104 × [13/25]). Furthermore, the genomic coverage of the constructed PAC library was calculated to be ∼19.3 genome equivalents. These results indicate that both the coverage of the PAC library and the average size of the inserted DNA fragments were sufficient for a genome-wide screen of drug-resistance genes.
Resistance genes can be identified from the PAC library by screening with the drug
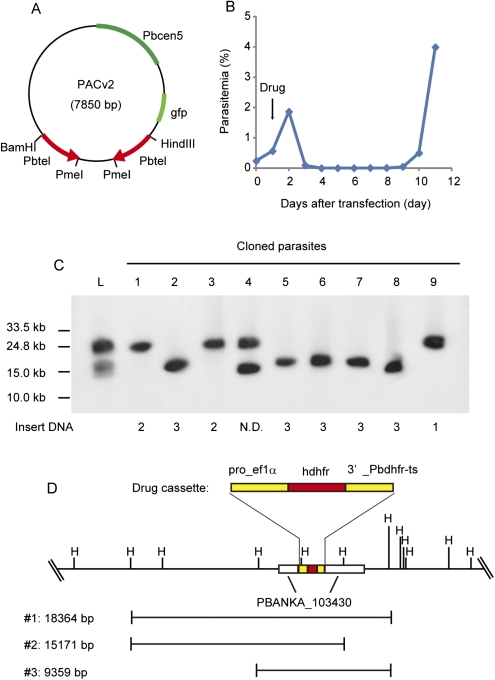
We further sought to determine whether a drug-resistance gene could be identified from a library made from the genomic DNA of a drug-resistant parasite. Briefly, genomic DNA was isolated from an artificial drug-resistant parasite, in which the hdhfr gene was integrated into the genetic locus of PBANKA_103430 (http://plasmodb.org/plasmo/) as a drug-resistance gene (Supplemental Fig. S1) and was used to prepare large DNA inserts. The purified large DNA fragments were incorporated into PACv2, which did not contain the drug-selectable marker gene (Fig. 4A, 7850 bp), and then directly introduced into the drug-sensitive (i.e., wild-type) parasites. Twenty-four hours after injecting the transfected parasites into rats, drug screening using pyrimethamine was conducted. In the presence of the drug, only parasites that acquired resistance from the introduction of the PACv2 that contained the hdhfr gene were expected to survive. The drug-resistant parasites were first observed in Giemsa-stained blood smears from the rats 6 d after transfection, and the percentage of parasitemia was >4% by day 11 (Fig. 4B). In contrast, no drug-resistant parasites were observed in the presence of the drug with the control parasite library, which was made from the genomic DNA of the wild-type (i.e., drug-sensitive) parasites. Southern blot analysis of the resultant drug-resistant parasites using the hdhfr gene as the probe revealed that this gene was present in the selected parasites, demonstrating that a drug-resistance gene could be screened from a PACv2 library using this approach (Fig. 4C, lane L). The selected parasites were further cloned using the limiting dilution procedure followed by Southern blot analysis, showing that all of the cloned parasites had the PACv2 construct containing the hdhfr gene (Fig. 4C). Sequence analyses of the DNA fragments inserted into PACv2 showed that these parasites were derived from at least three independent clones: PACv2 with 18,364-bp, 15,171-bp, and 9359-bp DNA fragments containing the resistance gene (Fig. 4D). Finally, we were able to identify the target drug-resistance gene (i.e., the hdhfr gene) by comparing the sequences of the inserted DNA fragments. We performed three additional independent experiments and were able to identify the drug-resistance gene every time (Supplemental Fig. S2), demonstrating the reproducibility of our approach.
Figure 4.
The identification of a drug-resistance gene from a PAC library by drug screening. (A) A schematic drawing of PACv2. (B) Parasites were transfected with the PACv2 library, which was made from the genomic DNA of the artificial drug-resistant parasite. The parasitemia was monitored every 24 h after transfection. (Arrow) Initiation of the drug treatment of the parasites. (C) Southern blot analysis of PACv2 (including DNA fragments) was performed using the hdhfr gene as the probe. (Lane L) PACv2 library in the total population of the transfected parasites. (Lanes 1–9) Cloned drug-resistant parasites. The inserted DNA fragments in PACv2 from each clone are numbered according to Figure 4D, as shown on the bottom. (D) A schematic drawing of the inserted DNA fragments 1–3 in PACv2 from the selected drug-resistant parasites. (Left) Lengths of the DNA. (Top) Genomic sequence surrounding PBANKA_103430, with the HindIII recognition sites.
A drug-resistance gene can be identified from the PAC libraries made from other Plasmodium species
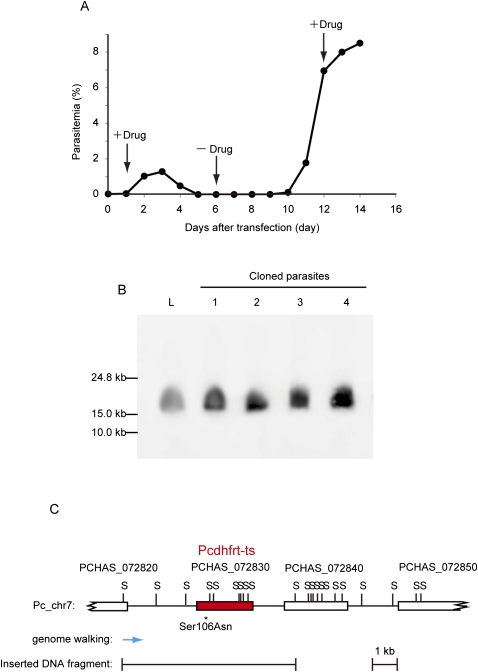
To examine the usefulness of our method, we next attempted to clone the drug-resistance gene from a PAC library made from the drug-resistant Plasmodium chabaudi AS(50S/P) strain. This P. chabaudi strain evolved in the laboratory from the drug-sensitive P. chabaudi AS-sens strain and is resistant to both pyrimethamine and sulphadoxine (Hayton et al. 2002; Culleton et al. 2005). The genomic DNA of P. chabaudi AS(50S/P) was partially digested with Sau3AI, and then DNA fragments ranging from 10–50 kb were purified. The obtained large DNA fragments were ligated into the BamHI site of PACv2 (Fig. 4A) and directly introduced into the wild-type P. berghei. Drug treatment with pyrimethamine was initiated 24 h after the transfection (Fig. 5A). The parasites completely disappeared from the rat 4 d after the initiation of the drug treatment (Fig. 5A), suggesting that all drug-sensitive parasites were eliminated. Then, the drug treatment was stopped, and the parasites were maintained in the absence of the drug (Fig. 5A). The emergence of the parasites was confirmed in the Giemsa-stained blood smears 4 d after the withdrawal of the drug, and the percentage of parasitemia ultimately reached ∼7% (Fig. 5A). All of the selected parasites expressed GFP, showing that they stably maintained PACv2. Subsequently, to confirm if the parasites were actually resistant to pyrimethamine, they were again treated with the drug, and the progression of their parasitemia was monitored. As shown in Figure 5A, the increase in parasitemia continued in the presence of the drug, indicating that the selected parasites carrying PACv2 acquired resistance to pyrimethamine. To identify the DNA fragments in the PACv2 constructs, the selected parasites were further cloned using the limiting dilution procedure followed by Southern blot analysis using the gfp gene as a probe. Both Southern blot analyses of the total selected parasite population and the clonal parasites gave identical single signals at a size of ∼15 kb, suggesting that the single parasite line that carried a PACv2 bearing a DNA fragment had been selected by the drug screening (Fig. 5B). Given the size of PACv2 (7850 bp), these results indicated that ∼7 kb of DNA was incorporated into the PACv2 construct. To further identify the drug-resistance gene in the DNA fragment inserted into PACv2, we performed a genome walking analysis using the genomic DNA purified from the cloned parasites. In brief, the purified genomic DNA was digested with SspI and ligated with the adaptor DNA, and then the end of the inserted DNA fragment was amplified by nested PCR using a set of adaptor- and PACv2-specific primers. Single PCR products were amplified from the genomic DNA of all cloned parasites, and a sequence similarity search showed that they were located at the genomic locus from 991819–992249 on chromosome 7 of P. chabaudi and encoded the N-terminal region of PCHAS_072820 (Fig. 5C, blue arrow). On chromosome 7, the dihydrofolate reductase-thymidylate synthase gene (dhfr-ts: PCHAS_072830), the mutation of which confers pyrimethamine resistance to parasites, is located next to PCHAS_072820. Given the length of the inserted DNA fragment in PACv2, it was expected that the 7 kb of the inserted DNA fragment covered the genomic region from PCHAS_072820 to PCHAS_072840 and thus contained the entire sequence of PCHAS_072830, the dhfr-ts gene (Fig. 5C). Therefore, we subsequently amplified the dhfr-ts gene of P. chabaudi from the DNA fragment inserted into PACv2 and determined its sequence. The result clearly showed that the DNA fragment inserted into PACv2 contained the open reading frame (ORF) of the dhfr-ts gene of P. chabaudi and that its amino acid at position 106 had changed from Ser to Asn (Fig. 5C). Because this mutation is known to be associated with the pyrimethamine resistance of parasites, this mutated dhfr-ts gene was identified as a pyrimethamine-resistance gene of the P. chabaudi AS(50S/P) strain. Consequently, these results from the drug-resistant P. chabaudi demonstrated the utility of our method using the PAC.
Figure 5.
The identification of a drug-resistance gene from the PACv2 library made from pyrimethamine-resistant P. chabaudi. (A) Wild-type P. berghei was transfected with the PACv2 library made from pyrimethamine-resistant P. chabaudi. The parasitemia was monitored every 24 h after transfection. The arrows indicate the initiation and the termination of the drug treatment. (B) CHEF electrophoresis of the selected parasites by screening with pyrimethamine was performed, followed by Southern blot analysis using the gfp gene as the probe. (Lane L) PACv2 library in the total population of drug-resistant parasites. (Lanes 1–4) Clonal drug-resistant parasites with the PACv2 construct containing the resistance gene. (C) The inserted DNA fragment in the PACv2 from the drug-resistant parasites obtained from the PACv2 library is schematically shown. (Top) Genomic sequence surrounding PCHAS_072830, the dhfr-ts gene, on chromosome 7 of P. chabaudi, and the Sau3AI recognition sites are also shown as S. (Blue arrow) Genomic locus of the end of the inserted DNA fragment, which was identified by genome walking analysis.
Discussion
The high coverage of the PAC library facilitated a genome-wide screen for drug-resistance genes in terms of the phenotype (resistance) of transgenic parasites, rather than the genotype, in one experimental trial. This advantage, which is only achieved using the PAC, allows for the quick and rigorous identification of drug-resistance genes from parasites. The drug-resistance gene could be identified within a few weeks, and because the identified genes conferred resistance to the parasites, functional verification was not necessary. In addition, a single copy of the PAC is useful for quantitatively evaluating the effects of mutations in drug-resistance genes. In contrast to the PAC, a plasmid that does not include the centromere forms a concatemer in the parasites, resulting in an increase in its copy number. Because an increased number of copies of the drug-resistance gene would result in an apparent enhancement of resistance, such a plasmid is not suitable for the quantitative evaluation of the drug-resistance gene. The quantitative analysis of drug-resistance genes using the PAC would reveal the relationship between mutations and resistance, thereby providing information for the development of new anti-malarial drugs that bypass or overcome resistance mechanisms.
Our method using the PAC will be useful for the identification of the drug-resistance genes from field-isolated parasites, the genomic sequences of which are highly diverse. Recently, whole-genome sequencing and microarray technologies were used for the identification of the specific SNPs that confer drug resistance (Hunt et al. 2010; Rottmann et al. 2011), but it might be hard to identify such SNPs from field-isolated parasites because both technologies can detect numerous SNPs (Dharia et al. 2010). In contrast, our method allows for the screening of drug-resistance genes based on their function, with no reference to the SNPs in the genome sequence. Therefore, the drug-resistance genes would be more straightforwardly identified from field-isolated parasites by our method than by the other two technologies.
Our future aim is to identify the drug-resistance gene in human malaria parasites isolated from patients; we plan to develop a similar method for P. falciparum, which is the most deadly human parasite. To this end, it will be necessary to develop a PAC for P. falciparum. The present PAC containing the centromere of P. berghei cannot be used for P. falciparum because the centromeres of Plasmodium spp. function in a species-specific manner (Iwanaga et al. 2010). The centromere of P. falciparum has already been predicted, and the telomere sequences are conserved between Plasmodium spp. (Gardner et al. 1998; Bowman et al. 1999; Hall et al. 2002); thus, the PAC for P. falciparum can be developed in a similar manner to our previous strategy (Iwanaga et al. 2010). Indeed, we have already cloned the centromere of P. falciparum and then successfully constructed the centromere plasmid for this parasite. This constructed centromere plasmid can segregate into daughter cells with high efficiency during multiple rounds of cell divisions as well as into an actual chromosome of P. falciparum. Now we will subsequently attempt to generate the linearized PAC for P. falciparum by incorporating the telomeric DNA fragments into this centromere plasmid. The transfection technique of P. falciparum would also need to be improved upon for the generation of its PAC library. Currently, plasmids are introduced into P. falciparum using DNA-preloaded red blood cells (Deitsch et al. 2001). These preloaded red blood cells are infected with the parasite, and then the parasites spontaneously uptake the plasmid, resulting in transgenic parasites. Because the plasmids are introduced indirectly into the parasite by this method, its transfection efficiency would be too low to generate a high coverage of the PAC library. Therefore, a highly efficient direct transfection technique is needed for the construction of a P. falciparum PAC library. As shown in our previous study (Iwanaga et al. 2010), the linearization of the artificial chromosome can significantly improve the transfection efficiency. Thus, a high-efficient transfection of P. falciparum could be achieved by using the linear form of the PAC.
In the present study, we identified the dhfr-ts gene, which included a single-amino-acid mutation at position 106, as a pyrimethamine-resistance gene of P. chabaudi from the PAC library. This point mutation in the dhfr-ts gene confers a weak resistance to the parasite, as demonstrated in P. falciparum (Sirawaraporn et al. 1997). In addition, the promoter activity of the P. chabaudi dhfr-ts gene is low in P. berghei, so the transcription of the mutated dhfr-ts gene might be insufficient for parasites to have strong resistance to pyrimethamine. Therefore, these data suggested that even drug-resistance genes that confer weak resistance to parasites could be identified by our method. Furthermore, the results clearly demonstrated that our method can be used for the identification of drug-resistance genes from other Plasmodium spp. Even if the products of the drug-resistance gene and its promoter did not function fully in P. berghei, the drug-resistance genes could be identified from the PAC library by appropriate screening conditions. Therefore, our method might be directly utilized for the identification of the drug-resistance genes from human malaria parasites, such as P. falciparum and Plasmodium vivax.
Our high-coverage library with the PAC will allow us to identify not only the genes that correspond to drug resistance but also biologically relevant mutant phenotypes. These mutant parasite strains have been isolated from the field or established in the laboratory; however, most of them have not been analyzed genetically. For example, a functional complementation study of sexual-deficient parasites could be performed using the PAC library, resulting in the identification of genes involved in the sexual commitment of the parasite. Our approach using the PAC will open a new avenue in previously unexplored molecular genetic research of mutant parasites.
The artificial chromosome has been thought to be unstable when it is <50 kb. For example, the YAC cannot be stably maintained in the cell if its total size is <50 kb (Murray and Szostak 1983). However, our results clearly demonstrated that a PAC of <50 kb can be stably maintained in parasite cells. This result indicates that a DNA construct including only a centromere, telomere, and replication origin can behave like an actual chromosome, regardless of its size. The differences in the behaviors of the artificial chromosomes of Plasmodium parasites and budding yeast suggest that budding yeast have an unknown molecular mechanism to eliminate small chromosomes from the cell. The elucidation of the mechanism by which Plasmodium parasites stably maintain a small chromosome could be used to develop the perfect artificial chromosomes in other eukaryotes and promote the genetic engineering of eukaryotes.
In conclusion, we have demonstrated a robust method using a PAC for the identification of drug-resistance genes. The drug-resistance genes could be utilized as molecular markers for the development of an accurate and rapid diagnostic method, which could be used to select the appropriate drugs for an individual patient and produce more effective treatments. Furthermore, drug-resistance genes from newly identified resistant parasites, such as artemisinin-resistant parasites, could be identified before dissemination of the parasite, forestalling a new threat. We anticipate that our developed method using the PAC will contribute to the identification of the drug-resistance genes from malaria parasites.
Methods
Parasites
The P. berghei ANKA strain, which is sensitive to pyrimethamine, was used as the wild-type parasite for this study. An artificial pyrimethamine-resistant parasite, in which the hdhfr gene is integrated into the PBANKA_103430 genomic locus (http://plasmodb.org/plasmo/), was used for the pilot experiment to screen for a resistance gene from the PAC library. The transcription of the hdhfr gene was controlled by the P. berghei elongation factor 1α promoter. To integrate the hdhfr gene into the parasite genome, two fragments of the PBANKA_103430 gene were amplified by PCR using genomic DNA as a template with the two primer pairs 5′-ACCACGATAAAGGAGGCATGTAAC-3′ (primer 1) and 5′-CTCATCTACAAGCATCGTCGACTCATCGTCTCCTTTCCTC-3′ (primer 2) and 5′-CCTTCAATTTCGGATCCACTAGGTAATGTTGAAAGCGACAG-3′ (primer 3) and 5′-GTTGTATTATCAACTTGAGCAGTTTC-3′ (primer 4). The resultant fragments were annealed to either side of the drug-resistant cassette (including the hdhfr gene) by PCR with primers 1 and 4 and introduced into wild-type P. berghei by electroporation, as described below. The parasites containing the integrated drug-resistant cassette were cloned by a limiting dilution method and used in the study. The pyrimethamine-resistant P. chabaudi AS(50S/P) line was previously generated (Hayton et al. 2002) and obtained from MR4 (http://www.mr4.org/).
Parasite transfection
To transfect P. berghei with the Nucleofector II device (Lonza), mature schizonts were purified according to the procedure described by Waters et al. (1997). The schizonts (0.5 × 108–1.0 × 108) were mixed with 100 μL T-cell Nucleofector solution containing the PAC or the genomic libraries and transfected using the U-033 program. Immediately after transfection, 100 μL of complete culture medium (RPMI 1640 containing 20% fetal bovine serum) was added, and a total of 200 μL of the solution was injected intravenously into a single Wistar rat (3 wk old). Twenty-four hours after transfection, the rat was treated with pyrimethamine via drinking water. The pyrimethamine was first dissolved in DMSO (7 mg/mL) and diluted 100× with water at pH 3.5–4.0 (adjusted with 1 M HCl).
Determination of the transfection efficiency
To determine the transfection efficiency of the PAC, P. berghei was transfected with various amounts of the PAC, as described above. After transfection, the parasitemia of each transfected parasite was monitored every 24 h, and the PAC multiplication rate was calculated to be 6.3 per day in the presence of the drug, based on the average daily increase in parasitemia. Similar experiments were performed with the control plasmid pbGFPcon, and the multiplication rate of the parasite containing the control plasmid was determined to be 4.1 per day under the same conditions. The multiplication rates of the parasites with the PAC and pbGFPcon constructs were different in the presence of the drug because their segregation efficiencies were different. The transfection efficiencies were determined as the initial number of transfected parasites per microgram of DNA. The number (N) of transfected parasites was calculated using the following equation: N = (T × P/100) × 1/MD, where M is the multiplication rate of the parasites (PAC, 6.3 per day; pbGFPcon, 4.1 per day), P is the percentage of parasitemia, D is the day when drug-resistant parasites were first observed during drug selection, and T is the total number of red blood cells in whole rat blood (4.0 × 1010 RBCs per rat). In this study, we used the following parasitemias: 1 ng PAC, 0.15% at day 7; 10 ng PAC, 0.19% at day 5; 100 ng PAC, 2.3% at day 4; 1 μg pbGFPcon, 0.24% at day 6; and 10 μg pbGFPcon, 0.21% at day 4. Finally, the transfection efficiencies were determined by normalizing the number of transfected parasites to the various amounts of DNA constructs.
Purification of genomic DNA
Whole blood collected from rats that were infected with the parasites was subjected to ion-exchange chromatography using CF11 cellulose twice to remove the leukocytes. The parasites were purified by lysing the eluted red blood cells in red blood cell lysis buffer (1.5 M NH4Cl, 0.1 M KHCO3, and 0.01 M EDTA) and then dissolved in HNE buffer (10 mM Tris-HCl, 150 mM NaCl, 10 mM EDTA at pH 8.0, 0.1% SDS), followed by proteinase K digestion (final concentration, 40 μg/mL). After digestion, the genomic DNA was extracted with phenol/chloroform/isoamyl alcohol (PCI) and precipitated using ethanol. The purity of the obtained genomic DNA was estimated by the A260/280 and A260/230 ratios calculated from spectrophotometric readings.
Construction of the genomic library using the PAC
Ninety micrograms of genomic DNA from wild-type parasites was partially digested with 72 units of the restriction enzyme HindIII for 30 min at 37°C and separated by electrophoresis using a 0.75% low-melting Agarose gel (Lonza). The gel section that contained DNA fragments of ∼10–50 kb was excised and digested with β-agarase (NIPPON GENE). After digestion, the DNA fragments were extracted with PCI and precipitated with ethanol. PACv1 was completely digested with HindIII and subsequently dephosphorylated with shrimp alkaline phosphatase. Three micrograms of the purified DNA fragments was ligated overnight with the same amount of HindIII-digested PACv1 using the Takara ligation kit version 2 (Takara). The ligated sample (i.e., the PACv1 library) was extracted by PCI treatment followed by ethanol precipitation. To linearize PACv1, the PACv1 library was digested with the restriction enzyme PmeI followed by PCI treatment and ethanol precipitation. Finally, the PACv1 library was introduced into the wild-type P. berghei ANKA strain by electroporation with the Nucleofector II device, as described above. The library from the artificial drug-resistant parasite with PACv2 was similarly constructed. In addition, a PAC library was made from the pyrimethamine-resistant P. chabaudi AS(50S/P) line in an essentially similar manner, as described above. In brief, 100 μg genomic DNA from the P. chabaudi AS(50S/P) line was partially digested with 2.25 units of the restriction enzyme Sau3AI for 15 min at 37°C, and then DNA fragments ranging from 10–50 kb were purified from the low-melting Agarose gel. The purified DNA fragments were ligated into the BamHI site on PACv2, and then the ligated sample was digested with the restriction enzyme PmeI. The linearized PACv2 library was finally introduced into the P. berghei ANKA strain, as described above. The GFP expression of the parasites containing PACv1 or PACv2 was monitored using fluorescence microscopy (Olympus). Each transfected parasite was cloned using the limiting dilution procedure. The coverage (C) of the constructed PAC library was calculated using the following equation: C = Avinsert × Nindependent / G, where G is the estimated genome size (∼25 Mb) of P. berghei, Avinsert is the average size of the DNA fragments that were inserted into the PAC, and Nindependent is the number of independent parasite clones.
CHEF electrophoresis and Southern hybridization of the PAC
The infected red blood cells were collected after leukocyte removal using CF11 cellulose and were then treated with red blood cell lysis buffer. After red blood cell lysis, the parasites were collected, washed with PBS, and mixed into a 2% low-melting Agarose gel at a ratio of 1:1 (v/v). The resulting Agarose block containing the parasites was placed in SE buffer (0.5 M EDTA at pH 8.0, and 1% sarcosyl), treated overnight with proteinase K (final concentration, 100 μg/mL), and set into a 1.5% Agarose gel (PFC gel, Bio-Rad). The PAC constructs containing the DNA fragments were separated from the parasite chromosomes using CHEF electrophoresis and a CHEF-DRIII apparatus (Bio-Rad) in 0.5 M TBE buffer under the following conditions: initial switching time of 1.0 sec, final switching time of 6.0 sec, angle of 120°, voltage gradient of 6 V/cm, run time of 20 h, and temperature of 14°C. After CHEF electrophoresis, the DNA was transferred onto a nitrocellulose membrane. Southern hybridization was performed using the DIG-labeled hdhfr gene and gfp gene as the probes at 42°C and 37°C, respectively. After washing, chemical luminescence signals were detected using the LAS3000 mini lumino-image analyzer (FujiFilm).
Sequence analysis of the inserted DNA fragments in the PAC
Ten primers that were specific to the genomic sequence of P. berghei were designed according to the available sequence information from contig berg10, which includes the gene encoding PBANKA_103430. The primers were designed to amplify every 5 kb upstream of and downstream from the genomic locus in which the hdhfr gene was integrated into the artificial drug-resistant parasite. In addition, two specific primers were designed for PACv2. All of the primers are listed in Supplemental Table S1. To determine the sequences of both ends of the DNA fragments that were inserted into PACv2 from the selected drug-resistant parasites, PCR was performed with the genomic DNA isolated from these parasites as the template using sets of primers for contig berg10 and PACv2, and the sequences of the amplified products were determined. The lengths of the DNA fragments, including the drug-resistance gene in PACv2, were estimated by comparing the sequences of contig berg10 and the amplified PCR products.
Genomic walking analysis of the inserted DNA fragments in the PAC
The genomic walking analysis of the DNA fragment inserted into the PACv2 was performed using the GenomeWalker Universal Kit (Clontech) essentially using the manufacturer's protocol. Genomic DNA was purified from the clonal parasites carrying PACv2 containing the drug-resistance gene, and 2.5 μg was completely digested with the restriction enzyme SspI followed by the ligation with the DNA adaptors. The first PCR was performed using the ligated DNA as a template with a pair of adaptor- and PACv2-specific primers, 5′-GTAATACGACTCACTATAGGGC-3′ (adaptor primer 1) and 5′-TACCGCCTTTGAGTGAGCTGATACC-3′ (PACv2-specific primer 1), respectively. Subsequently, the second PCR was performed using the first PCR product as a template with the primer pair 5′-ACTATAGGGCACGCGTTGGT-3′ (adaptor primer 2) and 5′- AACGACCGAGCGCAGCGAGTCAGTG-3′ (PACv2-specific primer 2). Finally, the amplified DNA fragments were cloned into the pMD20-T plasmid (TAKARA), and their sequences were determined.
Furthermore, to identify the mutation of the dhfr-ts gene of P. chabaudi in the selected parasites, the DNA fragment including its ORF was amplified using the genomic DNA isolated from clonal drug-resistant parasites carrying PACv2 as a template with the PACv2-specific primer 2 and a P. chabaudi dhfr-ts gene-specific primer (5′-ATATGCACACAATATACCCAATTTTACG-3′). The DNA sequence of the amplified PCR product was determined as described above.
Data access
All materials, including the PAC and the PAC libraries, used in this study are available upon request from the corresponding author (S.I.) with a material transfer agreement.
Acknowledgments
This work was supported in part by Grants-In-Aid (grant nos. 09009279 and 09001016 to S.I. and grant no. 08090906 to M.Y.); Special Coordination Funds for Promoting Science and Technology (grant no. 10100609 to M.Y.); the NEXT program (grant no. LS057 to S.I.) from the Ministry of Education, Science, Culture, and Sports of Japan; and the Strategic International Cooperative Program of the Japan Science and Technology Agency (grant no. 10102762 to S.I.).
Authors' contributions. S.I. constructed genomic libraries using the PAC and analyzed the resultant parasites after the drug screening. I.K. performed all transfection experiments. S.I. and M.Y. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.124164.111.
References
- Bowman S, Lawson D, Basham D, Brown D, Chillingworth T, Churcher CM, Craig A, Davies RM, Devlin K, Feltwell T, et al. 1999. The complete nucleotide sequence of chromosome 3 of Plasmodium falciparum. Nature 400: 532–538 [DOI] [PubMed] [Google Scholar]
- Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, et al. 2008. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455: 757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culleton R, Martinelli A, Hunt P, Carter R 2005. Linkage group selection: Rapid gene discovery in malaria parasites. Genome Res 15: 92–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch K, Driskill C, Wellems T 2001. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res 29: 850–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharia NV, Bright AT, Westenberger SJ, Barnes SW, Batalov S, Kuhen K, Borboa R, Federe GC, McClean CM, Vinetz JM, et al. 2010. Whole-genome sequencing and microarray analysis of ex vivo Plasmodium vivax reveal selective pressure on putative drug resistance genes. Proc Natl Acad Sci 107: 20045–20050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Dicko A, Su XZ, Nomura T, Fidock DA, et al. 2001a. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med 344: 257–263 [DOI] [PubMed] [Google Scholar]
- Djimde A, Doumbo OK, Steketee RW, Plowe CV 2001b. Application of a molecular marker for surveillance of chloroquine-resistant falciparum malaria. Lancet 358: 890–891 [DOI] [PubMed] [Google Scholar]
- Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, et al. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361: 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Tettelin H, Carucci DJ, Cummings LM, Aravind L, Koonin EV, Shallom S, Mason T, Yu K, Fujii C, et al. 1998. Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum. Science 282: 1126–1132 [DOI] [PubMed] [Google Scholar]
- Hall N, Pain A, Berriman M, Churcher C, Harris B, Harris D, Mungall K, Bowman S, Atkin R, Baker S, et al. 2002. Sequence of Plasmodium falciparum chromosomes 1, 3–9 and 13. Nature 419: 527–531 [DOI] [PubMed] [Google Scholar]
- Hayton K, Ranford-Cartwright LC, Walliker D 2002. Sulfadoxine-pyrimethamine resistance in the rodent malaria parasite Plasmodium chabaudi. Antimicrob Agents Chemother 46: 2482–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt P, Martinelli A, Modrzynska K, Borges S, Creasey A, Rodrigues L, Beraldi D, Loewe L, Fawcett R, Kumar S, et al. 2010. Experimental evolution, genetic analysis and genome re-sequencing reveal the mutation conferring artemisinin resistance in an isogenic lineage of malaria parasites. BMC Genomics 11: 499 doi: 10.1186/1471-2164-11-499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H 1998. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol 16: 431–439 [DOI] [PubMed] [Google Scholar]
- Iwanaga S, Khan SM, Kaneko I, Christodoulou Z, Newbold C, Yuda M, Janse CJ, Waters AP 2010. Functional identification of the Plasmodium centromere and generation of a Plasmodium artificial chromosome. Cell Host Microbe 7: 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse CJ, Carlton JM, Walliker D, Waters AP 1994. Conserved location of genes on polymorphic chromosomes of four species of malaria parasites. Mol Biochem Parasitol 68: 285–296 [DOI] [PubMed] [Google Scholar]
- Murray AW, Szostak JW 1983. Construction of artificial chromosomes in yeast. Nature 305: 189–193 [DOI] [PubMed] [Google Scholar]
- Rottmann M, McNamara C, Yeung BK, Lee MC, Zou B, Russell B, Seitz P, Plouffe DM, Dharia NV, Tan J, et al. 2011. Spiroindolones, a potent compound class for the treatment of malaria. Science 329: 1175–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirawaraporn W, Sathitkul T, Sirawaraporn R, Yuthavong Y, Santi DV 1997. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc Natl Acad Sci 94: 1124–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Kirkman LA, Fujioka H, Wellems TE 1997. Complex polymorphisms in an approximately 330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell 91: 593–603 [DOI] [PubMed] [Google Scholar]
- Valderramos SG, Valderramos JC, Musset L, Purcell LA, Mercereau-Puijalon O, Legrand E, Fidock DA 2010. Identification of a mutant PfCRT-mediated chloroquine tolerance phenotype in Plasmodium falciparum. PLoS Pathog 6: e1000887 doi: 10.1371/journal.ppat.1000887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AP, Thomas AW, van Dijk MR, Janse CJ 1997. Transfection of malaria parasites. Methods 13: 134–147 [DOI] [PubMed] [Google Scholar]
- Wellems TE, Walker-Jonah A, Panton LJ 1991. Genetic mapping of the chloroquine-resistance locus on Plasmodium falciparum chromosome 7. Proc Natl Acad Sci 88: 3382–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR 2002. Epidemiology of drug-resistant malaria. Lancet Infect Dis 2: 209–218 [DOI] [PubMed] [Google Scholar]
- World Health Organization. 2010. Global report on antimalarial drug efficacy and drug resistance: 2000–2010. World Health Organization, Geneva.