The Drosophila midblastula transition (MBT) is accompanied by a dramatic change in replication timing, when replication lengthens nearly fourfold. Duronio discusses the implications of recent work (Farrell et al. in the April 1, 2012, issue) identifying Cdk1 down-regulation as a key trigger in replication control and S-phase lengthening during development.
Keywords: Drosophila, midblastula transition, Cdc25, Cdk1, DNA replication, late replication
Abstract
The duration of S phase in early embryos is often short, and then increases as development proceeds because of the appearance of late-replicating regions of the genome. In the April 1, 2012, issue of Genes & Development, Farrell and colleagues (pp. 714–725) demonstrate that the down-regulation of cyclin-dependent kinase 1 (Cdk1) activity triggers the onset of late-replicating DNA and an increase in S-phase length in Drosophila embryos, revealing an unexpected role for Cdk1 in replication control during development.
Developmental biologists are generally interested in how cells acquire the diversity of shape, behavior, and function needed to produce the vast array of different organismal forms observed in nature. This cellular diversity applies to the cell cycle, for which evolution has crafted regulatory systems capable of supporting a high degree of plasticity during development. A prime example is early embryogenesis, which often begins with rapid cleavage cycles consisting of S phase and mitosis but lacking G1 and G2 phases (Foe and Alberts 1983; Etkin 1988). Cleavage cycles occur in many organisms used for the study of development and that span several phyla, including chordates (e.g., frogs), echinoderms (e.g., sea urchins), and arthropods (e.g., fruit flies). Cleavage cycles result in the production of many cells in a short period of time, a likely adaptation for rapid embryonic development. A key feature of cleavage cycles is an exceedingly short S phase, which in some organisms can be completed in a matter of minutes rather than hours (Blumenthal et al. 1974). However, such speed comes with a cost because it precludes other events necessary for development. For instance, cleavage cycles may not be compatible with the dramatic cellular movements and shape changes needed for the formation of complex tissues (Mata et al. 2000; Seher and Leptin 2000). In addition, for most cells, the maintenance of genome stability involves developmental and physiological inputs that occur during G1 and G2 phases and regulate the transition into S phase and mitosis, respectively (Medema and Macurek 2011). Thus, new regulatory capabilities arise when new cell cycle controls (e.g., G1/S regulation) appear as development proceeds. Determining mechanisms for the appearance of new cell cycle controls during development should impact our understanding of basic cell biology and perhaps also cancer, since the progression to neoplasia could be considered (albeit in an oversimplified way) as the step-by-step loss of cell cycle controls that were sequentially acquired during early development.
S phase is under developmental control
The first big changes in cell cycle control are observed at the midblastula transition (MBT), a dramatic demarcation during embryogenesis associated with the switch from maternal to zygotic control of gene expression as well as the onset of the cellular movements of gastrulation and the beginning of morphogenesis (Edgar and Schubiger 1986; Etkin 1988). An interesting feature of cell cycle changes at the MBT is an increase in the length of S phase. Changes in S-phase regulation are a common phenomenon during development, and understanding the mechanisms should provide insight into fundamental controls of DNA replication (Nordman and Orr-Weaver 2012). How might changes in the duration of S phase arise? To address this question, we have to consider some basic principles of replication. DNA synthesis initiates at sites in chromosomes called origins of replication, which “fire” during S phase, resulting in bidirectional replication forks (Fig. 1). Direct observations of S-phase chromosomes in Drosophila embryos indicated that the regions of DNA replicated from a single origin (called replicons) increase in size as the length of S phase increases (McKnight and Miller 1977). This makes intuitive sense: One way to make S phase short is to have small replicons by triggering the firing of a large number of origins. Moreover, if many origins are fired concurrently, rather than sequentially, S phase will be shorter. Reducing the total number of origins that fire or making origin firing asynchronous will increase the length of S phase. Because the rate of replication fork movement in S phases of different durations is constant (Walter and Newport 1997), the increase in replicon size associated with longer S phase must be due to origin control—i.e., the length of S phase increases because fewer origins fire and/or the timing of origin firing is asynchronous during S phase, resulting in “early-firing” and “late-firing” origins.
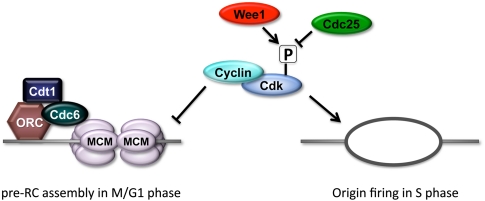
Figure 1.
Cdks are regulated by tyrosine phosphorylation and control pre-RC assembly and firing. Cdk activity plays opposing roles in controlling DNA replication at different times in the cell cycle, both inhibiting pre-RC assembly and triggering origin firing, leading to bidirectional replication fork movement. Cdk1 activity requires binding to cyclin and is regulated by the opposing activities of Cdc25 phosphatase and Wee1 kinase, which phosphorylates a tyrosine in the Cdk1 active site that inhibits ATP binding.
What mechanisms regulate the dynamics of origin firing during S phase, and how might developmental processes influence these mechanisms? For these questions, we need to consider some of the fundamental molecular features of DNA replication. A group of proteins composed of the six-subunit origin recognition complex (ORC), Cdt1, and Cdc6 assembles at origins of replication and functions to load onto the DNA the core components of the replicative helicase, consisting of hexamers of MCM proteins. The resulting prereplicative (pre-RC) complex marks the origin as “licensed” to be fired and initiate DNA synthesis (Fig. 1). Origin licensing and firing is complex and involves the recruitment and activity of many additional factors, and I refer the reader to a recent review for details (Diffley 2011). The observation that origin activity requires the recruitment of specific factors leads to the possibility that S-phase length increases during development because pre-RC factors become limiting as nuclei and DNA exponentially increase in abundance (note that early embryos lack zygotic transcription and thus do not increase the pool of available pre-RC factors). In this case, there would be fewer licensed origins and a resulting increase in replicon size and S-phase length. There is some evidence that limiting amounts of replication factors controls replication timing in budding yeast (Mantiero et al. 2011). However, in Drosophila, pre-RC factors are provided to the egg in abundance by maternal loading (Gossen et al. 1995), and measurements of replication in frog egg extracts indicated that the increase in replicon size at the MBT is not due to limiting amounts of pre-RC factors (Walter and Newport 1997).
So what else could be responsible? This is where Farrell et al. (2012) make a novel contribution by examining replication control in early Drosophila embryos, where the cell cycle program is known in great detail, and the length of S phase in each cycle can be measured down to the minute. The first 13 cycles in a Drosophila embryo are nuclear cleavage cycles that occur in a syncytium. S phase in these cycles is as short as 3–4 min (Blumenthal et al. 1974). The MBT occurs in cycle 14 and is accompanied by an increase in the duration of S phase to 40 min. This increase in S-phase length results from the appearance of late-replicating repetitive satellite DNA, as nicely demonstrated by another recent O'Farrell laboratory study (Shermoen et al. 2010). Because late-replicating DNA tends to be heterochromatic, and chromatin structure can influence origin firing (Eaton et al. 2011), one possibility is that changes in the chromatin landscape during development result in differential timing of origin firing during S phase and a consequent change in S-phase length (Gilbert et al. 2010). However, several features of heterochromatin appear prior to the onset of late-replicating DNA and the increase in S-phase length (Shermoen et al. 2010), suggesting that chromatin structure might not underlie the change. The investigators therefore turned their attention to cyclin-dependent kinase (Cdk) activity.
Cdk1 regulation underlies developmental changes in S phase
A fundamental and universal principle of cell cycle regulation is that Cdks control origins of replication in two opposing ways: Cdk activity is required to fire licensed origins during S phase and also inhibits pre-RC assembly, thus ensuring that origins do not fire more than once in a given S phase (Fig. 1A; Diffley 2011). Elimination of Cdk activity during mitosis creates a window of opportunity for new pre-RC assembly prior to the next S phase. In general, Cdk activity is constitutively high during the nuclear cleavage cycles of the early Drosophila embryo and then becomes regulated in subsequent cycles, providing the basis for the acquisition of new cell cycle controls as development progresses. The first Cdk that is developmentally down-regulated in Drosophila embryos is the essential mitotic Cdk (Cdk1), and this creates the appearance of G2 phase during cycle 14. There are several mechanisms that contribute to Cdk1 down-regulation, but the key for our discussion is the elimination of maternal Cdc25 phosphatase (encoded by the string and twine genes in Drosophila), which is responsible for removing an inhibitory phosphate on Y15 of Cdk1 that was placed there by Wee1 kinase (Fig. 1; Edgar and Datar 1996; Stumpff et al. 2004). When maternal Cdc25 mRNA and protein are destroyed just prior to cycle 14, mitotic Cyclin/Cdk1 complexes accumulate in a Y15-phosphorylated, inactive form. At subsequent stages of development, this pool of Cdk1 kinase is rapidly activated by zygotic transcription of string in G2 of cycle 14, driving cells into mitosis (Edgar and O'Farrell 1990). In contrast, the activity of Cyclin E/Cdk2, the essential S-phase kinase in Drosophila (Knoblich et al. 1994), is constitutively active during early embryonic development, including during G2 phase (Sauer et al. 1995; White et al. 2007). This is why embryonic cycles 1–16 lack a G1 phase, and only upon down-regulation of Cyclin E/Cdk2 does G1 phase first appear in cycle 17 (Knoblich et al. 1994).
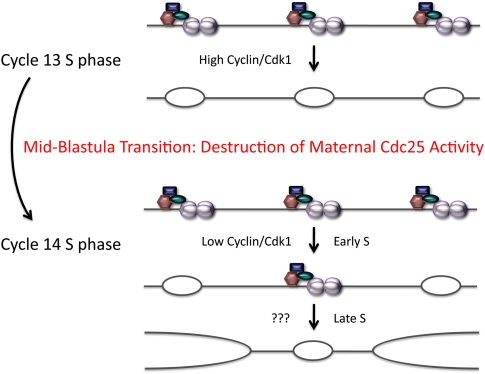
Because of these features of Cdk regulation in Drosophila embryos, the investigators hypothesized that the down-regulation of Cdk1 resulting from the elimination of maternal Cdc25 phosphatase activity is responsible for the appearance of late-replicating DNA and thus the increase in S-phase length during cycle 14. It has been known for some time that precocious activation of string expression after the establishment of G2 in cycle 14 triggers precocious entry into mitosis 14 (Edgar and O'Farrell 1990; Mata et al. 2000; Seher and Leptin 2000), but would the failure to eliminate maternal Cdk1 activity affect other aspects of the cell cycle that affect interphase length; namely, DNA replication? To test this idea, the investigators used time-lapse spinning-disc confocal microscopy of live embryos to examine the effects on S-phase length when Cdc25 was ectopically expressed by microinjection of mRNA into cycle 13 or early cycle 14, effectively preventing the developmentally controlled down-regulation of Cdc25. The investigators demonstrated that these treatments reduce the length of S-phase 14 because late-replicating DNA, which is already specified as late-replicating DNA by early cycle 14, now replicates early, leading to an early 14th cell division. This manipulation effectively converts cycle 14 into another cleavage cycle. Similar results were obtained by injecting active forms of Cyclin B/Cdk1 kinase. Cyclin E/Cdk2 injection did not shift the cycle 14 S-phase pattern to one more similar to the earlier syncytial cleavage cycles, consistent with the constitutively high level of activity of this kinase in early embryos. The somewhat surprising conclusion is that the onset of late-replicating DNA and the consequent increase in S-phase duration result from developmental regulation of the activity of the mitotic kinase Cdk1 (Fig. 2).
Figure 2.
Down-regulation of the mitotic kinase activator Cdc25 triggers the onset of late origin firing during S phase of early Drosophila development. The MBT in Drosophila embryos occurs between embryonic cycles 13 and 14 and is associated with the destruction of maternal Cdc25 mRNA and protein. Because of constitutive Wee1 kinase activity, the down-regulation of maternal Cdc25 leads to the accumulation of inactive Cyclin/Cdk1 complexes, the onset of late-replicating heterochromatin, and the lengthening of S phase in cycle 14. How Cdk1 triggers origin firing in pre-MBT cycles (e.g., cycle 13) and how late origins are specified and fired in cycle 14 (???) are unknown.
These mRNA injection experiments are gain-of-function experiments and indicate that down-regulation of Cdk1 is required for the onset of a temporal pattern of replication within S phase and the consequent increase in the length of S phase in cycle 14. However, it does not necessarily follow from these experiments that the converse is true: that continuously high Cdk1 activity in the earlier S/M cleavage cycles keeps S phase short. Importantly, the investigators also tested this idea by reducing Cdk1 activity through RNAi depletion of mitotic cyclins, which resulted in a modest extension of S phase in cycle 13, although not as dramatic as what normally occurs in cycle 14. This result suggests that either mechanisms other than high Cdk1 activity contribute to short S phase prior to cycle 14 or Cdk1 activity was not fully depleted in these experiments (or both). Finally, as an interesting additional observation, the investigators provided evidence that a low cyclin level in early cycle 14 functions as a novel checkpoint mechanism that limits the ability of Cdk1 to induce mitosis during ongoing DNA synthesis. Together, these results suggest different thresholds for Cdk1 function: Low Cdk1 activity converts late-firing origins to early-firing origins but cannot stimulate mitosis, and high Cdk1 activity triggers mitosis.
While the observation that Cdk1 regulation underlies the changing replication landscape in early Drosophila development is quite interesting, in retrospect, there are precedents in the cell cycle field. Genetic manipulation of cyclins and Cdks in many organisms from yeast to mammalian cells has revealed a tremendous capability of different Cdks to individually support all cell cycle events. The seminal studies of cell cycle control in budding and fission yeast showed early on that a single Cdk drives both S phase and mitosis in these organisms (albeit in partnership with different cyclins) (Nurse 2002). Moreover, human Cdk1 is capable of substituting for the mitotic and S-phase functions of Schizosaccharomyces pombe Cdk1 (Lee and Nurse 1987). Mouse knockout experiments also clearly indicate that Cdk1 can support S phase (Aleem et al. 2005). Thus, while different Cdks can substitute for one another, multiple Cdks may have arisen to perform different jobs within S phase (e.g., temporal control of origin firing) that likely fine-tune the cell cycle in important ways (Satyanarayana and Kaldis 2009). The early development of organisms like Drosophila has evolved to take advantage of this plasticity to shape alterations to cell cycle progression.
What next?
The Farrell et al. (2012) experiments raise a number of interesting questions. What ensures the late replication of heterochromatin in cycle 14? The most likely answer is late-firing origins located in or very near heterochromatin. Cyclin E/Cdk2 is highly active throughout S-phase 14 and stimulates origin firing early in S phase. Thus, any late-firing origins must be refractory to Cyclin E/Cdk2 in cycle 14, or something else triggers their firing. While high Cdk1 activity can trigger early replication of heterochromatin prior to cycle 14, in cycle 14, something else must perform this function, since Cdk1 becomes inhibited by Y15 phosphorylation, ultimately causing the appearance of G2. Does Cdk1 regulation affect replication timing in other cycles or at other stages of development? Would precocious activation shorten S phase in other cell types? If not, then perhaps other mechanisms exist that control replication timing, and these may be conserved in other organisms.
Does the acquisition of late-replicating DNA play a specific role during development? As noted above, adding controls to the cell cycle creates opportunity for new regulation that might facilitate developmental events. For instance, the acquisition of G2 regulation may accommodate the dramatic cell movements that occur during gastrulation because these movements require cytoskeletal rearrangements and cell shape changes that are precluded by the events of mitosis and cytokinesis. Similarly, the acquisition of G1 phase provides an opportunity to introduce growth control at later stages of development. But why lengthen interphase by extending the duration of S phase with the introduction of late-replicating regions of the genome? The phenomenon of early- and late-firing origins leading to a temporal program of DNA replication during S phase is common in eukaryotic cells, suggesting an important functional role. Indeed, forcing late origins to fire early in budding yeast causes growth defects (Mantiero et al. 2011). One possibility is that changes in the replication program help support the formation of specific aspects of chromatin structure that affects gene expression programs. While correlations between replication timing, chromatin organization within the nucleus, and gene expression certainly exist and can be observed in animal cells, the functional consequences of these complex relationships can be difficult to discern (Gondor and Ohlsson 2009; Hiratani et al. 2009; Gilbert et al. 2010).
Finally, what is the molecular mechanism by which Cdk1 controls origin firing? A recent report suggested that cyclin A controls early/late replication dynamics in S phase of Drosophila mechanosensory bristle cells via regulating heterochromatic localization of Orc2 (Salle et al. 2012). Cyclin A is also required for the rereplication of heterochromatin that occurs upon depletion of geminin, an inhibitor of Cdt1 (Ding and MacAlpine 2010). These are likely Cdk1 effects, since Cyclin A does not bind Cdk2 in flies (Knoblich et al. 1994). In addition, reduced levels of Orc binding correlate with underreplicated regions of the genome in polyploid salivary gland cells, suggesting that origin firing can be controlled developmentally (Sher et al. 2012). Cytological experiments performed in fly tissues cannot easily discern the precise molecular mechanism by which Cdk1 controls origin firing. Nevertheless, studies like Farrell et al. (2012) provide a wonderful example of the power of curiosity-driven research and how it results in unanticipated discovery that sets the stage for new, hypothesis-driven inquiries. Moreover, I would like to stress that “mechanism” is in the eye of the beholder: While the molecular mechanism by which Cdk1 kinase activity triggers replication of heterochromatic DNA in early S phase remains unknown at present, the Farrell et al. (2012) study established that the developmental mechanism causing the increase in the length of S phase is the down-regulation of Cdk1 activity via the elimination of maternal Cdc25 phosphatase. My distinction of mechanism here is meant to highlight the value of exploring S-phase control using a wide array of experimental approaches in different organisms and biological contexts.
Acknowledgments
I thank Jean Cook, Lindsay Rizzardi, and Joy Meserve for comments and literature suggestions, and Jean Cook for allowing me to use her drawings in the figures. The cell cycle work in my laboratory is funded by GM57859 from the NIH.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.191171.112.
References
- Aleem E, Kiyokawa H, Kaldis P 2005. Cdc2–cyclin E complexes regulate the G1/S phase transition. Nat Cell Biol 7: 831–836 [DOI] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS 1974. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol 38: 205–223 [DOI] [PubMed] [Google Scholar]
- Diffley JF 2011. Quality control in the initiation of eukaryotic DNA replication. Philos Trans R Soc Lond B Biol Sci 366: 3545–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, MacAlpine DM 2010. Preferential re-replication of Drosophila heterochromatin in the absence of geminin. PLoS Genet 6: e1001112. doi: 10.1371/journal.pgen.1001112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM 2011. Chromatin signatures of the Drosophila replication program. Genome Res 21: 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Datar SA 1996. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila's early cell cycle program. Genes Dev 10: 1966–1977 [DOI] [PubMed] [Google Scholar]
- Edgar BA, O'Farrell PH 1990. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell 62: 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Schubiger G 1986. Parameters controlling transcriptional activation during early Drosophila development. Cell 44: 871–877 [DOI] [PubMed] [Google Scholar]
- Etkin LD 1988. Regulation of the mid-blastula transition in amphibians. Dev Biol 5: 209–225 [DOI] [PubMed] [Google Scholar]
- Farrell JA, Shermoen AW, Yuan K, O'Farrell PH 2012. Embryonic onset of late replication requires Cdc25 down-regulation. Genes Dev 26: 714–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE, Alberts BM 1983. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci 61: 31–70 [DOI] [PubMed] [Google Scholar]
- Gilbert DM, Takebayashi SI, Ryba T, Lu J, Pope BD, Wilson KA, Hiratani I 2010. Space and time in the nucleus: Developmental control of replication timing and chromosome architecture. Cold Spring Harb Symp Quant Biol 75: 143–153 [DOI] [PubMed] [Google Scholar]
- Gondor A, Ohlsson R 2009. Replication timing and epigenetic reprogramming of gene expression: A two-way relationship? Nat Rev Genet 10: 269–276 [DOI] [PubMed] [Google Scholar]
- Gossen M, Pak DT, Hansen SK, Acharya JK, Botchan MR 1995. A Drosophila homolog of the yeast origin recognition complex. Science 270: 1674–1677 [DOI] [PubMed] [Google Scholar]
- Hiratani I, Takebayashi S, Lu J, Gilbert DM 2009. Replication timing and transcriptional control: Beyond cause and effect–part II. Curr Opin Genet Dev 19: 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA, Sauer K, Jones L, Richardson H, Saint R, Lehner CF 1994. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell 77: 107–120 [DOI] [PubMed] [Google Scholar]
- Lee MG, Nurse P 1987. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature 327: 31–35 [DOI] [PubMed] [Google Scholar]
- Mantiero D, Mackenzie A, Donaldson A, Zegerman P 2011. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J 30: 4805–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Curado S, Ephrussi A, Rorth P 2000. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell 101: 511–522 [DOI] [PubMed] [Google Scholar]
- McKnight SL, Miller OL Jr 1977. Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell 12: 795–804 [DOI] [PubMed] [Google Scholar]
- Medema RH, Macurek L 2011. Checkpoint control and cancer. Oncogene. doi: 10.1038/onc.2011.451 [DOI] [PubMed] [Google Scholar]
- Nordman J, Orr-Weaver TL 2012. Regulation of DNA replication during development. Development 139: 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P 2002. Cyclin dependent kinases and cell cycle control (Nobel lecture). ChemBioChem 3: 596–603 [DOI] [PubMed] [Google Scholar]
- Salle J, Campbell SD, Gho M, Audibert A 2012. CycA is involved in the control of endoreplication dynamics in the Drosophila bristle lineage. Development 139: 547–557 [DOI] [PubMed] [Google Scholar]
- Satyanarayana A, Kaldis P 2009. Mammalian cell-cycle regulation: Several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 28: 2925–2939 [DOI] [PubMed] [Google Scholar]
- Sauer K, Knoblich JA, Richardson H, Lehner CF 1995. Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes Dev 9: 1327–1339 [DOI] [PubMed] [Google Scholar]
- Seher TC, Leptin M 2000. Tribbles, a cell-cycle brake that coordinates proliferation and morphogenesis during Drosophila gastrulation. Curr Biol 10: 623–629 [DOI] [PubMed] [Google Scholar]
- Sher N, Bell GW, Li S, Nordman J, Eng T, Eaton ML, Macalpine DM, Orr-Weaver TL 2012. Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res 22: 64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen AW, McCleland ML, O'Farrell PH 2010. Developmental control of late replication and S phase length. Curr Biol 20: 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Duncan T, Homola E, Campbell SD, Su TT 2004. Drosophila Wee1 kinase regulates Cdk1 and mitotic entry during embryogenesis. Curr Biol 14: 2143–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport JW 1997. Regulation of replicon size in Xenopus egg extracts. Science 275: 993–995 [DOI] [PubMed] [Google Scholar]
- White AE, Leslie ME, Calvi BR, Marzluff WF, Duronio RJ 2007. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol Biol Cell 18: 2491–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]