Glioblastoma is both the most common and lethal primary malignant brain tumor. Extensive multiplatform genomic characterization has provided a higher-resolution picture of the molecular alterations underlying this disease. These comprehensive data sets give a clearer picture of glioblastoma as a heterogeneous collection of distinct diseases. In this review by Hahn and colleagues, the recent efforts to classify glioblastoma based on these new data and novel insights into the molecular pathways regulating glioblastoma are discussed.
Keywords: glioblastoma, invasion, angiogenesis, tumor-initiating cells, IDH1, glioma classification
Abstract
Glioblastoma is both the most common and lethal primary malignant brain tumor. Extensive multiplatform genomic characterization has provided a higher-resolution picture of the molecular alterations underlying this disease. These studies provide the emerging view that “glioblastoma” represents several histologically similar yet molecularly heterogeneous diseases, which influences taxonomic classification systems, prognosis, and therapeutic decisions.
In his effort in the 1920s to cure two comatose patients with extensive glioblastomas, neurosurgeon Walter Dandy took the radical step of removing the entire affected hemisphere in each of these patients. Despite these heroic interventions, both of these patients succumbed to their disease due to involvement of the contralateral hemisphere (Dandy 1928). More than 80 years later, we continue to face the same clinical challenges illustrated by this vignette from the early 20th Century. Glioblastoma represents the most common primary intrinsic malignant brain tumor diagnosed each year in the United States; there are ∼10,000 new diagnoses annually, and >50,000 patients are currently living with the disease (Davis et al. 2001; Porter et al. 2010; CBTRUS 2011). The clinical hallmarks of glioblastoma are its aggressive growth and inexorable recurrence despite multimodal therapy with surgery followed by radiation and temozolomide therapy. Unfortunately, current standard-of-care therapy results in a median survival of only 12–15 mo (Stupp et al. 2005).
The emergence of a molecularly focused approach to cancers represents a profound shift in our approach to the diagnosis and treatment of malignancy. This framework centers on a new taxonomy that describes cancers by their molecular alterations and the identification of inhibitors that target these cancer-specific changes. Indeed, therapies targeting HER2-amplified breast cancer (Slamon et al. 2001), CML harboring the BCR–ABL translocation (Druker et al. 2001), mutant EGFR lung cancer (Lynch et al. 2004), lung cancer harboring the EML4–ALK translocation (Kwak et al. 2010), and BRAF mutant melanoma (Chapman et al. 2011) provide clear proof of principle for this approach. Histopathologic diagnosis is increasingly being supplemented with annotation of amplification or mutational status where relevant (MacConaill et al. 2009; Dias-Santagata et al. 2010), and these data subsequently inform therapeutic decision-making, all in a time frame from target discovery to therapy that is becoming progressively shorter (Chabner 2011). However, despite clinical progress across many cancer types and extensive characterization of genomic alterations in glioblastoma, we still have not identified and exploited clinically meaningful tumor dependencies in this dreaded disease.
The recent characterization of the genome (Beroukhim et al. 2007; The Cancer Genome Atlas Research Network 2008; Parsons et al. 2008) and transcriptome (Phillips et al. 2006; Verhaak et al. 2010) of glioblastoma provides a high-resolution picture of the glioblastoma landscape that has revealed the major structural and expression alterations that may drive disease pathogenesis and biology. These comprehensive data sets reveal “glioblastoma” as a heterogeneous collection of distinct diseases with multiple dependencies both within and across each particular subtype. Here we summarize recent efforts to catalog the structural genomic landscape of glioblastoma and focus on emerging insights into critical molecular pathways central to glioblastoma pathobiology.
Current histopathological classification
As described by the World Health Organization (WHO) classification (Louis et al. 2007), malignant diffuse gliomas are comprised of astrocytic, oligodengroglial, and mixed oligoastrocytic neoplasms based solely on morphology and are further subdivided by tumor grade based on additional histologic features present in the tumor. Nuclear atypia and mitotic activity are required criteria for grade III lesions, and the presence of necrosis or microvascular proliferation is required for the diagnosis of grade IV astrocytoma, glioblastoma (Miller and Perry 2007). While 90%–95% of glioblastomas arise de novo and are considered “primary,” ∼5%–10% arise from lower-grade gliomas in younger patients and are termed “secondary” (Biernat et al. 2004; Ohgaki and Kleihues 2005). Together with grade III anaplastic astrocytoma, which has an incidence of >1500 cases annually (CBTRUS 2011), these tumors comprise the clinical entity termed “malignant glioma.” However, as discussed below, emerging classification schemes are likely to incorporate molecular differences that distinguish these tumors (von Deimling et al. 2011).
Large-scale genomic studies of glial neoplasms
Glioblastoma, like other cancers, is the product of accumulated genetic and epigenetic alterations (Vogelstein and Kinzler 2004; Weir et al. 2004; Stratton et al. 2009), and the application of genome-scale approaches to enumerate these genetic alterations has uncovered both molecular subclasses and common pathways mutated in this disease. To date, glioblastoma has been subjected to the most extensive genomic profiling of any cancer, and thus we review here our current understanding of the critical underlying molecular pathology in this disease that has come from an integrated view of somatic copy number alterations (SCNAs) and sequence mutations and also review recent discoveries in lower-grade astrocytomas.
Integrated approaches to characterize the genomic landscape of glioblastoma
Malignant gliomas, like other human cancers, are characterized by genetic instability and complex alterations in chromosome structure and copy number. The SCNAs found in malignant gliomas include broad or regional alterations spanning segments or whole arms of entire chromosomes as well as focal events involving one or a few genes. The development of high-resolution, array-based comparative genomic hybridization (CGH) using bacterial artificial chromosomes, oligomers, and single-nucleotide polymorphism (SNP) arrays made possible the systematic analysis of cancer genomes and defined many new recurrent SCNAs in cancer (Chin et al. 2011). To fully explore these data, several groups have also developed bioinformatic tools to highlight the cancer-derived genomic alteration signals from among the background noise (Beroukhim et al. 2007; Greenman et al. 2007; Wiedemeyer et al. 2008; Riddick and Fine 2011). These methodologies, including Genomic Identification of Significant Targets in Cancer (GISTIC) (Beroukhim et al. 2007; Mermel et al. 2011) and Genomic Topography Scan (GTS) (Wiedemeyer et al. 2008), were first deployed in glioblastoma and subsequently across thousands of cancer samples (Beroukhim et al. 2010; http://www.broadinstitute.org/tumorscape). Combined work using these tools and others (Bredel et al. 2005; Kotliarov et al. 2006) identified overlapping presumed targets of amplification (including EGFR, MET, PDGFRA, MDM4, MDM2, CCND2, PIK3CA, MYC, CDK4, and CDK6) and deletion (including CDKN2A/B, CDKN2C, PTEN, and RB1) in glioblastoma.
This work in SCNAs preceded two recent studies that further clarified our understanding of the glioblastoma genomic landscape (The Cancer Genome Atlas Research Network 2008; Parsons et al. 2008). The Cancer Genome Atlas (TCGA) pilot project, the first of many ongoing comprehensive TCGA consortium-based cancer genome analyses (http://cancergenome.nih.gov), applied multiplatform profiling to systematically and comprehensively define the genomic landscape of glioblastoma. This approach used targeted Sanger sequencing to investigate 601 genes in 91 samples as well as array-based platforms to analyze copy number, mRNA expression, and the epigenetic state of ∼200 tumors, most of which were untreated, primary glioblastomas. This larger effort confirmed prior SCNA analyses on smaller numbers of tumors and also identified novel lesions, such as homozygous deletions of PARK2 and amplification of AKT3. Moreover, separate work also showed that NF-κB inhibitor α (NFKBIA) is heterozygously deleted in a mutually exclusive manner to EGFR amplification (Bredel et al. 2011).
The most significantly somatically mutated genes (false discovery rate <0.1) among the 601 genes analyzed were TP53 (42%), PTEN (33%), neurofibromatosis-1 (NF1) (21%), EGFR (18%), RB1 (11%), PIK3R1 (10%), and PIK3CA (7%). However, this effort demonstrated that the integration of multiple types of genomic analysis permitted the projection of identified alterations onto known pathways (Fig. 1). This approach revealed the high incidence of p53, Rb, and receptor tyrosine kinase (RTK)/Ras/phosphoinositide 3-kinase (PI3K) pathway dysregulation, confirming previous work that had delineated lesions in these critical cascades (Ekstrand et al. 1991; Henson et al. 1994; Louis 1994, 2006; Reifenberger et al. 1994; Schmidt et al. 1994; Ueki et al. 1996). Specifically, p53 signaling was impaired in 87% of the samples through CDKN2A deletion (49%), MDM2 (14%) and MDM4 (7%) amplification, and mutation and deletion of TP53 (35%). Likewise, Rb signaling was impaired in 78% of the samples through CDKN2 family deletion; amplification of CDK4 (18%), CDK6 (1%), and CCND2 (2%); and mutation or deletion of RB1 (11%). Additional work also showed that CDK6 is an oncogene, and particular patterns of CDKN2 loss can predict cellular dependency on CDK4 and CDK6 inhibition (Wiedemeyer et al. 2010). Finally, evidence of RTK/RAS/PI3K activation was found in 88% of tumors, including contributions from unexpected mutations or deletions in NF1 (18%) and PIK3R1, which encodes the p85α regulatory subunit of PIK3CA. Building on this pilot glioblastoma effort, TCGA is profiling primary glioblastoma samples with the goal of achieving complete genomic characterization of >500 tumors. As next-generation sequencing technologies (Meyerson et al. 2010) are incorporated into the TCGA pipeline, many of these glioblastoma specimens will undergo whole-exome, transcriptome, and/or whole-genome sequencing, providing higher-resolution detail of lower-frequency somatic alterations, including intrachromosomal and interchromosomal translocations. A subset of these data from nearly 500 tumors is available through the TCGA portal (http://tcga-data.nci.nih.gov/tcga).
Figure 1.
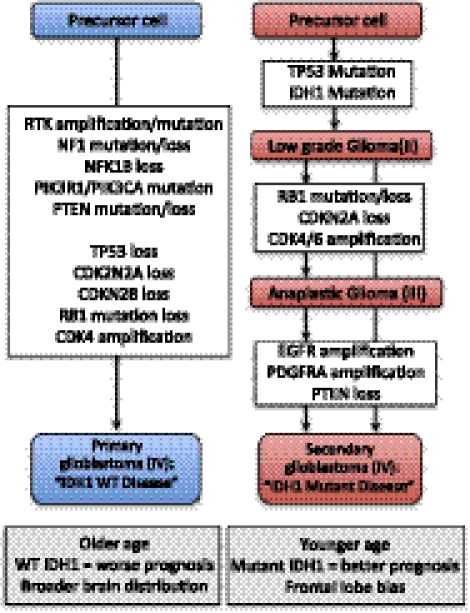
Genomic alterations underlying gliomagenesis. Both primary and secondary glioblastomas arise from precursor cells that may be distinct. Primary glioblastomas arise de novo and exhibit p53 and Rb pathway dysfunction as well as RTK/Ras/PI3K signaling dysregulation, leading to tumors that arise in older patients with a worse prognosis, likely owing to the predominant wild-type IDH1 genotype. In contrast, secondary glioblastomas are preceded by lower-grade II lesions, which progress either through grade III lesions or directly to glioblastoma. These tumors occur in younger patients and are dominated by a mutant IDH1 genotype that confers a better prognosis and is associated with a more restricted frontal lobe location.
In parallel to TCGA, the Vogelstein laboratory (Parsons et al. 2008) pursued a complementary strategy to characterize glioblastoma genomes composed of sequence, copy number, and expression analysis of a greater number of genes in fewer tumors—specifically, 23,219 transcripts from 20,661 protein-coding genes in 22 malignant gliomas were analyzed by Sanger-based sequencing. Strikingly, five out of 22 tumors, which included one “high-grade” glioma and one secondary glioblastoma, harbored recurrent R132H-encoded substitutions in the isocitrate dehydrogenase 1 (IDH1) gene. These findings were extended to 18 out of 149 malignant gliomas, which included mostly primary glioblastomas but also a proportion of secondary tumors. Three IDH isoforms exist in humans: IDH1 is a cytosolic protein, whereas IDH2 and IDH3 are located within the mitochondria. IDH1 and IDH2 are known to catalyze the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG), leading to the production of NADPH, in the tricarboxylic acid (TCA) cycle, a biochemical sequence critical in sugar, lipid, and amino acid metabolism (Raimundo et al. 2011). Although a missense mutation in IDH1, encoding IDH1 R132C, was first identified in one patient with colon cancer in a sequencing analysis of the coding regions in breast and colon cancer (Sjoblom et al. 2006), this study provided the first evidence of recurrent mutations in IDH1.
Subsequent work from several groups has provided a comprehensive picture of the IDH status in brain tumors (Balss et al. 2008; Hartmann et al. 2009; Ichimura et al. 2009; Yan et al. 2009). IDH1/2 is mutated in grade II and III gliomas as well the secondary glioblastomas that arise from prior low-grade tumors, with most mutations found in the IDH1 gene. Importantly, these mutations usually occur at conserved residues and are virtually never homozygous. Specifically, whereas only 3%–7% of primary glioblastomas harbor IDH1 mutations, the majority (50%–80%) of secondary glioblastomas express mutant IDH1. Furthermore, most lower-grade gliomas harbor IDH1 mutations; although grade I pilocytic astrocytomas usually express wild-type IDH1, ∼60%–80% of grade II and III astrocytomas, oligodendrogliomas, and oligoastrocytomas express mutant IDH1, with the R132H mutation representing the majority of mutations observed. In addition, ∼3% of these tumors that express wild-type IDH1 were found to express IDH2 R172 mutations (Balss et al. 2008; Hartmann et al. 2009; Ichimura et al. 2009; Yan et al. 2009), although this mutation in IDH2 has only been documented in a single glioblastoma in the literature (Hartmann et al. 2010). Other CNS tumors found to harbor IDH1 mutations include gangliogliomas, giant cell glioblastomas, and primitive neuroectodermal tumors, although small numbers of these tumors have been studied (Balss et al. 2008). Whereas mutations in other TCA cycle enzymes, such as fumarate hydratase in leiomyomas and renal cell cancer and succinate dehydrogenase in paragangliomas, have been identified (Kaelin 2011; Raimundo et al. 2011), mutations in these genes have not been found in gliomas.
IDH1/2 mutations have also been identified in 12%–17% of acute myeloid leukemias (AMLs) (Mardis et al. 2009; Paschka et al. 2010; Ward et al. 2010; Graubert and Mardis 2011), the majority of central and periosteal cartilaginous tumors (Amary et al. 2011a), and also 23% of cholangiocarcinomas (Borger et al. 2012). Interestingly, somatic mosaic IDH1/2 mutations were found to be the likely genetic basis of Ollier disease and Maffuci syndrome (Amary et al. 2011b; Pansuriya et al. 2011). These rare, nonfamilial conditions, both characterized by the early development of multiple cartilaginous tumors, have also been reported to manifest concomitant glioma or AML, thereby providing an intriguing demonstration of the likely causal role that mutant IDH1/2 plays in these three distinct tumor types (Rawlings et al. 1987). Finally, 50% of patients with D-2-hydroxyglutaric aciduria (D-2-HGA), a rare inherited neurometabolic disorder, have been found to carry IDH2 mutations (Kranendijk et al. 2010). Indeed, the discovery of IDH1/2 mutations is one of the major novel findings to emerge from genome annotation studies and has stimulated renewed attention to altered metabolism in cancer biology.
The genetic basis of oligodendrogliomas and pediatric gliomas
In addition to the frequency of IDH1 mutations in grade II glioma, cancer sequencing studies have provided new insights into the genetic basis of other lower-grade glial neoplasms. Specifically, >50% of oligodendrogliomas display loss of heterozygosity (LOH) at chromosomes 1p and 19q (Cairncross et al. 1998), although the targets of these deletions have remained elusive. However, Bettegowda et al. (2011) recently used next-generation sequencing to analyze the exomes of seven anaplastic oligodendrogliomas (WHO grade III) and found novel recurrent inactivating mutations affecting FUBP1 (far-upstream element [FUSE]-binding protein 1; five out of 34 tumors), a regulator of MYC signaling located on chromosome 1p, and the homolog of Drosopila capicua, CIC (18 out of 34 tumors), a downstream transcriptional repressor of RTK/MAPK signaling located on chromosome 19q. Yip et al. (2012) confirmed the high incidence of CIC mutation with concurrent 1p/19q loss and IDH1 mutation in their series.
Recent work has also identified a high incidence of specific mutations in two types of pediatric gliomas. First, several studies have revealed BRAF alterations in lower-grade pediatric tumors. Copy number analysis of WHO grade I pilocytic astroctyomas identified a tandem duplication at chromosome 7q34 resulting in a novel oncogenic BRAF fusion gene, KIAA1549:BRAF, in >60% of these tumors (Bar et al. 2008; Jones et al. 2008; Pfister et al. 2008). Together with other identified fusion events such as SRGAP3:RAF1 (Jones et al. 2009), RAF fusion events occur in >80% of pilocytic astrocytomas (von Deimling et al. 2011). Furthermore, BRAFV600E mutations have been found most commonly in WHO grade II pleomorphic xanthoastrocytomas (66%) (Dias-Santagata et al. 2011; Schindler et al. 2011) as well as WHO grade I gangliogliomas (18%) (MacConaill et al. 2009; Schindler et al. 2011). In addition, Wu et al. (2012) used whole-genome sequencing to identify recurrent mutations in H3FA, which encodes the H3.3 protein, and the closely related HIST1H3B gene, which encodes the H3.1 protein isoform, in pediatric diffuse pontine gliomas. Mutations in these two genes were found in 78% of these tumors, 22% of nonbrainstem pediatric glioblastomas, and virtually no other CNS tumors evaluated. Together, these findings have clear implications for taxonomic classification and, in the case of BRAF alterations, potential targeted therapies.
Transcriptional profiling: identification of subtypes and biological programs in malignant glioma
Classification
The genome-wide analysis of mRNA expression to identify molecular subclasses (Golub et al. 1999) has led to a fundamental shift in our understanding of glioblastoma subtypes. Indeed, the identification of multiple subtypes within glioblastoma has underscored the heterogeneity of diseases that all share the same WHO histopathological grade. This approach has revealed that the glioma transcriptome is highly structured and reflects tumor histology, molecular alterations, and clinical outcome (Nutt et al. 2003; Freije et al. 2004; Phillips et al. 2006; Verhaak et al. 2010; Brennan 2011; Huse et al. 2011).
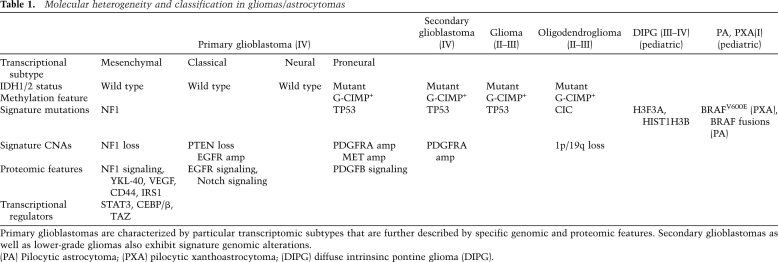
Although expression profiling of glioblastoma has been used by many groups (for review, see Brennan 2011; Huse et al. 2011), two studies have provided the foundation for classification of glioblastoma subtypes (Phillips et al. 2006; Verhaak et al. 2010). Using unsupervised hierarchical clustering analysis, Verhaak et al. (2010) classifed 200 TCGA glioblastoma samples into four subtypes (Table 1), which were subsequently validated using previously published data from 260 independent samples. Each of the four subtypes was ultimately defined by a minimum list of 210 genes (http://tcga-data.nci.nih.gov/docs/publications/gbm_exp). By incorporating the available copy number and sequence data, three of the four subtypes were found to harbor distinct molecular alterations. Specifically, the proneural subtype was enriched for amplifications of PDGFRA, CDK6, CDK4, and MET; 11 out of 12 IDH1 mutations found in the TCGA samples; PIK3CA/PIK3R1 mutations; and mutation or LOH of TP53. Of note, this subtype contained the highest percentage of young patients, likely due in part to the high number of IDH1 mutant tumors in this category. The classical subtype was enriched for amplification of EGFR and loss of PTEN and CDKN2A, whereas the mesenchymal subtype harbored mutations and/or loss of NF1, TP53, and CDKN2A. To date, no unique genetic alterations define the neural class from the other classes.
Table 1.
Molecular heterogeneity and classification in gliomas/astrocytomas
In contrast to Verhaak et al. (2010), Phillips et al. (2006) identified three distinct glioblastoma subtypes based on their differences of expression of a panel of genes most strongly correlated with survival. This approach delineated subtypes that were termed proneural, mesenchymal, and proliferative. Although the number of subtypes identified by the Verhaak et al. (2010) and Phillips et al. (2006) studies differs, the proneural and mesenchymal classifications identified using distinct methodologies and sample sets are the most robust and concordant (Huse et al. 2011); genes defining the Verhaak and Phillips subclasses are listed together in Supplemental Table S1B of Bhat et al. (2011,). For instance, both groups identified proneural class expression of DLL3 and OLIG2 and mesenchymal class expression of CD40 and CHI3L1/YKL-40, the latter of which appears to be a potential serum protein marker of prognosis in glioblastoma patients (Iwamoto et al. 2011). Moreover, a subset of the genes represented in these subtypes is represented in a nine-gene panel shown to predict outcome in glioblastoma, as increased expression of mesenchymal genes such as CHI3L1/YKL-40 and LGALS3 combined with decreased expression of a proneural gene, OLIG2, were associated with typical short-term survival compared with longer-term survivors (Colman et al. 2010). Considering that other groups have used alternative methodologies to derive nonoverlapping prognostically significant gene sets (Bredel et al. 2009), further work in large, relatively uniform sample sets will be necessary to resolve subtype classification systems and validate multiple prognostic gene sets.
To determine whether these genomically identified subtypes predict biological or clinical differences in glioblastoma, Brennan et al. (2009) used a targeted proteomics approach to determine whether glioblastomas also segregated into distinct classes by activation of signal transduction pathways. Unsupervised clustering of 57 proteins or protein modifications assessed in 20 glioblastoma samples identified distinct tumor subgroups defined by EGFR-related signaling, PDGF-related signaling, or proteins associated with decreased NF1 expression. Moreover, analysis of TCGA glioblastoma expression profiles showed that SCNAs or mutations of EGFR, PDGFRA, and NF1 were largely mutually exclusive, suggesting that the identified signaling nodes are nonoverlapping. Interestingly, because PDGF core tumors often expressed high PDGF protein but low PDGF mRNA and no PDGFRA SCNAs, it is possible that PDGF/PDGFRA-mediated biology is underestimated in purely mRNA-based expression schemes. Moreover, the finding that EGFR core tumors harbored evidence of active Notch signaling not reflected in copy number, expression, or sequence data was additional evidence that not all biologically important information can be gleaned from genomic data. This study supports the integration of protein-based biomarker assessment, perhaps by immunohistochemistry for a targeted biomarker panel, into the broader subtype classification architecture currently defined by genomic profiling.
Network analysis of expression profiles
Additional work has extended the utility of mRNA profiling by using computational network analysis to uncover the causal regulatory modules underlying particular transcriptomically defined subtypes. Broadly, these approaches attempt to infer the upstream biological programs, or networks, that coordinately produce a given transcriptional state. Recently, two groups used a reverse engineering-based algorithm called ARACNe (algorithm for the reconstruction of accurate cellular networks) (Basso et al. 2005) to identify transcription factors that regulate the mesenchymal expression state (Carro et al. 2010; Bhat et al. 2011). Using ARACNe and a novel master regulatory algorithm, Carro et al. (2010) identified six transcription factors—STAT3, C/EBPβ, RUNX1, FOSL, bHLH-B2, and ZNF-238—that are likely to control the majority of mesenchymal subtype tumors. Overexpression of STAT3 and C/EBPβ both induced a mesenchymal phenotype and were individually required for orthotopic xenograft growth. Bhat et al. (2011) also applied the ARACNe method to TCGA samples to identify transcription factors that regulated the mesenchymal subtype and identified several distinct components, including TAZ, YAP, MAFB, and HCLS1. Follow-up work demonstrated a pivotal involvement of TAZ and dysregulated Hippo pathway signaling in driving mesenchymal biology; TAZ tended to be hypermethylated in proneural tumors, could drive a mesenchymal phenotype when overexpressed, and cooperated with PDGF-B to induce malignant mesenchymal-type gliomas in the RCAS/N-tva mouse model. Together, these network analyses of gene expression data demonstrate the utility of these data sets not only to classify distinct tumor states, but also to provide clues to the underlying biology.
Signature molecular lesions in glioma: key drivers, tumor suppressors, and the IDH proteins
Combined work from genomic and proteomic analyses has focused attention on critical pathways—driven by the dysregulation of driver and checkpoint proteins—that contribute to gliomagenesis (Phillips et al. 2006; The Cancer Genome Atlas Research Network 2008; Parsons et al. 2008; Brennan et al. 2009; Verhaak et al. 2010). In particular, it is clear that RTKs (including EGFR, PDGFRA, and MET), the PI3K pathway, signaling pathways activated by PTEN and NF1 loss, and the mutant IDH proteins play central roles in the pathobiology of glioblastoma.
EGFR/EGFRvIII
EGFR amplification is observed in ∼50% of primary glioblastomas and is associated with poor prognosis (Hurtt et al. 1992; Jaros et al. 1992; Schlegel et al. 1994). Furthermore, ∼50% of EGFR-amplified cells harbor the EGFRvIII mutant, which is an intragenic gene rearrangement generated by an in-frame deletion of exons 2–7 that encode part of the extracellular region. The expression of EGFRvIII has been determined to confer a worse prognosis than wild-type EGFR expression alone (Shinojima et al. 2003; Heimberger et al. 2005). Experimentally, ectopic overexpression of EGFRvIII in glioma cell lines induces constitutive autophosphorylation, activation of the Shc–Grb2–Ras and class I PI3K pathways (Huang et al. 1997; Narita et al. 2002), enhanced tumorigenicity (Huang et al. 1997), increased cell proliferation (Narita et al. 2002), and resistance to apoptosis induced by DNA-damaging agents through modulation of Bcl-XL expression (Nagane et al. 1998). Notably, the downstream effects of EGFRvIII overexpresson are not recapitulated by overexpression of wild-type EGFR. For example, wild-type EGFR cannot substitute for EGFRvIII in driving infiltrative glioma formation in genetically engineered mice (Hesselager and Holland 2003; Zhu et al. 2009) or in Ink4a/Arf−/− murine neural stem cells or astrocytes (Holland et al. 1998; Bachoo et al. 2002), except when EGF ligand is infused at a high concentration into the injection site of wild-type EGFR-transduced cells (Bachoo et al. 2002). Both EGFRvIII and wild-type EGFR/ErbB family proteins have been identified in the nucleus and are thought to drive proliferation and DNA damage repair through both transcriptional and signaling functions (Wang and Hung 2009). Moreover, the observation that EGFR also translocates to the mitochondria (Boerner et al. 2004) provides further evidence that the contributions of EGFR malignancy may not be limited to its conventional cell membrane location and merit further study.
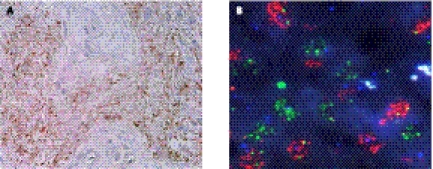
Despite the well-recognized proproliferative functions of EGFRvIII, its expression in human glioblastoma is heterogeneous and is most often observed only in a subpopulation of cells (Fig. 2A; Nishikawa et al. 2004). Recent observations support a model of functional heterogeneity in which a minority of EGFRvIII-expressing cells not only drive their own intrinsic growth, but also potentiate the proliferation of adjacent wild-type EGFR-expressing cells in a paracrine fashion through the cytokine coreceptor gp130 (Inda et al. 2010). Even though these results illustrate that cytokines produced from EGFRvIII expression can be drivers of heterogeneity, there are likely additional cytokine-inducing mechanisms at work. For example, it has been recently shown that NFKB1A, which encodes IκBα, a critical negative regulator of canonical NF-κB activation, was found to undergo monoallelic loss in glioblastomas that lack EGFR amplification (Bredel et al. 2011), suggesting that NF-κB plays physiologically relevant roles downstream from EGFR/EGFRvIII that include IL-8 production (Bonavia et al. 2011). These results suggest that intraclonal cooperativity drives the persistence of intratumoral heterogeneity, which has implications for both our basic understanding of gliomagenesis and also drug sensitivity profiles of these tumors (Yao et al. 2010).
Figure 2.
Molecular heterogeneity in glioblastoma. (A) Immunohistochemistry for the mutant EGFR receptor EGFRvIII demonstrates a heterogeneous staining pattern within the tumor. Images from Nishikawa et al. (2004) used with permission. (B) Multicolor FISH reveals distinct subpopulations of either EGFR (red) or PDGFRA (green) amplification within a glioblastoma specimen. Images obtained from Cameron Brennan.
In addition, although most human glioma cell lines fail to faithfully recapitulate the EGFR amplification and EGFRvIII expression observed in primary tumor specimens, recent studies have reported successful passage of EGFRvIII-expressing glioblastoma xenografts in vivo as well as in vitro when grown in stem cell culture conditions (Stockhausen et al. 2011), suggesting that durable EGFRvIII expression may be linked to differentiation and/or development. Moreover, it is now clear that the type of genetic alterations involving EGFR in glioblastoma are distinct from those observed in other EGFR-altered cancers, such as non-small-cell lung cancer (NSCLC). In glioma, focal EGFR amplification occurs at an extremely high level (>20 copies). In addition, the vast majority of other mutations, including the vIII mutant as well as missense mutations (Lee et al. 2006b; The Cancer Genome Atlas Research Network 2008), are found within the extracellular domain, while most mutations in other nonglioma cancers are found in the intracellular domain (Janne et al. 2005). Although EGFRvIII expression is sufficient to rescue the knockdown of an endogenous kinase domain mutant EGFR in NSCLC cells (Rothenberg et al. 2008), it is not clear whether EGFR mutant glioma cells drive similar downstream signaling and/or confer the same “addiction” to EGFR activation as is the case in NSCLC (Sharma and Settleman 2007).
PDGFR
Nearly 30% of human gliomas show expression patterns that are correlated with PDGFR signaling (Brennan et al. 2009) and genes involved in oligodendrocyte development (OLIG2, NKX2-2, and PDGF), both hallmarks of the proneural glioblastoma subtype. PDGFRA amplification is found in 15% of all tumors and is enriched in the proneural subtype (Phillips et al. 2006; Verhaak et al. 2010). Of those tumors harboring gene amplification, recent work showed that 40% harbor an intragenic deletion, termed PDGFRAΔ8,9 (Clarke and Dirks 2003), in which an in-frame deletion of 243 base pairs (bp) of exons 8 and 9 leads to a truncated extracellular domain (Ozawa et al. 2010). In addition, in-frame gene fusion of the extracellular domain of KDR/VEGFR-2 and the kinase and intracellular domains of PDGFRA has also been identified, and both the PDGFRAΔ8,9 and KDR-PDGFRA mutant proteins were constitutively active and transforming and could be inhibited with inhibitors of PDGFRA. Point mutations in PDGFRA are associated with amplification but, unlike EGFR, are generally rare events (The Cancer Genome Atlas Research Network 2008). Of the additional ways to activate PDGFR signaling, PDGF ligands (A–D) are up-regulated in ∼30% of glioma surgical samples and cell lines, while PDGFRB expression appears to be restricted to proliferating endothelial cells in glioblastoma (Fleming et al. 1992; Hermanson et al. 1992; Di Rocco et al. 1998; Smith et al. 2000; Lokker et al. 2002). The intratumoral coexpression of PDGF and PDGFR suggests that both autocrine and paracrine loops play roles in glioblastoma. This possibility is supported by the demonstration that stimulation of PDGFRB-expressing endothelial cells by tumor-derived PDGF can drive VEGF-mediated angiogenesis within the local microenvironment (Guo et al. 2003).
Similar to the case of EGFR/EGFRvIII described above, cell and receptor heterogeneities appear to be other distinct mechanisms by which PDGF/PDGFR promotes aggressive glioma growth. For example, transduction of cells of the subventricular zone (SVZ) of the lateral ventricle of neonatal rat pups with a retrovirus expressing PDGF yielded large, diffusely infiltrating tumors resembling glioblastoma (Assanah et al. 2006, 2009). The tumors that formed contained a massive proliferation of both infected and uninfected PDGFRα+-expressing progenitors, suggesting that PDGF was driving tumor formation through both autocrine and paracrine signaling, leading to the recruitment of non-PDGF-expressing resident progenitors and the evolution of cellularly heterogeneous malignant gliomas. Further work has demonstrated that these recruited cells become transformed, overtake PDGF-induced gliomas, and can be serially transplanted (Fomchenko et al. 2011), and that topotecan delivery to these tumors results in ablation of both tumor-initiating cells and recruited glial progenitors (Lopez et al. 2011). Although these results still remain to be validated within the context of human tumors, they raise the possibility that cells distinct from the initial transformed cells of origin within the tumor environment can be corrupted to become bona fide tumor cells. This model of glioma evolution is distinct from the generally established view of linear gliomagenesis (Fomchenko et al. 2011).
c-Met
The c-Met RTK is amplified in ∼5% of glioblastomas, has been found to be overexpressed in 18 out of 62 (29%) glioblastoma samples with shorter median survival (Kong et al. 2009), and is rarely mutated (The Cancer Genome Atlas Research Network 2008). Moreover, c-MET was found to be coactivated in glioblastoma cells with increased levels of EGFR/EGFRvIII (Huang et al. 2007; Stommel et al. 2007; Pillay et al. 2009) and represents a critical dependency in MET-amplified cells (Beroukhim et al. 2007). In this setting, activated EGFR can associate with c-Met, leading to the activation of this receptor in the absence of its ligand, HGF (Jo et al. 2000). Conversely, HGF transcriptionally activates the expression of the EGFR ligands TGF-α- and heparin-binding EGF and can therefore activate EGFR (Reznik et al. 2008). Blocking EGFRvIII activity with an EGFR-specific monoclonal antibody, panitumumab, can result in a switch to HGF-mediated c-Met activation, which could be prevented by cotreatment with AMG102, a neutralizing antibody to HGF (Pillay et al. 2009). Similar results were achieved with combined c-Met and EGFR small molecule inhibitors (Huang et al. 2007; Stommel et al. 2007) and neutralizing anti-HGF monoclonal antibody combined with erlotinib (Lal et al. 2009). The relationship of c-Met with EGFR is also not surprising in light of prior studies showing that overexpression of Met can lead to gefitinib resistance in mutant EGFR lung cancers, often by activating ERBB3 (Engelman et al. 2007). Concurrent activation of c-Met with PDGFR has also been detected in glioblastoma and has been suspected to be another mechanism for resistance to EGFR kinase inhibitors. In this case, inhibition of three RTKS—EGFR inhibition with erlotinib, c-Met inhibition with SU11274, and PDGFR inhibition with imatinib—significantly inhibited the in vitro growth of glioblastoma cell lines, compared with single drugs alone (Stommel et al. 2007), possibly by attenuating downstream PI3K signaling. In contrast, in scenarios in which MET is amplified in isolation, one report showed that treatment with crizotinib, which inhibits ALK and also c-Met (Christensen et al. 2007), can induce radiographic and clinical improvement (Chi et al. 2012).
Src family kinases (SFKs)
SRC and SFKs are frequently activated in glioblastoma patient samples and cell lines (Stettner et al. 2005; Du et al. 2009) and are widely expressed in glioblastoma (Lu et al. 2009). SFKs mediate signaling from growth factor receptors that are commonly overexpressed in glioblastoma, providing a potential explanation for SFK activation. Bead-based profiling of tyrosine kinase activation in 130 human cancer cells showed that the most frequently activated tyrosine kinases were EGFR, fibroblast growth factor receptor 3 (FGFR3), protein tyrosine kinase 2 (PTK2, also known as focal adhesion kinase, or FAK), and SFKs including SRC, LYN, and LCK (Du et al. 2009). Moreover, screening of 31 primary glioblastomas samples showed SRC activation in 61% of samples (Du et al. 2009) and that the SRC inhibitor dasatinib inhibited cell viability and migration in vitro and tumor growth in vivo, nominating SRC/SFK as potential therapeutic targets in a subset of glioblastomas.
RTK cooperativity and heterogeneity
One potential explanation for the failure of EGFR and PDGFRA inhibitors to elicit significant clinical outcomes (De Witt Hamer 2010) is that additional RTKs may cooperate to provide an integrated signaling threshold that is not sufficiently attenuated through the inactivation of any single RTK (Huang et al. 2007; Stommel et al. 2007). Indeed, Stommel et al. (2007) demonstrated that three or more RTKs were activated in a majority of glioblastoma cell lines and patient specimens. This discovery of concomitant receptor expression and coactivation suggests that tumor RTK profiling may be an important step in the development of a personalized glioblastoma therapeutic regimen and that cross-talk between the receptors could be targeted with specific inhibitors to both, resulting in enhanced cytotoxicity.
Recent studies provide additional evidence that glioblastomas are composed of heterogeneous subpopulations within tumors. Indeed, varied expression patterns of several proteins have been described: Wild-type EGFR and EGFRvIII, as described above (Nishikawa et al. 2004; Inda et al. 2010); PDGFRA (Hermanson et al. 1992); c-Met (Nabeshima et al. 1997); angiogenic factors (Koga et al. 2001); and adhesion molecules (Bello et al. 2001b) all exhibit heterogeneous distributions. Two recent studies provide further compelling examples of clonal heterogeneity in glioblastoma. Both Snuderl et al. (2011) and Szerlip et al. (2012) observed that 5%–7% of all glioblastomas—and nearly 13% of glioblastomas with EGFR, PDGFRA, or MET amplification—harbored multiple RTK amplifications. Moreover, while a minority of cells harbored amplification of multiple RTKs (Szerlip et al. 2012), the predominant pattern was mosaic amplification such that tumors were comprised of individual cell populations harboring isolated amplification of a single RTK (Fig. 2B). This point was further demonstrated in a remarkable FISH analysis of tumor sections from a whole-brain autopsy of an untreated patient with bilateral multifocal glioblastoma in which there was a striking anatomic distribution of EGFR- and PDGFRA-amplified cells (Snuderl et al. 2011). It is likely that this pattern of clonal RTK heterogeneity is a late event in gliomagenesis (Snuderl et al. 2011; Szerlip et al. 2012) and is influenced by several factors, including binomial segregation of unstable amplicons or local microenvironment selection (Szerlip et al. 2012). It is likely that somatic mutations in cancer genes will demonstrate a pattern of heterogeneity similar to that seen with SCNAs, but this possibility awaits additional study at this point. Together, these studies provide further molecular evidence that glioblastomas are often heterogeneous, which has major biologic and therapeutic implications.
PI3K pathway
Activating kinases
The PI3K signaling pathway is dysregulated in many cancers (Yuan and Cantley 2008), including glioblastomas. Some of the major genomic alterations discussed above—RTK amplification/mutation, PIK3CA and PIK3R1 mutation, and PTEN loss—all activate this pathway (The Cancer Genome Atlas Research Network 2008). Of the three PI3K classes, the class IA kinases are likely to play a direct role in cell transformation and are composed of both a catalytic subunit isoform (p110α, p110β, p110δ, and p110γ) and a regulatory subunit isoform (p85α, p55α, p50α, p85β, and p55γ). Broadly, activation of the pathway can be initiated by GTP-bound Ras (Rodriguez-Viciana et al. 1994, 1996) and through RTK signaling, which recruits PI3K to the cell membrane, whereupon the lipid phosphatidylinositol (PtdIns)-4,5-bisphosphate (PIP2) is phosphorylated to PtdIns-3,4,5-bisphosphate (PIP3). This reaction is antagonized by the major glioma tumor suppressor and PtdIns(3,4,5)P3 phosphatase PTEN. PIP3 subsequently recruits the serine/threonine kinase AKT to the plasma membrane, where it is fully activated by its main dual inputs—Thr 308 phosphorylation by PDK1, and Ser 473 phosphorylation by the mammalian target of rapamycin (mTOR) complex 2 (mTORC2) (Sarbassov et al. 2005). Elevated AKT phosphorylation has been observed in up to 85% of glioblastoma cell lines and patient samples (Wang et al. 2004). RTK-independent activation of this pathway in glioblastoma can occur via mutation or amplification of PIK3CA (p110α) (Gallia et al. 2006; Kita et al. 2007; The Cancer Genome Atlas Research Network 2008), and PIK3CD (p110δ) is also overexpressed in some gliomas (Mizoguchi et al. 2004). Moreover, the TCGA study revealed recurrent mutations in the gene encoding the p85α regulatory subunit PIK3R1, which likely drive PIK3CA activation through decreased SH2 domain-mediated inhibition (Sun et al. 2010).
PI3K drives many glioma-relevant processes, including survival, proliferation, migration, and invasion (Engelman et al. 2006). In addition to its well-known functions (Lino and Merlo 2011), recent work has revealed several novel mechanisms underlying PI3K function in glioma. Specifically, CD95 promotes invasion through interaction with PI3K and the SFK Yes, triggering activation of GSK3β and induction of matrix metalloproteinase (MMP) expression (Kleber et al. 2008). The RTK EphA2, which is highly expressed in glioblastoma (Wykosky et al. 2005) and binds PI3K upon ligand stimulation (Pandey et al. 1994), induces glioblastoma cell migration in an AKT-dependent manner (Miao et al. 2009). In addition, the insulin-like growth factor 2 (IGF2) promotes aggressive growth in glioblastoma lacking EGFR amplification or overexpression through IGF receptor 1 (IGFR1) and PIK3R3 (Soroceanu et al. 2007). The PI3K pathway also maintains glioblastoma tumor-initiating cells, as direct silencing of mTOR or inactivation with rapamycin reduces neurosphere formation and expression of neural stem cell progenitor markers (Sunayama et al. 2010) as well as growth of patient-derived tumor-initiating cells (Gallia et al. 2009). In addition, AKT activation due to PTEN loss likely contributes to RTK inhibitor insensitivity in glioblastoma (Mellinghoff et al. 2005, 2007).
Effectively inhibiting the PI3K signaling pathway is challenging because the cascade and its feedback regulation remain incompletely understood. Specifically, RTK-activated PI3K signaling does not always require AKT, as EGFR can also signal to mTORC1 through protein kinase C (PKC) independently of AKT (Fan et al. 2009), perhaps through PDK1 (Dutil et al. 1998; Le Good et al. 1998). Indeed, a novel PI3K/PDK1/PKCι pathway can regulate phosphorylation and inactivation of the proapoptotic protein Bad, thereby increasing glioma cell survival (Desai et al. 2011). Additional work has revealed the existence of an AKT-independent, mutant PIK3CA signaling pathway leading to PDK1-mediated activation of a critical downstream effector, SGK3, in PTEN intact tumor cells (Vasudevan et al. 2009). PI3K pathway inhibition is thought be cytostatic, rather than cytotoxic, potentially due to G1 cell cycle arrest (Paternot and Roger 2009; Fan et al. 2010). Combination mTOR and MEK inhibition has therefore been used that suppresses CDK4 phosphorylation in a synergistic manner (Paternot and Roger 2009). Due to complex feedback pathways, other combination therapies being explored to target PI3K signaling in glioma include inhibition of PI3K and mTOR with inhibitors such as PI-103 (Fan et al. 2006), as well as simultaneous inhibition of mTORC1 and mTORC2 (Q Liu et al. 2011). Monotherapy with the mTORC1 inhibitor rapacmycin disrupts an IRS-1-mediated negative feedback loop and can actually increase AKT activity (Fan et al. 2006; Cloughesy et al. 2008) through mTORC2-mediated phosphorylation (Gulati et al. 2009). Interestingly, dual PI3K/mTOR inhibition with PI-103 induces autophagy, which, when also inhibited pharmacologically, leads to apoptosis (Fan et al. 2010). Thus, a combinatorial therapeutic strategy for targeting the PI3K pathway in glioblastoma will likely be necessary as more of its complex biology and regulation are revealed (Akhavan et al. 2010).
PTEN
PTEN directly antagonizes PI3K signaling and is one of the most frequently altered genes in cancer. It undergoes genomic loss, mutation, or epigenetic inactivation in 40%–50% of gliomas, resulting in high levels of PI3K activity and downstream signaling (Koul 2008). As the PI3K pathway is a driving force in gliomas, even small changes in the expression or function of its critical negative regulator, PTEN, have profound effects on tumor cell behavior. The stability of PTEN is regulated post-translationally by GSK3-mediated phosphorylation at Thr 366 (Maccario et al. 2007) and by proteasomal degradation through polyubiquitination by the HECT domain ubiquitin ligase NEDD4-1 (Wang et al. 2007). NEDD4-1 up-regulation has been recently associated with overexpression of the FoxM1B transcription factor in gliomas (Dai et al. 2007). Moreover, a multiprotein tumor suppressor network exists in glioblastoma comprised of PTEN, the adaptor protein Na+/H+ exchanger regulatory factor (NHERF1), and the pleckstrin homology domain leucine-rich repeat protein phosphatase 1, which form a heterotrimeric complex that undergoes disruption in high-grade tumors (Molina et al. 2012). Thus, in gliomas where PTEN is not deleted, mutated, or epigenetically silenced, mechanisms such as aberrant up-regulation of NEDD4-1 or loss of NHERF1 could contribute to suppressing PTEN function. These mechanisms raise the possibility that the level of PTEN dysfunction is underestimated solely on the basis of genomic data.
The physiologic relevance of PTEN loss was underscored using several genetically engineered mouse models. CNS-specific GFAP-Cre-p53lox/loxPTENlox/+ mice developed malignant gliomas with short latency that was strikingly similar to human disease both histopathologically and molecularly, given the degree of concomitant RTK activation (Zheng et al. 2008). Moreover, dual loss of p53 and PTEN in this model promoted increased c-Myc activity, which led to impaired differentiation and stable tumorigenic capacity of glioma tumor-initiating cells. Mice conditionally lacking p53, PTEN, and Rb also formed glioblastoma with SCNAs recapitulating those seen in human tumors (Chow et al. 2011). Furthermore, successive loss of each PTEN allele in the NF1−/− p53−/− model of progressive astrocytoma accelerated formation of grade III astrocytomas and progression into glioblastoma (Kwon et al. 2008). Together, these models demonstrate the causal role of PTEN loss in glioblastoma and provide the means with which to study patient-relevant disease in tractable murine models.
Clinically, PTEN loss has been shown to confer resistance to EGFR inhibitors in patients harboring EGFRvIII-expressing glioblastoma in part due to its activation of downstream AKT (Mellinghoff et al. 2005) as well as loss of its RTK degradation function (Vivanco et al. 2010). PTEN has also been implicated in the ubiquitin-mediated control of protein stability with respect to apoptosis, as it influences the half-life of the anti-apoptotic protein FLIPS (Panner et al. 2009). With respect to development, PTEN loss may also promote a “side population” phenotype of glioma stem-like cells by driving expression of the drug transporter protein ABCG2 (Bleau et al. 2009). Furthermore, recent studies have described new roles for PTEN in metabolism and cellular homeostasis; in PTEN-null cells, the enzyme ectonucleoside triphosphate diphosphohydrolase 5 (ENTPD5) promotes protein N-glycosylation and folding in the setting of increased AKT-mediated anabolism, increases growth factor receptor levels, contributes to increased aerobic glycolysis (i.e., the Warburg effect), and is required for PTEN-null tumor cell growth (Fang et al. 2010), although this result awaits validation in glioma models. The nuclear localization of PTEN may also play an important role in tumor suppressor activity (Perren et al. 2000; Whiteman et al. 2002; Zhou et al. 2002). Specifically, nuclear PTEN interacts with APC/C to promote formation and enhance the tumor-suppressive ability of the APC–CDH1 complex in a phosphatase-independent manner (Song et al. 2011). This demonstrates that loss and mutation of PTEN are not synonymous, as cells in these respective states are differentially sensitive to pharmacological inhibition of APC–CDH1 targets. Further work is necessary to fully understand which of the many functions attributed to PTEN play key roles in human glioblastomas.
NF1
Neurofibromin, the product of the NF1 gene, is a potent tumor suppressor that negatively regulates Ras and mTOR signaling in astrocytes. Inactivation of NF1 has been observed in gliomas and can arise as a result of excessive proteasomal degradation mediated by hyperactivation of PKC (The Cancer Genome Atlas Research Network 2008; McGillicuddy et al. 2009) or by genetic loss or mutation, which were surprising findings from large-scale sequencing analyses (Parsons et al. 2008). NF1 mutations are most commonly found in the mesenchymal subtype of glioblastoma (Verhaak et al. 2010). Experiments using NF1-deficient primary murine astrocytes have revealed that NF1 loss results in increased cell proliferation and migration that is dependent on Ras-mediated hyperactivation of mTOR. In this setting, mTOR induces rapamycin-sensitive activation of Rac1 GTPase that is independent of elongation factor 4E-binding protein 1(4EBP-1)/S6 kinase (S6K) (Sandsmark et al. 2007). In addition, NF1 deficiency causes hyperproliferation and impacts glioma formation in a manner that is independent of the tuberous sclerosis complex (TSC)/Ras homolog enriched in brain (Rheb) control of mTOR (Banerjee et al. 2011). Other potential downstream targets of neurofibromin include Stat3, which was identified in a chemical library screen using NF1-deficient malignant peripheral nerve sheath tumor cells (Banerjee et al. 2010). In this case, Stat3 is regulated in an mTORC1 and Rac1-dependent manner and increases cyclinD1 expression. Additionally, it will be important to determine whether NF1-deficient gliomas are as susceptible to proteoxicity through HSP0/mTOR inhibition as in other NF1-deficient cancers (De Raedt et al. 2011).
Genetically engineered mouse models have demonstrated that targeted homozygous loss of NF1 in astrocytes, while sufficient to increase cell growth in vitro and in vivo, is not sufficient to induce glioma formation (Bajenaru et al. 2002). Interestingly, NF1−/− astrocytes develop optic gliomas in the context of an NF1+/− brain environment (Bajenaru et al. 2003; Zhu et al. 2005), in part through paracrine factors including hyaluronidase (Daginakatte and Gutmann 2007). In addition, it has been found that low levels of cAMP expression in the stroma are sufficient to induce optic glioma formation in genetically engineered mouse models of NF1 such that local depletion of cAMP resulted in glioma formation in regions of the brain otherwise not observed to develop tumors (Warrington et al. 2010). These findings emphasize the importance of heterogeneity and the cell type-specific effects of various genetic alterations in tumorigenesis. Other genetically engineered mouse models have demonstrated that NF1 loss in glial cells, in combination with a germline p53 mutation, results in fully penetrant malignant astrocytomas (Zhu et al. 2005), which progress to glioblastoma upon deletion of PTEN (Kwon et al. 2008). More recent work has revealed that the same combination of genetic alterations in these tumor suppressor genes in neural stem/progenitor cells is necessary and sufficient to induce astrocytoma formation (Alcantara Llaguno et al. 2009).
Mutant IDH1/2 proteins
Mechanistic basis of mutant IDH1 biology
Mutant IDH1/2 have been shown to catalyze a neomorphic function (Dang et al. 2010). Specifically, while wild-type IDH1 catalyzes the NADP+-dependent oxidation of isocitrate, mutant IDH1 catalyzes the NADPH-dependent reduction of α-KG to the (R)-enantiomer of 2-hydroxyglutarate (2-HG), which is the same stereoisomer of 2-HG that is seen in D-2-HGA. Although wild-type IDH1 can also catalyze this particular reaction (Pietrak et al. 2011), mutant enzymes perform this reaction with much higher efficiency because the R132 substitutions modify the active site to increase α-KG and NADPH binding (Dang et al. 2010; Pietrak et al. 2011). Dang et al. (2010) showed that mutant cells contained extremely high levels of 2-HG, which was also found in primary IDH1 mutant gliomas and in the serum of IDH mutant AML patients (Gross et al. 2010; Ward et al. 2010).
Work on the downstream biological effects of IDH1/2 mutation expression has focused largely on the inhibition of α-KG-dependent dioxygenases by 2-HG. This diverse group of enzymes controls a broad range of physiological processes, including hypoxic sensing, histone demethylation, demethylation of hypermethylated DNA, fatty acid metabolism, and collagen modification, among others (Loenarz and Schofield 2008). Several studies have provided evidence to demonstrate that several of these functions are influenced by IDH1/2 mutation expression. IDH1 mutant gliomas exhibit a global DNA hypermethylation state, termed the glioma CpG island methylator phenotype (G-CIMP) (Noushmehr et al. 2010). This state was also observed in IDH1/2 mutant AML (Figueroa et al. 2010). Mutant IDH1 expression was sufficient to produce a G-CIMP phenotype in engineered normal human astrocytes that was highly concordant with that seen in IDH mutant human tumor samples. Furthermore, this methylation phenotype correlated with a gene expression signature comprised of a limited set of down-regulated genes that discriminated between IDH1 mutant and wild-type proneural tumors. This hypermethylation state may be caused in part by the 2-HG-mediated inhibition of the α-KG-dependent TET2 enzyme (Xu et al. 2011; Turcan et al. 2012); the resultant decrease in 5-hydroxymethylcytosine was also observed in glioblastoma specimens (Xu et al. 2011). Moreover, mutant IDH1/2 cells displayed impaired hematopoietic differentiation, suggesting that a hypermethylated epigenetic landscape contributed to a persistent dedifferentiated state (Figueroa et al. 2010). The inhibition of histone demethylases in IDH1 mutant cells may also impair differentiation (Chowdhury et al. 2011; Xu et al. 2011; Lu et al. 2012). Repressive histone methylation was shown to be associated with impaired mutant IDH1-expressing astrocyte differentiation, and the accumulation of these histone marks preceded significant DNA hypermethylation in engineered IDH1 mutant cells (Lu et al. 2012). Furthermore, Lai et al. (2011) showed that global expression profiles of IDH1 mutant glioblastomas more closely resembled lineage-committed neural precursors, whereas wild-type counterparts appear to resemble neural stem cells.
The production of 2-HG also appears to influence HIF biology in several ways. First, 2-HG may stabilize HIF-1 under some conditions (Zhao et al. 2009). However, the HIF-1 response to hypoxia in IDH mutant cells is attenuated (Koivunen et al. 2012). Specifically, the (R)-enantiomer of 2-HG stimulates the EGLN prolyl 4-hydroxylases, which mark HIF for degradation. Either expression of mutant IDH1, suppression of HIF-1α, or overexpression of EGLN1 was sufficient to stimulate colony formation by immortalized human astrocytes, and HIF-regulated genes were down-regulated in IDH1 mutant proneural tumors. Hypoxic cells drive lipogenesis via reduction of glutamine to α-KG by wild-type IDH1 (Metallo et al. 2012). This observation suggests a mechanism by which cells can survive under hypoxic or pseudohypoxic conditions and points to a physiological selection pressure to maintain a copy of the wild-type IDH1 gene in cancer cells. Finally, mutant IDH1/2 expression or 2-HG administration can potently inhibit multiple histone demethylases, likely altering transcriptional programs. Additional work has shown that cells expressing the IDH1 R132H mutation display metabolomics alterations in amino-free and branched chain amino acid levels and also choline phospholipid synthesis (Reitman et al. 2011). Thus, together, these data have pointed to several biological processes affected by mutant IDH1/2 expression that, collectively, may promote tumor growth by integrating changes in development, global transcriptional programs, metabolism, and responses to hypoxia.
Several studies suggest that IDH1/2 mutation may be an early event in IDH1/2 mutant neoplasms. When patients with diffuse astrocytoma or oligoastrocytoma were subjected to serial biopsies, there were seven patients who carried only IDH1 mutations at the first biopsy but acquired either TP53 mutation or 1p19q loss at the second biopsy, suggesting a temporal sequence of mutation acquisition. This possibility is also supported by recent sequence analysis of IDH1 and p53 genes in a separate study by Lai et al. (2011). Analysis of a large panel of grade II–IV astrocytomas showed a higher propensity for Arg-to-Cys substitutions at position 273 in p53 compared with the high rate of Arg-to-His substitutions at position 132 in IDH1, which would be consistent with a strand asymmetry mechanism (Rodin and Rodin 1998) in which C → T mutations took place on the transcribed strand in IDH1 but on the nontranscribed strand in p53. The result of this mutational asymmetry would be that mutant IDH1 could be expressed in a nonreplicating clone, whereas mutant p53 could be expressed only after DNA replication in S phase. Together, these studies provide potential insights into the evolution of IDH1 mutant cancers and highlight the importance of serial tissue analysis and the need for careful clonal analysis to fully clarify how these cancers progress.
Translational relevance of IDH1 status
Despite our incomplete understanding of mutant IDH biology, the mutant status of the IDH1/2 genes may serve as an important prognostic indicator. Specifically, patients with anaplastic astrocytoma (Parsons et al. 2008; Sanson et al. 2009; Yan et al. 2009; Hartmann et al. 2010) and glioblastoma (Yan et al. 2009) harboring mutant IDH1 demonstrate a significantly longer overall survival compared with wild-type IDH1 counterparts and are younger at presentation, and this survival benefit has also been observed in grade II gliomas (Sanson et al. 2009). Patients with G-CIMP+ tumors also experience a similar survival benefit (Noushmehr et al. 2010). In addition, a comprehensive genomic and clinical analysis of glioblastomas harboring mutant and wild-type IDH1 suggests that, while histopathologically similar, these tumors may represent disease processes far more disparate than has been appreciated. Specifically, IDH1 mutant tumors display less contrast enhancement, less peritumoral edema, larger initial size, greater cystic components, and a greater likelihood of frontal lobe involvement compared with wild-type tumors (Lai et al. 2011). In addition, several methods have been developed to assess IDH1 mutant protein status (Capper et al. 2009) or its 2-HG by-product (Sahm et al. 2012) in clinical settings. While 2-HG is easily detected in the serum of AML patients, the correlation between serum 2-HG and tumor mutation status may be less specific in the setting of glioma (Capper et al. 2011). Finally, it may also be possible to monitor the presence of 2-HG noninvasively using magnetic resonance spectroscopy (MRS) imaging of the brain (Pope et al. 2012). Together, these studies provide overwhelming evidence for the clinical relevance of IDH1 status in gliomas from grades II to IV and support the proposal that its status be incorporated into the current WHO histopathological scheme for every glioma analyzed (Hartmann et al. 2010). Despite their histological similarities, IDH1 mutant and wild-type glioblastomas are clearly distinct diseases (Fig. 1), and understanding the biological basis behind the differences in their natural histories will surely be a major area of focus in the field.
Tumor biological hallmarks in glioma: invasion, angiogenesis, and tumor-initiating cells
Glioma cell invasion
The ability of glioblastoma cells to invade adjacent brain tissue contributes to the major clinical problem in achieving disease control. The majority of glioblastomas treated initially with standard therapy will recur within several centimeters of the initial tumor location (Hochberg and Pruitt 1980; Chamberlain 2011). In addition, 10%–20% of patients may harbor “macroscopic” evidence of invasion at the time of presentation, including multifocal disease—which can even be bihemispheric in the so-called “butterfly” pattern—at noncontiguous sites in the brain or involving spread along white matter tracts and through subependymal and/or subarachnoid spaces (Parsa et al. 2005; Chamberlain 2011). It is well established that histologically identifiable tumors invariably greatly exceed the area of disease detected radiographically (Burger et al. 1988). The intrinsic capacity of glioblastoma to invade was well illustrated by classic autopsy studies, which demonstrated that 25%–50% of untreated glioma patients examined harbored histological evidence of bilateral disease (Scherer 1940; Matsukado et al. 1961). Notably, even grade II, or “diffuse,” gliomas exhibit invasive capacity such that cells derived from this tumor grade can be observed histologically at least 2 cm from the main tumor mass when “supratotal” surgical resections have been attempted (Yordanova et al. 2011). Thus, glioma cell invasiveness is a clinical problem in the majority of diffuse gliomas.
The process of glioblastoma invasion likely involves sequential adhesion to the extracellular matrix (ECM), degradation of the ECM, and altered cell contractility (Giese and Westphal 1996; Nakada et al. 2007; Tate and Aghi 2009; Onishi et al. 2011). Unlike non-CNS cancers, clinically manifest hematogenous or lymphatic spread of glioblastoma outside the brain is exceedingly rare (Hoffman and Duffner 1985). Also, the ECM composition and distribution in the brain is distinct from other extra-CNS tissue sites. A typical collagen-rich basement membrane ECM exists in the glia externa limitans, which covers the cortical surface, and also surrounding cerebral blood vessels (Rutka et al. 1988; Louis 2006; Gritsenko et al. 2012). However, the brain parenchyma harbors a unique ECM structure, the perineuronal network, which is a meshwork composed predominantly of hyaluronan sulfate proteoglycans as well as chondroitin sulfates proteoglycans, tenascins, and link proteins (Kwok et al. 2011; Gritsenko et al. 2012). Moreover, distinct brain regions, such as the neurogenic areas of the SVZs, are more enriched for chondroitin and heparan sulfate proteoglycans (Sirko et al. 2007).
Glioma cells adhere to the ECM using several mechanisms. The immunoglobulin superfamily member CD44 and the hyaluronan-mediated motility receptor (RHAMM), both receptors for hyaluronan, are expressed in glioblastoma (Akiyama et al. 2001). CD44 is cleaved by both ADAM proteases (Murai et al. 2004) and MMP-9 (Chetty et al. 2012) in a process that promotes motility via cytoskeletal reorganization (Murai et al. 2004; Bourguignon 2008); it is likely that myosin II is also important for glioma cell contractility during invasion (Beadle et al. 2008). Strikingly, CD44 and RHAMM are both suppressed by p53 (Godar et al. 2008; Sohr and Engeland 2008), suggesting that early cellular progression through canonical checkpoints and the ability to migrate/invade are linked, although this possibility remains to be validated in glioma models. Integrins, particularly the αvβ3 and αvβ5 heterodimers (Bello et al. 2001a), also likely contribute to glioma cell adherence to the ECM in a process that activates cytoskeletal rearrangement through cytoplasmic mediators, including FAK (Rutka et al. 1999; Riemenschneider et al. 2005) and/or Pyk2 (Lipinski et al. 2008). Their putative involvement in glioma pathobiology (D'Abaco and Kaye 2007; Desgrosellier and Cheresh 2010) has prompted the testing of the αvβ3 and αvβ5 integrin inhibitor cilengitide in an ongoing phase III CENTRIC clinical trial for newly diagnosed glioblastoma in combination with radiation therapy and temozolomide (Reardon et al. 2011a).
Several MMPs have been implicated in modifying the ECM in the local microenvironment to promote invasion (Rao 2003). Recent work revealed a novel mechanism of EGFR-mediated invasion in which EGFR-dependent up-regulation induced high expression of the IFN-regulated factor guanylate-binding protein 1, leading to increased glioma invasion through MMP-1 expression (Li et al. 2011). MMP-2 and MMP-9 also drive glioma invasion (Forsyth et al. 1999) and are regulated through several molecular cascades. Both enzymes promoted invasion when up-regulated via a CD95-mediated activation of AKT1 involving recruitment of Src family member Yes and p85. Importantly, CD95L expression was demonstrated at the leading glioma edge in clinical samples (Kleber et al. 2008). MMP-2 and MMP-9 were also coordinately up-regulated by the low-density lipoprotein receptor-related protein 1 in an ERK-dependent promigratory process (Song et al. 2009). Further work has revealed additional regulators of MMP-2 expression, including the PTEN-regulated insulin growth factor-binding protein 2 (IGFBP2) (Wang et al. 2003; Levitt et al. 2005) and the forkhead transcription factor FoxM1B. FoxM1B not only up-regulated MMP-2 and was overexpressed in human glioblastoma specimens (Dai et al. 2007), but also transformed engineered, immortalized normal human astrocyte cells into invasive glioblastoma cells via a pathway involving PTEN degradation and AKT activation (Dai et al. 2010). Notably, AKT also regulates the actin-binding protein Girdin, which directs neural cell migration and contributes to tumor-initiating cell invasiveness (Natsume et al. 2011); because several invasive mechanisms can be regulated by the PTEN/PI3K/AKT pathway, its contribution to this phenotype deserves further investigation. Given that the MMP inhibitor marimistat did not show efficacy in a randomized clinical trial, albeit as a monotherapy without temozolomide (Levin et al. 2006), it will be important to further clarify both the critical proteases and their regulatory pathways that promote glioblastoma invasiveness.
A growing number of additional molecules have been implicated in invasion and are reviewed elsewhere (Nakada et al. 2007; Teodorczyk and Martin-Villalba 2010). Importantly, it will be critical to test candidate proinvasive targets and programs in physiologically relevant tumor models with aggressive growth behavior. Conventional malignant glioma cell lines do not tend to exhibit invasive growth in orthotopic xenograft models, but tumor-initiating cells not only invade (Singh et al. 2004; Wong et al. 2011), but also often phenocopy the type of invasion observed radiographically in patients from which the cells were derived (Wakimoto et al. 2012), thereby representing high-fidelity platforms to explore invasion (Bhat et al. 2011). Furthermore, the striking, but poorly understood, migratory capacity of neural and mesenchymal stem cells in transplant models (Carney and Shah 2011) affords another invasive model that can be used to interrogate this process.
Angiogenesis
Angiogenesis plays an important role in glioblastoma (Kargiotis et al. 2006), as evidenced by the presence of microvascular proliferation (Louis et al. 2007). The recent demonstration that therapeutic strategies to inhibit components that contribute to angiogenesis have been shown to have some efficacy in glioblastoma (Reardon et al. 2011b), including the recent approval of bevacizumab, provides the foundation for future studies in this disease.
Glioma cells require blood vessels for metabolic purposes, such as oxygen and nutrient delivery and waste removal, and also for the creation of a vascular niche that may selectively support glioma stem cells (Calabrese et al. 2007; Gilbertson and Rich 2007). The development of glioma vasculature may occur through several mechanisms (Carmeliet and Jain 2011a): angiogenesis, the formation of new blood vessels from the existing vasculature (Folkman 1971; Kerbel 2008); vasculogenesis, which involves the recruitment of bone marrow-derived endothelial progenitor cells (Patenaude et al. 2010); recruitment of tumor cells directly into the vascular wall; or the differentiation of tumor stem cells directly into vascular endothelium (Ricci-Vitiani et al. 2010; Wang et al. 2010). The end result of these blood vessel-producing processes is an intratumoral vasculature that is highly aberrant, incomplete, and tortuous (Long 1970), creating areas of hypoxia, acidosis, and peritumoral edema.
Blood vessel formation is regulated by a balance between pro- and anti-angiogenic molecules that comprise an angiogenic switch (Bergers and Benjamin 2003). VEGF, acting through VEGFR-2/KDR, is believed to be the central proangiogenic factor and is induced by hypoxia via HIF-1α (Shweiki et al. 1992; Kaur et al. 2005) and several mitogenic pathways that are dysregulated in glioblastoma (Maity et al. 2000; Pore et al. 2003; Watnick et al. 2003; Phung et al. 2006). VEGFR-2 activation then regulates endothelial cell survival, proliferation, migration, and permeability (Hicklin and Ellis 2005). In addition to VEGF, there are a large number of other factors that stimulate angiogenesis in glioblastoma (Carmeliet and Jain 2011b), including PDGF, FGF, the ANG/TIE system, Notch signaling, integrins, ephrins, IL-8, and SDF-1α (Carmeliet and Jain 2011a; Weis and Cheresh 2011). These proangiogenic mediators are opposed by anti-angiogenic factors, including angiostatin, thrombospondins, endostatin, tumstatin, and interferons (Nyberg et al. 2005). When stimulatory factors outweigh inhibitory factors, the angiogenic switch favors blood vessel creation.
Several types of angiogenesis inhibitors have been developed for therapeutic use. Because it is the main driver of angiogenesis, most approaches target VEGF signaling by interfering with either the VEGF ligand (Vredenburgh et al. 2007b), its receptor (VEGFR-2/KDR) (Batchelor et al. 2010), or its downstream signaling cascade. The anti-VEGF antibody bevacizumab is now used heavily in the clinic after undergoing accelerated approval by the FDA for use in recurrent glioblastoma (Cohen et al. 2009). Two initial phase II trials of bevacizumab and irinotecan demonstrated a 60% radiographic response and an apparent doubling of 6-mo progression-free survival (PFS; 38%–46%) and median survival (40–42 wk) compared with historical controls (9%–15% and 22–26 wk, respectively) (Vredenburgh et al. 2007a,b). In addition, two follow-up phase II trials demonstrated a radiographic response (27%–38%), increased PFS (29%–50%), and a slightly prolonged overall survival (31–35 wk) compared with historic controls (Friedman et al. 2009; Kreisl et al. 2009); phase III trials, including the RTOG-0825 study, are currently under way (http://www.clinicaltrials.gov). Additional approaches, such as angiopoietin inhibitors, are reviewed elsewhere (Norden et al. 2008a; Reardon et al. 2011b).
However, several unanswered clinical questions remain that require an improved understanding of the basic mechanisms involved in glioma angiogenesis (Verhoeff et al. 2009). For example, anti-angiogenic agents reduce vasogenic edema and corticosteroid requirements by decreasing vascular permeability (Gerstner et al. 2009), thereby dramatically modifying the MRI appearance of glioblastoma (Wen et al. 2010). However, it is unclear whether these changes result from “vascular normalization” (Jain 2005; Carmeliet and Jain 2011b), actions on tumor cells, or other alterations in the blood–brain barrier (Bechmann et al. 2007). In addition, patients on these agents tend to progress rapidly when disease recurs after treatment with anti-angiogenic agents (Ellis and Hicklin 2008), with minimal response to subsequent chemotherapy (Quant et al. 2009), underscoring the importance of understanding the basis for treatment resistance. Due to compensatory up-regulation of alternative angiogenic (Batchelor et al. 2007; Sathornsumetee and Rich 2007) or vasculogenic (Du et al. 2008) pathways, combinations of inhibitors that target nonredundant vascular pathways may be necessary. In addition, there is some evidence that angiogenesis inhibitors may promote infiltrative glioma growth (Norden et al. 2008b; Iwamoto et al. 2009; Narayana et al. 2012). An improved understanding of the basic biology of angiogenesis, the mechanisms of resistance to inhibitors, and the contributions of tumor-initiating cell-derived vasculature to the microenvironment will be necessary to optimize angiogenesis inhibition as a therapeutic approach in glioblastoma.
Glioblastoma ontogeny: cellular origins and tumor-initiating cells
Cellular origins
The cellular origins of malignant gliomas continue to be a source of debate. As in other cancers, the continued interest in glioma ontogeny is stimulated by the possibility that an improved understanding of the normal cell of origin will help identify fundamental pathways and lineage dependencies that could represent novel diagnostic and therapeutic targets (Visvader 2011). Specifically, numerous studies in genetically engineered mouse models have provided evidence that gliomas arise from the normal reservoirs of cycling stem and progenitor cells within the brain, and numerous genetically engineered mouse models have supported this idea in that a diverse range of glioma-relevant mutations targeted to neural stem cells in vivo readily produce gliomas with high fidelity and penetrance (Bachoo et al. 2002; Zhu et al. 2005; Zheng et al. 2008). For instance, tamoxifen-inducible Cre-recombinase-mediated inactivation of p53 and NF1 in adult stem cells drove the consistent formation of astrocytomas (Alcantara Llaguno et al. 2009; Wang et al. 2009); interestingly, these tumors were not restricted to the SVZ neural stem cell niche location but were found within multiple other brain regions. This finding was extended by C Liu et al. (2011) using the mosaic analysis with double markers (MADM) technique (Zong et al. 2005). This approach enabled careful longitudinal, lineage tracing analysis of developing astrocytomas in mice with mosaic p53/NF1 mutant neural stem cells, which showed that the accelerated phase of tumor growth occurs not in the original cell of mutation, but within OLIG2+ oligodendroglial progenitor cells (OPCs) that migrate away from the niche zones (C Liu et al. 2011). The finding that “astrocytomas” can arise from OPCs in mice could help to explain the long-standing observation that human glioblastoma and all other astrocytomas are characterized by both expression of oligodendroglial markers to an even greater degree than astrocytic markers (Ligon et al. 2004; Phillips et al. 2006; Verhaak et al. 2010) and their dependency on the same lineage factors (e.g., OLIG2) as normal counterparts (Ligon et al. 2007). Distributed progenitor cells, such as OPCs, actually represent the largest pool of cycling cells in the brain and are defined by expression of OLIG2 and NG2 (Dawson et al. 2003; Geha et al. 2010). Targeting of these nonstem cell progenitors through transgenic or viral approaches has been shown to lead to malignant astrocytomas or oligodendrogliomas, depending on the combination of mutations and cell types targeted (Geha et al. 2010; Persson et al. 2010).
In addition, work in genetically engineered mice has demonstrated that gliomas may also arise from terminally differentiated cells, likely through a process of dedifferentiation. Bachoo et al. (2002) demonstrated that cultured mature Ink4a/Arf− astrocytes stimulated with EGF or expressing mutant EGFRvIII dedifferentiated to nestin-expressing progenitor cells capable of forming high-grade gliomas in vivo, providing evidence that both progenitor and lineage-restricted, mature cells were permissive contexts for glioma formation. Moreover, PDGF has also been shown to induce the dedifferentiation of mature astrocytes and induce glioma formation in GFAP-expressing Ink4a/Arf− cells in vivo (Dai et al. 2001). Together, these studies emphasize the influence of particular combinations of dysregulated pathways on glioma development and highlight the notion that specific genetic alterations, and not just the precise a priori developmental state of the cell, help to create permissive contexts for glioma formation.
Although murine studies have been helpful in clarifying glioma origins, it remains to be seen whether their findings can be fully extrapolated to human disease. For instance, human and mouse OPCs may be biologically similar, as recent work demonstrated conserved mechanisms for oligodendroglioma formation through disruption of asymmetric division of NG2+ OPCs in mouse verbB/p53+/−-induced oligodendrogliomas and human oligodendrogliomas (Sugiarto et al. 2011). However, in adult mice, the SVZ is a prominent regenerative zone, but the analogous region in adult human brains, the subventricular astrocyte ribbon, may not harbor similar numbers or types of stem cells (Sanai et al. 2011). In addition, the plasticity of cells in response to engineered oncogene and tumor suppressor alterations is well established and thus can confound deconvolution of tumor-initiating populations from established tumors. For instance, studies of the effects of Smo activation on development of a mouse model of medulloblastoma suggested that the same mutation at multiple stages of lineage commitment could result in dedifferentiation of committed cells to a common phenotypic end point, thereby obscuring the original origin of the tumors (Schuller et al. 2008). It will be important to continue to model human disease in mice given the powerful technologies enabling sophisticated genetic targeting. These approaches will be invaluable to explore clinical observations such as the distinct differences in location between IDH1 mutant and wild-type glioblastomas (Lai et al. 2011). Given the diverse number of histological subtypes, various subsets of molecular patterns and subclasses, and increasingly broad number of stem and progenitor cells in the brain, further studies will be required to clarify the origins of malignant gliomas and the programs that drive their subsequent evolution.
Tumor-initiating cells
Complementing these studies of the cell of origin in glioblastoma, several groups have shown that different cell populations within a tumor exhibit varied tumor-forming capacity. These observations suggest that stem/progenitor cells may serve as the cell of origin for many tumors. Irrespective of the ontogeny argument, however, the finding that distinct cellular subpopulations within glioblastomas harbor potent tumor-initiating capacity was yet another example that heterogeneity—morphological, molecular, and cellular—is the rule, rather than the exception, in this group of diseases. First described by several groups (Ignatova et al. 2002; Hemmati et al. 2003; Singh et al. 2003), the finding that tumor-initiating cancer cells can be isolated from glioblastomas and other cancers and propagated as neurospheres when grown in serum-free neural stem cell conditions, have self-renewal capacity, and recapitulate malignant glioma behavior in vivo (Singh et al. 2004) has transformed glioblastoma research. Moreover, these cells in malignant gliomas now represent well-characterized experimental models for the investigation of the cancer stem cell hypothesis (Dirks 2010), since these cells represent highly faithful glioma models when cultured as tumor-initiating cell lines or orthotopic xenografts. For instance, culturing tumor specimens in stem cell conditions has enabled the maintenance of lines carrying glioma-specific lesions such as EGFRvIII (Stockhausen et al. 2011) or mutant IDH1 expression (Luchman et al. 2012) that were not well-modeled under serum-containing conditions. In addition, abundant evidence confirms that these models recapitulate the phenotypes as well as genomic alterations found in patient tumors (Lee et al. 2006a; Wakimoto et al. 2012). The glioma stem cell hypothesis (Dirks 2010; Taylor et al. 2010; Lathia et al. 2011a) incorporates a model in which these tumor-initiating cells are the principal drivers of gliomagenesis. Specifically, these cells are thought to represent transformed neural progenitors that give rise to tumor cell progeny and also have the capacity to self-renew. Considering the high malignant potential of the tumor-initiating cell population, this hierarchical model implies that this subset, which may represent a minority of cells within the bulk tumor mass, is the critical subpopulation of glioblastoma that could be highly relevant clinically due to the potential for intrinsic therapeutic resistance (Bao et al. 2006). In contrast, an alternative view of gliomagenesis involves a less hierarchical model in which most glioblastoma cells are functionally and developmentally similar and have developed through a progressive process of clonal selection of transformed somatic cells. In this case, there may be a higher proportion of the tumor mass with malignant potential than in the stem cell model. Further work will be important to discern the physiological relevance and potential translational impact of these not mutually exclusive models.
Although the tumor-initiating cell subpopulation has been traditionally defined by the cell surface expression of CD133, it has become clear that this marker does not exclusively define tumor-initiating populations (Ogden et al. 2008; Chen et al. 2010; Beier and Beier 2011), and thus further work correlating cell surface markers (Lathia et al. 2011b) with phenotype is necessary to more clearly define these cells. Moreover, while substantial work has begun to clarify the programs—including PLAGL2, Olig2, and c-Myc (Ligon et al. 2007; Zheng et al. 2008, 2010; Guryanova et al. 2011)—that drive and maintain tumor-initiating characteristics, additional study is needed regarding the molecular pathways underlying cellular phenotype and hierarchies. Also, the degree to which these cells strictly preserve the phenotypes of all genotypes of original patient tumors remains to be examined in large numbers of patients in detail; reports of variation with respect to preservation of RTK, bias toward tumors with PTEN inactivation, and difficulties in modeling the vascular changes seen in human glioblastoma suggest that careful attention to the unique properties of each line will be required (Chen et al. 2010). Nevertheless, the use of glioblastoma tumor-initiating cell models in the basic research setting is now routine, and numerous emerging preclinical studies using patient-derived models will provide clues as to whether these lines gathered into patient cohorts might advance drug screening efforts (Pollard et al. 2009).
Future directions: areas of further study
Identifying dependencies and targets
Despite the tremendous progress in our understanding of the genetic basis of glioblastoma, targeted therapeutic approaches based on known genomic alterations have not proved to be efficacious to date (De Witt Hamer 2010). It is likely that intratumoral heterogeneity and target cooperativity (Stommel et al. 2007) conspire to create a “multiple dependency” state wherein single-target inhibitors are not sufficient to attenuate tumor growth. Moreover, transcriptionally defined subclasses may harbor unique dependencies. In addition, we have an incomplete understanding of the functional consequences of many of the genes found mutated in glioblastoma. Indeed, it will be necessary to approach functional space in glioblastoma in the systematic manner with which the structural space has been clarified (Boehm and Hahn 2011). To this end, genome-wide tools capable of loss- and gain-of-function studies will likely be invaluable in interrogating phenotypes particularly relevant to glioblastoma. Large-scale pooled shRNA screening of a panel of glioblastoma cell lines as part of a larger panel of multiple lineages led to the identification of a list of candidate dependencies (Cheung et al. 2011). These approaches should use panels of highly faithful models—murine and human—that recapitulate the salient in vivo behaviors of interest. In addition to survival, phenotypes such as invasion should also be assessed. Systematic approaches may also be useful in revealing synthetic-lethal relationships (Barbie et al. 2009), such as the role of ENTPD5 in the context of PTEN loss (Fang et al. 2010). In addition to systematic approaches, the computational methodologies involving gene expression data that have pointed to important biological roles for STAT3 (Carro et al. 2010; Bhat et al. 2011) and the Hippo pathways, among others, have shown great promise and should be explored further in preclinical contexts.
Understanding malignant transformation
The phenomenon of transformation from grade II glioma to malignant disease is poorly understood yet accounts for the mortality of low-grade disease. These patients are younger at diagnosis, and it is thought that 45%–70% will undergo disease progression to histological malignancy within 5 years (Ohgaki and Kleihues 2005). Although the entity of secondary glioblastoma represents <10% of all diagnosed glioblastomas (Ohgaki and Kleihues 2009), the existence of histologically proven antecedent lower-grade tissue presents a unique and as yet relatively underexploited opportunity to understand the programs that drive malignancy and also to explore the possibility of delaying clinical transformation. The study of cancers that progress from lower-grade lesions to higher-grade malignancies has provided critical insights into the genetic basis of cancer evolution. Specifically, analysis of the molecular changes along the histopathological spectrum in colorectal cancer (Vogelstein et al. 1988) pioneered comparative analyses of lower-grade lesions and their higher-grade counterparts. Together, the dissection of the chronological genomic events that drive disease progression not only provides invaluable fundamental mechanistic information on a cancer's natural history, but also may identify potential therapeutic targets that drive the transition from less aggressive to more malignant disease. Especially considering the comparatively young age of presentation in low-grade patients, the benefit to interrupting or delaying the transformation pathway would be significant. Moreover, despite the clear differences between IDH1 mutant secondary glioblastoma and IDH1 wild-type primary disease, the isolation of transformation programs identified by studying low-grade and matched malignant disease could be relevant to the programs, such as those driving angiogenesis, that contribute to the biology of primary disease. Earlier studies comparing primary and secondary glioblastomas (Maher et al. 2006; Tso et al. 2006), characterizing the expression of DCC in low- and high-grade tumors (Reyes-Mugica et al. 1997), and performing comparative array CGH between a small set of grade II/IV gliomas (Idbaih et al. 2008) point to the differences in these tumors.
Understanding the extent and impact of heterogeneity
The morphological heterogeneity that prompted the original description of high-grade glioma as “glioblastoma multiforme” (Mallory 1914; Bailey and Cushing 1926) has also extended to the molecular level. Through studies demonstrating variable intratumoral distributions of EGFRvIII-expressing cells (Inda et al. 2010) as well as mutually exclusive SCNAs within distinct populations of the same tumor (Snuderl et al. 2011; Szerlip et al. 2012), among others, we are beginning to have a better understanding of how molecular heterogeneity is manifest. While these studies illustrate examples of distinct alterations occurring in unique cells, there are also examples of concomitant kinase activation within the same cell (Stommel et al. 2007). In both cases, it is not entirely clear whether intermingled cell populations with unique genomic alterations behave as independent tumors or are interdependent with each other. Ultimately, it will be necessary to study the populations of a heterogeneous tumor mass at the single-cell level in order to understand co-occurring genetic alterations and accompanying protein activation states. Developing technology capable of assessing single-cell-scale genomes, isolated through laser capture microscopy or live cell-sorting techniques, is making these approaches feasible (Navin and Hicks 2011; Navin et al. 2011). Moreover, in addition to understanding the molecular basis of tumor cell heterogeneity, it will be critical to consider the contributions and phenotypes of the diverse nontumor cell types, such as stromal and immune/inflammatory cells (Dunn et al. 2007), that populate the glioma microenvironment.
Developmental oncobiology
Given the growing prominence of stem and progenitor cell biology in glioblastoma, further characterization of the heterogeneous cell populations found within these tumors will be critical in understanding their origins and evolution. Although it is clear that defects in developmental hierarchy play a key role in many tumors, further work is necessary to determine the molecular basis underlying the differentiation potential of the cells within each tumor. Specifically, comprehensive genomic, epigenomic, and transcriptional characterization of carefully isolated progenitor cells from tumors compared with their nonneoplastic counterparts may clarify the structural basis of particular cell states, whether there are distinct differences across the transcriptomic subtypes in glioblastoma, and how the neoplastic context influences phenotypes such as invasion and migration, resistance to therapy, and signaling dysregulation. Furthermore, these types of comparative studies may enable the development of therapeutic approaches that reduce the tumor-initiating/tumor-propagating capacities of glioma tumor-initiating cells by interrupting the cellular programs that likely contribute to their aggressiveness, such as self-renewal, maintenance of multipotentiality, and the phenotypic plasticity to dedifferentiate.
Therapeutic resistance
Given the inevitable progression of glioblastoma following standard-of-care surgery, temozolomide treatment, and radiation therapy (Stupp et al. 2005), it is likely that both intrinsic and acquired mechanisms of resistance play significant roles in glioblastoma. Currently, most patients continue to receive temozolomide as the main chemotherapeutic agent in their treatment regimen. In addition to the de novo resistance mechanism of MGMT hypomethlyation (Hegi et al. 2005), several groups have described additional mechanisms of treatment resistance that include loss of the mismatch repair gene MSH6 (Cahill et al. 2007), silencing of the base excision repair enzyme alkylpurine-DNA-N-glycosylase (Agnihotri et al. 2012), and also overexpression of both inhibitor of apoptosis proteins (Ziegler et al. 2008) and multidrug resistance protein family members (Shervington and Lu 2008). Although perturbation of recurrent resistance mechanisms could enhance the therapeutic efficacy of this current workhorse agent, these studies point to the broader pivotal framework of carefully studying pre- and post-intervention tumor samples, as has been demonstrated in other cancers. Comparative post-treatment annotation will be important for additional agents, such as bevacizumab, and perhaps every emerging targeted agent that enters clinical trials. This approach will also facilitate our understanding of acquired resistance and/or de novo insensitivity (Bao et al. 2006) to radiation therapy, as the vast majority of post-treatment tissue has been subjected to up-front radiation treatment. We anticipate that components of the tumor microenvironment may also contribute to resistance in a tumor cell-extrinsic manner. Specifically, the blood–brain barrier, formed by intracerebral capillary tight junctions and astrocytic foot processes, impairs the passive transit of molecules between the vascular lumen and brain parenchyma (Huber et al. 2001) and is highly dysregulated in glioblastoma (Long 1970). This structure is known to present a challenge to effective transit of therapeutic molecules to the brain, and thus continued study of its biology in both the normal and tumor settings could enhance the efficacy of drug delivery to glioblastoma.
Toward a molecular classification of glioma
The comprehensive molecular characterization of gliomas is now starting to transform their classification (Table 1), which currently follows the consensus WHO histopathological criteria instrumental in standardizing pathological classification of these challenging tumors (Louis et al. 2007). However, a high percentage of gliomas, such as mixed oligoastrocytomas and lower-grade gliomas, remain difficult to categorize reproducibly due to considerable histological overlap. Genomic approaches applied to clinically characterized patient cohorts now clearly show that combined molecular and histological classification offers great opportunities to significantly improve clinical predictive power over use of histology alone, even for individual patients. A clear example is the group including oligodendrogliomas, grade II and III astrocytomas, and mixed gliomas, which are best defined as a subset of gliomas harboring shared genomic features of mutations in IDH genes and are G-CIMP+ and MGMT methylated due to globally increased levels of methylation (Noushmehr et al. 2010).
Further subclassification of this group will likely use unique lesions in oligodendroglioma, such as 1p/19q whole-arm codeletion/translocation, CIC mutations, and 1p or 19q polysomy (Snuderl et al. 2009; Bettegowda et al. 2011; Yip et al. 2012), in the presence or absence of TP53 mutations to enable reliable identification of a group in a manner not possible by histology alone. Likewise, BRAF fusions (Horbinski et al. 2010; Hawkins et al. 2011) and V600E mutants characterize additional astrocytoma subsets with clinical relevance (Dias-Santagata et al. 2011; Schindler et al. 2011; Tian et al. 2011). In contrast to the mutant IDH/G-CIMP+ tumors, glioblastomas appear surprisingly distinct but significantly more complex genomically, making diagnostic subclassification schemes more challenging. Deeper sequencing efforts have yet to be fully completed and may yet reveal novel recurrent mutations that draw together what appears now to be a highly diverse group of tumors. Clearly, mutant IDH will be critical in discerning these diseases, and translation of expression profiling work into diagnostic tools is also under way (Colman et al. 2010). Additional diagnostic scenarios, such as distinction of pseudoprogression versus true tumor progression, will hopefully benefit from molecular pathology approaches. The rapid pace of genomic and epigenomic discovery in gliomas will likely necessitate the development of consensus mechanisms to deploy integrated molecular and histologic data to clinical practice to improve diagnosis and predictive capability and stratify response to therapy.
Conclusion
The commitment to genomic characterization of glioblastoma has fueled substantial progress in our understanding of this cancer in the last 5 years. The heterogeneous nature of this neoplasm, both within and across tumors, underscores the difficulty in developing efficacious treatments and provides a challenge both to annotate tumors to understand the disease and to stratify patients for trials and treatment. Already there are several efforts to connect the information obtained from retrospective, population-based genomic characterization to the individual patient in the form of multiplexed patient profiling on clinical samples (MacConaill et al. 2009; Sequist et al. 2011). Further work will be necessary to fully understand glioblastoma biology and will increasingly require integrated studies, including genomics, faithful animal models of disease (Lim et al. 2011; Wee et al. 2012), and the careful study of human tissue.
Acknowledgments
We thank Tanaz Sharifinia and Ravindra Uppaluri for comments on the manuscript. This work was supported by NIH Grants PO1 CA117969, RC2 CA148268, and PO1 CA142536 (to W.C.H., K.L., L.C., and R.A.D.); the Ben and Catherine Ivy Foundation (to L.C., R.A.D., and K.L.); an award from the Goldhirsh Foundation (to F.F. and K.L.); NIH (NINDS) Fellowship Award F32 NS066519 (to J.W.); NIH Grant P01 CA95616 (to C.B, K.L., L.C., W.C.H., W.K.C., F.F., and R.A.D.); NIH Grant NCI-MMHCC-CA84313-06 (to L.C. and R.A.D.); NIH Grant K08-NS062907 (to M.G.C.); an award from the Sontag Foundation (to K.L.); an ABTA Basic Research Fellowship (to G.P.D.); NINDS R25 award (to G.P.D.); and the Brain Science Foundation (to I.F.D.). W.K.C. is a fellow of the National Foundation for Cancer Research.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.187922.112.
References
- Agnihotri S, Gajadhar AS, Ternamian C, Gorlia T, Diefes KL, Mischel PS, Kelly J, McGown G, Thorncroft M, Carlson BL, et al. 2012. Alkylpurine-DNA-N-glycosylase confers resistance to temozolomide in xenograft models of glioblastoma multiforme and is associated with poor survival in patients. J Clin Invest 122: 253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhavan D, Cloughesy TF, Mischel PS 2010. mTOR signaling in glioblastoma: Lessons learned from bench to bedside. Neuro Oncol 12: 882–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama Y, Jung S, Salhia B, Lee S, Hubbard S, Taylor M, Mainprize T, Akaishi K, van Furth W, Rutka JT 2001. Hyaluronate receptors mediating glioma cell migration and proliferation. J Neurooncol 53: 115–127 [DOI] [PubMed] [Google Scholar]
- Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, Parada LF 2009. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 15: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O'Donnell P, Grigoriadis A, Diss T, et al. 2011a. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol 224: 334–343 [DOI] [PubMed] [Google Scholar]
- Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F, McCarthy S, Fantin VR, Straley KS, Lobo S, et al. 2011b. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet 43: 1262–1265 [DOI] [PubMed] [Google Scholar]
- Assanah M, Lochhead R, Ogden A, Bruce J, Goldman J, Canoll P 2006. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J Neurosci 26: 6781–6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assanah MC, Bruce JN, Suzuki SO, Chen A, Goldman JE, Canoll P 2009. PDGF stimulates the massive expansion of glial progenitors in the neonatal forebrain. Glia 57: 1835–1847 [DOI] [PubMed] [Google Scholar]
- Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, et al. 2002. Epidermal growth factor receptor and Ink4a/Arf: Convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1: 269–277 [DOI] [PubMed] [Google Scholar]
- Bailey P, Cushing H 1926. A classification of the tumors of the glioma group on a histogenetic basis with a correlated study of prognosis. Lippincott, Philadelphia [Google Scholar]
- Bajenaru ML, Zhu Y, Hedrick NM, Donahoe J, Parada LF, Gutmann DH 2002. Astrocyte-specific inactivation of the neurofibromatosis 1 gene (NF1) is insufficient for astrocytoma formation. Mol Cell Biol 22: 5100–5113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajenaru ML, Hernandez MR, Perry A, Zhu Y, Parada LF, Garbow JR, Gutmann DH 2003. Optic nerve glioma in mice requires astrocyte Nf1 gene inactivation and Nf1 brain heterozygosity. Cancer Res 63: 8573–8577 [PubMed] [Google Scholar]
- Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A 2008. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol 116: 597–602 [DOI] [PubMed] [Google Scholar]
- Banerjee S, Byrd JN, Gianino SM, Harpstrite SE, Rodriguez FJ, Tuskan RG, Reilly KM, Piwnica-Worms DR, Gutmann DH 2010. The neurofibromatosis type 1 tumor suppressor controls cell growth by regulating signal transducer and activator of transcription-3 activity in vitro and in vivo. Cancer Res 70: 1356–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Crouse NR, Emnett RJ, Gianino SM, Gutmann DH 2011. Neurofibromatosis-1 regulates mTOR-mediated astrocyte growth and glioma formation in a TSC/Rheb-independent manner. Proc Natl Acad Sci 108: 15996–16001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN 2006. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444: 756–760 [DOI] [PubMed] [Google Scholar]
- Bar EE, Lin A, Tihan T, Burger PC, Eberhart CG 2008. Frequent gains at chromosome 7q34 involving BRAF in pilocytic astrocytoma. J Neuropathol Exp Neurol 67: 878–887 [DOI] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. 2009. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A 2005. Reverse engineering of regulatory networks in human B cells. Nat Genet 37: 382–390 [DOI] [PubMed] [Google Scholar]
- Batchelor TT, Sorensen AG, di Tomaso E, Zhang WT, Duda DG, Cohen KS, Kozak KR, Cahill DP, Chen PJ, Zhu M, et al. 2007. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell 11: 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor TT, Duda DG, di Tomaso E, Ancukiewicz M, Plotkin SR, Gerstner E, Eichler AF, Drappatz J, Hochberg FH, Benner T, et al. 2010. Phase II study of cediranib, an oral pan-vascular endothelial growth factor receptor tyrosine kinase inhibitor, in patients with recurrent glioblastoma. J Clin Oncol 28: 2817–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle C, Assanah MC, Monzo P, Vallee R, Rosenfeld SS, Canoll P 2008. The role of myosin II in glioma invasion of the brain. Mol Biol Cell 19: 3357–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechmann I, Galea I, Perry VH 2007. What is the blood–brain barrier (not)? Trends Immunol 28: 5–11 [DOI] [PubMed] [Google Scholar]
- Beier CP, Beier D 2011. CD133 negative cancer stem cells in glioblastoma. Front Biosci 3: 701–710 [DOI] [PubMed] [Google Scholar]
- Bello L, Francolini M, Marthyn P, Zhang J, Carroll RS, Nikas DC, Strasser JF, Villani R, Cheresh DA, Black PM 2001a. αvβ3 and αvβ5 integrin expression in glioma periphery. Neurosurgery 49: 380–390 [DOI] [PubMed] [Google Scholar]
- Bello L, Lucini V, Carrabba G, Giussani C, Machluf M, Pluderi M, Nikas D, Zhang J, Tomei G, Villani RM, et al. 2001b. Simultaneous inhibition of glioma angiogenesis, cell proliferation, and invasion by a naturally occurring fragment of human metalloproteinase-2. Cancer Res 61: 8730–8736 [PubMed] [Google Scholar]
- Bergers G, Benjamin LE 2003. Tumorigenesis and the angiogenic switch. Nat Rev Cancer 3: 401–410 [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. 2007. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci 104: 20007–20012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettegowda C, Agrawal N, Jiao Y, Sausen M, Wood LD, Hruban RH, Rodriguez FJ, Cahill DP, McLendon R, Riggins G, et al. 2011. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 333: 1453–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, James JD, Gumin J, Diefes KL, Kim SH, et al. 2011. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev 25: 2594–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernat W, Huang H, Yokoo H, Kleihues P, Ohgaki H 2004. Predominant expression of mutant EGFR (EGFRvIII) is rare in primary glioblastomas. Brain Pathol 14: 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleau AM, Hambardzumyan D, Ozawa T, Fomchenko EI, Huse JT, Brennan CW, Holland EC 2009. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell 4: 226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm JS, Hahn WC 2011. Towards systematic functional characterization of cancer genomes. Nat Rev Genet 12: 487–498 [DOI] [PubMed] [Google Scholar]
- Boerner JL, Demory ML, Silva C, Parsons SJ 2004. Phosphorylation of Y845 on the epidermal growth factor receptor mediates binding to the mitochondrial protein cytochrome c oxidase subunit II. Mol Cell Biol 24: 7059–7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonavia R, Inda MM, Vandenberg S, Cheng SY, Nagane M, Hadwiger P, Tan P, Sah DW, Cavenee WK, Furnari FB 2011. EGFRvIII promotes glioma angiogenesis and growth through the NF-κB, interleukin-8 pathway. Oncogene doi: 10.1038/onc.2011.563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, Schenkein DP, Hezel AF, Ancukiewicz M, Liebman HM, et al. 2012. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 17: 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon LY 2008. Hyaluronan-mediated CD44 activation of RhoGTPase signaling and cytoskeleton function promotes tumor progression. Semin Cancer Biol 18: 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, Sikic BI 2005. High-resolution genome-wide mapping of genetic alterations in human glial brain tumors. Cancer Res 65: 4088–4096 [DOI] [PubMed] [Google Scholar]
- Bredel M, Scholtens DM, Harsh GR, Bredel C, Chandler JP, Renfrow JJ, Yadav AK, Vogel H, Scheck AC, Tibshirani R, et al. 2009. A network model of a cooperative genetic landscape in brain tumors. JAMA 302: 261–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, Yu IL, Carro MS, Dai F, Tagge MJ, et al. 2011. NFKBIA deletion in glioblastomas. N Engl J Med 364: 627–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C 2011. Genomic profiles of glioma. Curr Neurol Neurosci Rep 11: 291–297 [DOI] [PubMed] [Google Scholar]
- Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, Holland E 2009. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS ONE 4: e7752 doi: 10.1371/journal.pone.0007752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger PC, Heinz ER, Shibata T, Kleihues P 1988. Topographic anatomy and CT correlations in the untreated glioblastoma multiforme. J Neurosurg 68: 698–704 [DOI] [PubMed] [Google Scholar]
- Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB, Batchelor TT, Futreal PA, Stratton MR, Curry WT, et al. 2007. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res 13: 2038–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR, Ino Y, et al. 1998. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst 90: 1473–1479 [DOI] [PubMed] [Google Scholar]
- Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, et al. 2007. A perivascular niche for brain tumor stem cells. Cancer Cell 11: 69–82 [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capper D, Zentgraf H, Balss J, Hartmann C, von Deimling A 2009. Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol 118: 599–601 [DOI] [PubMed] [Google Scholar]
- Capper D, Simon M, Langhans CD, Okun JG, Tonn JC, Weller M, Deimling AV, Hartmann C 2011. 2-Hydroxyglutarate concentration in serum from patients with gliomas does not correlate with IDH1/2 mutation status or tumor size. Int J Cancer doi: 10.1002/ijc.26425 [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Jain RK 2011a. Molecular mechanisms and clinical applications of angiogenesis. Nature 473: 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Jain RK 2011b. Principles and mechanisms of vessel normalization for cancer and other angiogenic diseases. Nat Rev Drug Discov 10: 417–427 [DOI] [PubMed] [Google Scholar]
- Carney BJ, Shah K 2011. Migration and fate of therapeutic stem cells in different brain disease models. Neuroscience 197: 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. 2010. The transcriptional network for mesenchymal transformation of brain tumours. Nature 463: 318–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CBTRUS. 2011. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2004–2007. Central Brain Tumor Registry of the United States, Hinsdale, IL. [Google Scholar]
- Chabner BA 2011. Early accelerated approval for highly targeted cancer drugs. N Engl J Med 364: 1087–1089 [DOI] [PubMed] [Google Scholar]
- Chamberlain MC 2011. Radiographic patterns of relapse in glioblastoma. J Neurooncol 101: 319–323 [DOI] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. 2011. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 364: 2507–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, Greve JM, Soriano RH, Gilmour LL, Rivers CS, et al. 2010. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell 17: 362–375 [DOI] [PubMed] [Google Scholar]
- Chetty C, Vanamala SK, Gondi CS, Dinh DH, Gujrati M, Rao JS 2012. MMP-9 induces CD44 cleavage and CD44 mediated cell migration in glioblastoma xenograft cells. Cell Signal 24: 549–559 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, et al. 2011. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci 108: 12372–12377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi AS, Batchelor TT, Kwak EL, Clark JW, Wang DL, Wilner KD, Louis DN, Iafrate AJ 2012. Rapid radiographic and clinical improvement after treatment of a MET-amplified recurrent glioblastoma with a mesenchymal–epithelial transition inhibitor. J Clin Oncol 30: e30–e33 doi: 10.1200/JCO.2011.38.4586 [DOI] [PubMed] [Google Scholar]
- Chin L, Hahn WC, Getz G, Meyerson M 2011. Making sense of cancer genomic data. Genes Dev 25: 534–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LM, Endersby R, Zhu X, Rankin S, Qu C, Zhang J, Broniscer A, Ellison DW, Baker SJ 2011. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell 19: 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. 2011. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep 12: 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JG, Zou HY, Arango ME, Li Q, Lee JH, McDonnell SR, Yamazaki S, Alton GR, Mroczkowski B, Los G 2007. Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol Cancer Ther 6: 3314–3322 [DOI] [PubMed] [Google Scholar]
- Clarke ID, Dirks PB 2003. A human brain tumor-derived PDGFR-α deletion mutant is transforming. Oncogene 22: 722–733 [DOI] [PubMed] [Google Scholar]
- Cloughesy TF, Yoshimoto K, Nghiemphu P, Brown K, Dang J, Zhu S, Hsueh T, Chen Y, Wang W, Youngkin D, et al. 2008. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med 5: e8 doi: 10.1371/journal.pmed.0050008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MH, Shen YL, Keegan P, Pazdur R 2009. FDA drug approval summary: Bevacizumab (Avastin) as treatment of recurrent glioblastoma multiforme. Oncologist 14: 1131–1138 [DOI] [PubMed] [Google Scholar]
- Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A, Popoff S, Nutt CL, Louis DN, Cairncross JG, et al. 2010. A multigene predictor of outcome in glioblastoma. Neuro Oncol 12: 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Abaco GM, Kaye AH 2007. Integrins: Molecular determinants of glioma invasion. J Clin Neurosci 14: 1041–1048 [DOI] [PubMed] [Google Scholar]
- Daginakatte GC, Gutmann DH 2007. Neurofibromatosis-1 (Nf1) heterozygous brain microglia elaborate paracrine factors that promote Nf1-deficient astrocyte and glioma growth. Hum Mol Genet 16: 1098–1112 [DOI] [PubMed] [Google Scholar]
- Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC 2001. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev 15: 1913–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R, Huang S 2007. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene 26: 6212–6219 [DOI] [PubMed] [Google Scholar]
- Dai B, Pieper RO, Li D, Wei P, Liu M, Woo SY, Aldape KD, Sawaya R, Xie K, Huang S 2010. FoxM1B regulates NEDD4-1 expression, leading to cellular transformation and full malignant phenotype in immortalized human astrocytes. Cancer Res 70: 2951–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandy W 1928. Removal of right cerebral hemisphere for certain tumors with hemiplegia. JAMA 90: 823–825 [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. 2010. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 465: 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FG, Kupelian V, Freels S, McCarthy B, Surawicz T 2001. Prevalence estimates for primary brain tumors in the United States by behavior and major histology groups. Neuro Oncol 3: 152–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MR, Polito A, Levine JM, Reynolds R 2003. NG2-expressing glial progenitor cells: An abundant and widespread population of cycling cells in the adult rat CNS. Mol Cell Neurosci 24: 476–488 [DOI] [PubMed] [Google Scholar]
- De Raedt T, Walton Z, Yecies JL, Li D, Chen Y, Malone CF, Maertens O, Jeong SM, Bronson RT, Lebleu V, et al. 2011. Exploiting cancer cell vulnerabilities to develop a combination therapy for ras-driven tumors. Cancer Cell 20: 400–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai S, Pillai P, Win-Piazza H, Acevedo-Duncan M 2011. PKC-ι promotes glioblastoma cell survival by phosphorylating and inhibiting BAD through a phosphatidylinositol 3-kinase pathway. Biochim Biophys Acta 1813: 1190–1197 [DOI] [PubMed] [Google Scholar]
- Desgrosellier JS, Cheresh DA 2010. Integrins in cancer: Biological implications and therapeutic opportunities. Nat Rev Cancer 10: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Witt Hamer PC 2010. Small molecule kinase inhibitors in glioblastoma: A systematic review of clinical studies. Neuro Oncol 12: 304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias-Santagata D, Akhavanfard S, David SS, Vernovsky K, Kuhlmann G, Boisvert SL, Stubbs H, McDermott U, Settleman J, Kwak EL et al. 2010. Rapid targeted mutational analysis of human tumours: A clinical platform to guide personalized cancer medicine. EMBO Mol Med 2: 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias-Santagata D, Lam Q, Vernovsky K, Vena N, Lennerz JK, Borger DR, Batchelor TT, Ligon KL, Iafrate AJ, Ligon AH, et al. 2011. BRAF V600E mutations are common in pleomorphic xanthoastrocytoma: Diagnostic and therapeutic implications. PLoS ONE 6: e17948 doi: 10.1371/journal.pone.0017948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks PB 2010. Brain tumor stem cells: The cancer stem cell hypothesis writ large. Mol Oncol 4: 420–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rocco F, Carroll RS, Zhang J, Black PM 1998. Platelet-derived growth factor and its receptor expression in human oligodendrogliomas. Neurosurgery 42: 341–346 [DOI] [PubMed] [Google Scholar]
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, et al. 2001. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med 344: 1031–1037 [DOI] [PubMed] [Google Scholar]
- Du R, Lu KV, Petritsch C, Liu P, Ganss R, Passegue E, Song H, Vandenberg S, Johnson RS, Werb Z, et al. 2008. HIF1α induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell 13: 206–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Bernasconi P, Clauser KR, Mani DR, Finn SP, Beroukhim R, Burns M, Julian B, Peng XP, Hieronymus H, et al. 2009. Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nat Biotechnol 27: 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Dunn IF, Curry WT 2007. Focus on TILs: Prognostic significance of tumor infiltrating lymphocytes in human glioma. Cancer Immun 7: 12–28 [PMC free article] [PubMed] [Google Scholar]
- Dutil EM, Toker A, Newton AC 1998. Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1 (PDK-1). Curr Biol 8: 1366–1375 [DOI] [PubMed] [Google Scholar]
- Ekstrand AJ, James CD, Cavenee WK, Seliger B, Pettersson RF, Collins VP 1991. Genes for epidermal growth factor receptor, transforming growth factor α, and epidermal growth factor and their expression in human gliomas in vivo. Cancer Res 51: 2164–2172 [PubMed] [Google Scholar]
- Ellis LM, Hicklin DJ 2008. Pathways mediating resistance to vascular endothelial growth factor-targeted therapy. Clin Cancer Res 14: 6371–6375 [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 7: 606–619 [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. 2007. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Fan QW, Knight ZA, Goldenberg DD, Yu W, Mostov KE, Stokoe D, Shokat KM, Weiss WA 2006. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell 9: 341–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan QW, Cheng C, Knight ZA, Haas-Kogan D, Stokoe D, James CD, McCormick F, Shokat KM, Weiss WA 2009. EGFR signals to mTOR through PKC and independently of Akt in glioma. Sci Signal 2: ra4 doi: 10.1126/scisignal.2000014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan QW, Cheng C, Hackett C, Feldman M, Houseman BT, Nicolaides T, Haas-Kogan D, James CD, Oakes SA, Debnath J, et al. 2010. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal 3: ra81 doi: 10.1126/scisignal.2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Shen Z, Huang S, Zhao L, Chen S, Mak TW, Wang X 2010. The ER UDPase ENTPD5 promotes protein N-glycosylation, the Warburg effect, and proliferation in the PTEN pathway. Cell 143: 711–724 [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming TP, Saxena A, Clark WC, Robertson JT, Oldfield EH, Aaronson SA, Ali IU 1992. Amplification and/or overexpression of platelet-derived growth factor receptors and epidermal growth factor receptor in human glial tumors. Cancer Res 52: 4550–4553 [PubMed] [Google Scholar]
- Folkman J 1971. Tumor angiogenesis: Therapeutic implications. N Engl J Med 285: 1182–1186 [DOI] [PubMed] [Google Scholar]
- Fomchenko EI, Dougherty JD, Helmy KY, Katz AM, Pietras A, Brennan C, Huse JT, Milosevic A, Holland EC 2011. Recruited cells can become transformed and overtake PDGF-induced murine gliomas in vivo during tumor progression. PLoS ONE 6: e20605 doi: 10.1371/journal.pone.0020605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth PA, Wong H, Laing TD, Rewcastle NB, Morris DG, Muzik H, Leco KJ, Johnston RN, Brasher PM, Sutherland G, et al. 1999. Gelatinase-A (MMP-2), gelatinase-B (MMP-9) and membrane type matrix metalloproteinase-1 (MT1-MMP) are involved in different aspects of the pathophysiology of malignant gliomas. Br J Cancer 79: 1828–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, Mischel PS, Nelson SF 2004. Gene expression profiling of gliomas strongly predicts survival. Cancer Res 64: 6503–6510 [DOI] [PubMed] [Google Scholar]
- Friedman HS, Prados MD, Wen PY, Mikkelsen T, Schiff D, Abrey LE, Yung WK, Paleologos N, Nicholas MK, Jensen R, et al. 2009. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol 27: 4733–4740 [DOI] [PubMed] [Google Scholar]
- Gallia GL, Rand V, Siu I-M, Eberhart CG, James CD, Marie SKN, Oba-Shinjo SM, Carlotti CG, Caballero OL, Simpson AJG, et al. 2006. PIK3CA gene mutations in pediatric and adult glioblastoma multiforme. Mol Cancer Res 4: 709–714 [DOI] [PubMed] [Google Scholar]
- Gallia GL, Tyler BM, Hann CL, Siu IM, Giranda VL, Vescovi AL, Brem H, Riggins GJ 2009. Inhibition of Akt inhibits growth of glioblastoma and glioblastoma stem-like cells. Mol Cancer Ther 8: 386–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geha S, Pallud J, Junier MP, Devaux B, Leonard N, Chassoux F, Chneiweiss H, Daumas-Duport C, Varlet P 2010. NG2+/Olig2+ cells are the major cycle-related cell population of the adult human normal brain. Brain Pathol 20: 399–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner ER, Duda DG, di Tomaso E, Ryg PA, Loeffler JS, Sorensen AG, Ivy P, Jain RK, Batchelor TT 2009. VEGF inhibitors in the treatment of cerebral edema in patients with brain cancer. Nat Rev Clin Oncol 6: 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese A, Westphal M 1996. Glioma invasion in the central nervous system. Neurosurgery 39: 235–252 [DOI] [PubMed] [Google Scholar]
- Gilbertson RJ, Rich JN 2007. Making a tumour's bed: Glioblastoma stem cells and the vascular niche. Nat Rev Cancer 7: 733–736 [DOI] [PubMed] [Google Scholar]
- Godar S, Ince TA, Bell GW, Feldser D, Donaher JL, Bergh J, Liu A, Miu K, Watnick RS, Reinhardt F, et al. 2008. Growth-inhibitory and tumor-suppressive functions of p53 depend on its repression of CD44 expression. Cell 134: 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. 1999. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 286: 531–537 [DOI] [PubMed] [Google Scholar]
- Graubert TA, Mardis ER 2011. Genomics of acute myeloid leukemia. Cancer J 17: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. 2007. Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsenko PG, Ilina O, Friedl P 2012. Interstitial guidance of cancer invasion. J Pathol 226: 185–199 [DOI] [PubMed] [Google Scholar]
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, et al. 2010. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med 207: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati N, Karsy M, Albert L, Murali R, Jhanwar-Uniyal M 2009. Involvement of mTORC1 and mTORC2 in regulation of glioblastoma multiforme growth and motility. Int J Oncol 35: 731–740 [DOI] [PubMed] [Google Scholar]
- Guo P, Hu B, Gu W, Xu L, Wang D, Huang HJ, Cavenee WK, Cheng SY 2003. Platelet-derived growth factor-B enhances glioma angiogenesis by stimulating vascular endothelial growth factor expression in tumor endothelia and by promoting pericyte recruitment. Am J Pathol 162: 1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryanova OA, Wu Q, Cheng L, Lathia JD, Huang Z, Yang J, MacSwords J, Eyler CE, McLendon RE, Heddleston JM, et al. 2011. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell 19: 498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, et al. 2009. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1,010 diffuse gliomas. Acta Neuropathol 118: 469–474 [DOI] [PubMed] [Google Scholar]
- Hartmann C, Hentschel B, Wick W, Capper D, Felsberg J, Simon M, Westphal M, Schackert G, Meyermann R, Pietsch T, et al. 2010. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: Implications for classification of gliomas. Acta Neuropathol 120: 707–718 [DOI] [PubMed] [Google Scholar]
- Hawkins C, Walker E, Mohamed N, Zhang C, Jacob K, Shirinian M, Alon N, Kahn D, Fried I, Scheinemann K, et al. 2011. BRAF–KIAA1549 fusion predicts better clinical outcome in pediatric low-grade astrocytoma. Clin Cancer Res 17: 4790–4798 [DOI] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. 2005. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352: 997–1003 [DOI] [PubMed] [Google Scholar]
- Heimberger AB, Hlatky R, Suki D, Yang D, Weinberg J, Gilbert M, Sawaya R, Aldape K 2005. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res 11: 1462–1466 [DOI] [PubMed] [Google Scholar]
- Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI 2003. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci 100: 15178–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JW, Schnitker BL, Correa KM, von Deimling A, Fassbender F, Xu HJ, Benedict WF, Yandell DW, Louis DN 1994. The retinoblastoma gene is involved in malignant progression of astrocytomas. Ann Neurol 36: 714–721 [DOI] [PubMed] [Google Scholar]
- Hermanson M, Funa K, Hartman M, Claesson-Welsh L, Heldin CH, Westermark B, Nister M 1992. Platelet-derived growth factor and its receptors in human glioma tissue: Expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer Res 52: 3213–3219 [PubMed] [Google Scholar]
- Hesselager G, Holland EC 2003. Using mice to decipher the molecular genetics of brain tumors. Neurosurgery 53: 685–695 [DOI] [PubMed] [Google Scholar]
- Hicklin DJ, Ellis LM 2005. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol 23: 1011–1027 [DOI] [PubMed] [Google Scholar]
- Hochberg FH, Pruitt A 1980. Assumptions in the radiotherapy of glioblastoma. Neurology 30: 907–911 [DOI] [PubMed] [Google Scholar]
- Hoffman HJ, Duffner PK 1985. Extraneural metastases of central nervous system tumors. Cancer 56: 1778–1782 [DOI] [PubMed] [Google Scholar]
- Holland EC, Hively WP, DePinho RA, Varmus HE 1998. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev 12: 3675–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbinski C, Hamilton RL, Nikiforov Y, Pollack IF 2010. Association of molecular alterations, including BRAF, with biology and outcome in pilocytic astrocytomas. Acta Neuropathol 119: 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, Huang CM, Gill GN, Wiley HS, Cavenee WK 1997. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem 272: 2927–2935 [DOI] [PubMed] [Google Scholar]
- Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM 2007. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci 104: 12867–12872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JD, Egleton RD, Davis TP 2001. Molecular physiology and pathophysiology of tight junctions in the blood–brain barrier. Trends Neurosci 24: 719–725 [DOI] [PubMed] [Google Scholar]
- Hurtt MR, Moossy J, Donovan-Peluso M, Locker J 1992. Amplification of epidermal growth factor receptor gene in gliomas: Histopathology and prognosis. J Neuropathol Exp Neurol 51: 84–90 [DOI] [PubMed] [Google Scholar]
- Huse JT, Phillips HS, Brennan CW 2011. Molecular subclassification of diffuse gliomas: Seeing order in the chaos. Glia 59: 1190–1199 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Pearson DM, Kocialkowski S, Backlund LM, Chan R, Jones DT, Collins VP 2009. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol 11: 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idbaih A, Carvalho Silva R, Criniere E, Marie Y, Carpentier C, Boisselier B, Taillibert S, Rousseau A, Mokhtari K, Ducray F, et al. 2008. Genomic changes in progression of low-grade gliomas. J Neurooncol 90: 133–140 [DOI] [PubMed] [Google Scholar]
- Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA 2002. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia 39: 193–206 [DOI] [PubMed] [Google Scholar]
- Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, Brennan C, Johns TG, Bachoo R, Hadwiger P, et al. 2010. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev 24: 1731–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto FM, Abrey LE, Beal K, Gutin PH, Rosenblum MK, Reuter VE, DeAngelis LM, Lassman AB 2009. Patterns of relapse and prognosis after bevacizumab failure in recurrent glioblastoma. Neurology 73: 1200–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto FM, Hottinger AF, Karimi S, Riedel E, Dantis J, Jahdi M, Panageas KS, Lassman AB, Abrey LE, Fleisher M, et al. 2011. Serum YKL-40 is a marker of prognosis and disease status in high-grade gliomas. Neuro Oncol 13: 1244–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RK 2005. Normalization of tumor vasculature: An emerging concept in antiangiogenic therapy. Science 307: 58–62 [DOI] [PubMed] [Google Scholar]
- Janne PA, Engelman JA, Johnson BE 2005. Epidermal growth factor receptor mutations in non-small-cell lung cancer: Implications for treatment and tumor biology. J Clin Oncol 23: 3227–3234 [DOI] [PubMed] [Google Scholar]
- Jaros E, Perry RH, Adam L, Kelly PJ, Crawford PJ, Kalbag RM, Mendelow AD, Sengupta RP, Pearson AD 1992. Prognostic implications of p53 protein, epidermal growth factor receptor, and Ki-67 labelling in brain tumours. Br J Cancer 66: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo M, Stolz DB, Esplen JE, Dorko K, Michalopoulos GK, Strom SC 2000. Cross-talk between epidermal growth factor receptor and c-Met signal pathways in transformed cells. J Biol Chem 275: 8806–8811 [DOI] [PubMed] [Google Scholar]
- Jones DT, Kocialkowski S, Liu L, Pearson DM, Backlund LM, Ichimura K, Collins VP 2008. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res 68: 8673–8677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Kocialkowski S, Liu L, Pearson DM, Ichimura K, Collins VP 2009. Oncogenic RAF1 rearrangement and a novel BRAF mutation as alternatives to KIAA1549:BRAF fusion in activating the MAPK pathway in pilocytic astrocytoma. Oncogene 28: 2119–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG Jr 2011. Cancer and altered metabolism: Potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb Symp Quant Biol doi: 10.1101/sqb.2011.76.010975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargiotis O, Rao JS, Kyritsis AP 2006. Mechanisms of angiogenesis in gliomas. J Neurooncol 78: 281–293 [DOI] [PubMed] [Google Scholar]
- Kaur B, Khwaja FW, Severson EA, Matheny SL, Brat DJ, Van Meir EG 2005. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro Oncol 7: 134–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerbel RS 2008. Tumor angiogenesis. N Engl J Med 358: 2039–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita D, Yonekawa Y, Weller M, Ohgaki H 2007. PIK3CA alterations in primary (de novo) and secondary glioblastomas. Acta Neuropathol 113: 295–302 [DOI] [PubMed] [Google Scholar]
- Kleber S, Sancho-Martinez I, Wiestler B, Beisel A, Gieffers C, Hill O, Thiemann M, Mueller W, Sykora J, Kuhn A, et al. 2008. Yes and PI3K bind CD95 to signal invasion of glioblastoma. Cancer Cell 13: 235–248 [DOI] [PubMed] [Google Scholar]
- Koga K, Todaka T, Morioka M, Hamada J, Kai Y, Yano S, Okamura A, Takakura N, Suda T, Ushio Y 2001. Expression of angiopoietin-2 in human glioma cells and its role for angiogenesis. Cancer Res 61: 6248–6254 [PubMed] [Google Scholar]
- Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, et al. 2012. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature doi: 10.1038/nature10898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong DS, Song SY, Kim DH, Joo KM, Yoo JS, Koh JS, Dong SM, Suh YL, Lee JI, Park K, et al. 2009. Prognostic significance of c-Met expression in glioblastomas. Cancer 115: 140–148 [DOI] [PubMed] [Google Scholar]
- Kotliarov Y, Steed ME, Christopher N, Walling J, Su Q, Center A, Heiss J, Rosenblum M, Mikkelsen T, Zenklusen JC, et al. 2006. High-resolution global genomic survey of 178 gliomas reveals novel regions of copy number alteration and allelic imbalances. Cancer Res 66: 9428–9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul D 2008. PTEN signaling pathways in glioblastoma. Cancer Biol Ther 7: 1321–1325 [DOI] [PubMed] [Google Scholar]
- Kranendijk M, Struys EA, van Schaftingen E, Gibson KM, Kanhai WA, van der Knaap MS, Amiel J, Buist NR, Das AM, de Klerk JB, et al. 2010. IDH2 mutations in patients with D-2-hydroxyglutaric aciduria. Science 330: 336. [DOI] [PubMed] [Google Scholar]
- Kreisl TN, Kim L, Moore K, Duic P, Royce C, Stroud I, Garren N, Mackey M, Butman JA, Camphausen K, et al. 2009. Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma. J Clin Oncol 27: 740–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, et al. 2010. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363: 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok JC, Dick G, Wang D, Fawcett JW 2011. Extracellular matrix and perineuronal nets in CNS repair. Dev Neurobiol 71: 1073–1089 [DOI] [PubMed] [Google Scholar]
- Kwon CH, Zhao D, Chen J, Alcantara S, Li Y, Burns DK, Mason RP, Lee EY, Wu H, Parada LF 2008. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res 68: 3286–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A, Kharbanda S, Pope WB, Tran A, Solis OE, Peale F, Forrest WF, Pujara K, Carrillo JA, Pandita A, et al. 2011. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol 29: 4482–4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal B, Goodwin CR, Sang Y, Foss CA, Cornet K, Muzamil S, Pomper MG, Kim J, Laterra J 2009. EGFRvIII and c-Met pathway inhibitors synergize against PTEN-null/EGFRvIII+ glioblastoma xenografts. Mol Cancer Ther 8: 1751–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Heddleston JM, Venere M, Rich JN 2011a. Deadly teamwork: Neural cancer stem cells and the tumor microenvironment. Cell Stem Cell 8: 482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Venere M, Rao MS, Rich JN 2011b. Seeing is believing: Are cancer stem cells the Loch Ness monster of tumor biology? Stem Cell Rev 7: 227–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. 2006a. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9: 391–403 [DOI] [PubMed] [Google Scholar]
- Lee JC, Vivanco I, Beroukhim R, Huang JH, Feng WL, DeBiasi RM, Yoshimoto K, King JC, Nghiemphu P, Yuza Y, et al. 2006b. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med 3: e485 doi: 10.1371/journal.pmed.0030485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Good JA, Ziegler WH, Parekh DB, Alessi DR, Cohen P, Parker PJ 1998. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science 281: 2042–2045 [DOI] [PubMed] [Google Scholar]
- Levin VA, Phuphanich S, Yung WK, Forsyth PA, Maestro RD, Perry JR, Fuller GN, Baillet M 2006. Randomized, double-blind, placebo-controlled trial of marimastat in glioblastoma multiforme patients following surgery and irradiation. J Neurooncol 78: 295–302 [DOI] [PubMed] [Google Scholar]
- Levitt RJ, Georgescu MM, Pollak M 2005. PTEN-induction in U251 glioma cells decreases the expression of insulin-like growth factor binding protein-2. Biochem Biophys Res Commun 336: 1056–1061 [DOI] [PubMed] [Google Scholar]
- Li M, Mukasa A, Del-Mar Inda M, Zhang J, Chin L, Cavenee W, Furnari F 2011. Guanylate binding protein 1 is a novel effector of EGFR-driven invasion in glioblastoma. J Exp Med 208: 2657–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon KL, Alberta JA, Kho AT, Weiss J, Kwaan MR, Nutt CL, Louis DN, Stiles CD, Rowitch DH 2004. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol 63: 499–509 [DOI] [PubMed] [Google Scholar]
- Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, et al. 2007. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron 53: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SK, Llaguno SR, McKay RM, Parada LF 2011. Glioblastoma multiforme: A perspective on recent findings in human cancer and mouse models. BMB reports 44: 158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino MM, Merlo A 2011. PI3Kinase signaling in glioblastoma. J Neurooncol 103: 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA, Tran NL, Viso C, Kloss J, Yang Z, Berens ME, Loftus JC 2008. Extended survival of Pyk2 or FAK deficient orthotopic glioma xenografts. J Neurooncol 90: 181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, et al. 2011. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell 146: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang J, Kang SA, Thoreen CC, Hur W, Ahmed T, Sabatini DM, Gray NS 2011. Discovery of 9-(6-aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)benzo[h][1,6]naphthyr idin-2(1H)-one (Torin2) as a potent, selective, and orally available mammalian target of rapamycin (mTOR) inhibitor for treatment of cancer. J Med Chem 54: 1473–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenarz C, Schofield CJ 2008. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol 4: 152–156 [DOI] [PubMed] [Google Scholar]
- Lokker NA, Sullivan CM, Hollenbach SJ, Israel MA, Giese NA 2002. Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells: Evidence that the novel PDGF-C and PDGF-D ligands may play a role in the development of brain tumors. Cancer Res 62: 3729–3735 [PubMed] [Google Scholar]
- Long DM 1970. Capillary ultrastructure and the blood–brain barrier in human malignant brain tumors. J Neurosurg 32: 127–144 [DOI] [PubMed] [Google Scholar]
- Lopez KA, Tannenbaum AM, Assanah MC, Linskey K, Yun J, Kangarlu A, Gil OD, Canoll P, Bruce JN 2011. Convection-enhanced delivery of topotecan into a PDGF-driven model of glioblastoma prolongs survival and ablates both tumor-initiating cells and recruited glial progenitors. Cancer Res 71: 3963–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN 1994. The p53 gene and protein in human brain tumors. J Neuropathol Exp Neurol 53: 11–21 [DOI] [PubMed] [Google Scholar]
- Louis DN 2006. Molecular pathology of malignant gliomas. Annu Rev Pathol 1: 97–117 [DOI] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P 2007. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KV, Zhu S, Cvrljevic A, Huang TT, Sarkaria S, Ahkavan D, Dang J, Dinca EB, Plaisier SB, Oderberg I, et al. 2009. Fyn and SRC are effectors of oncogenic epidermal growth factor receptor signaling in glioblastoma patients. Cancer Res 69: 6889–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. 2012. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature doi: 10.1038/nature10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchman HA, Stechishin OD, Dang NH, Blough MD, Chesnelong C, Kelly JJ, Nguyen SA, Chan JA, Weljie AM, Cairncross JG, et al. 2012. An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro Oncol 14: 184–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. 2004. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139 [DOI] [PubMed] [Google Scholar]
- Maccario H, Perera NM, Davidson L, Downes CP, Leslie NR 2007. PTEN is destabilized by phosphorylation on Thr366. Biochem J 405: 439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacConaill LE, Campbell CD, Kehoe SM, Bass AJ, Hatton C, Niu L, Davis M, Yao K, Hanna M, Mondal C, et al. 2009. Profiling critical cancer gene mutations in clinical tumor samples. PLoS ONE 4: e7887 doi: 10.1371/journal.pone.0007887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher EA, Brennan C, Wen PY, Durso L, Ligon KL, Richardson A, Khatry D, Feng B, Sinha R, Louis DN, et al. 2006. Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities. Cancer Res 66: 11502–11513 [DOI] [PubMed] [Google Scholar]
- Maity A, Pore N, Lee J, Solomon D, O'Rourke DM 2000. Epidermal growth factor receptor transcriptionally up-regulates vascular endothelial growth factor expression in human glioblastoma cells via a pathway involving phosphatidylinositol 3′-kinase and distinct from that induced by hypoxia. Cancer Res 60: 5879–5886 [PubMed] [Google Scholar]
- Mallory F. 1914. The principles of pathologic histology. Saunders, Philadelphia. [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukado Y, Maccarty CS, Kernohan JW 1961. The growth of glioblastoma multiforme (astrocytomas, grades 3 and 4) in neurosurgical practice. J Neurosurg 18: 636–644 [DOI] [PubMed] [Google Scholar]
- McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, Johnson BW, Williams SM, Nghiemphu P, Liau LM, et al. 2009. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell 16: 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. 2005. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med 353: 2012–2024 [DOI] [PubMed] [Google Scholar]
- Mellinghoff IK, Cloughesy TF, Mischel PS 2007. PTEN-mediated resistance to epidermal growth factor receptor kinase inhibitors. Clin Cancer Res 13: 378–381 [DOI] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G 2011. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 12: R41 doi: 10.1186/gb-2011-12-4-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. 2012. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481: 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Gabriel S, Getz G 2010. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 11: 685–696 [DOI] [PubMed] [Google Scholar]
- Miao H, Li DQ, Mukherjee A, Guo H, Petty A, Cutter J, Basilion JP, Sedor J, Wu J, Danielpour D, et al. 2009. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell 16: 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CR, Perry A 2007. Glioblastoma. Arch Pathol Lab Med 131: 397–406 [DOI] [PubMed] [Google Scholar]
- Mizoguchi M, Nutt CL, Mohapatra G, Louis DN 2004. Genetic alterations of phosphoinositide 3-kinase subunit genes in human glioblastomas. Brain Pathol 14: 372–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina JR, Agarwal NK, Morales FC, Hayashi Y, Aldape KD, Cote G, Georgescu MM 2012. PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene 31: 1264–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai T, Miyazaki Y, Nishinakamura H, Sugahara KN, Miyauchi T, Sako Y, Yanagida T, Miyasaka M 2004. Engagement of CD44 promotes Rac activation and CD44 cleavage during tumor cell migration. J Biol Chem 279: 4541–4550 [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Shimao Y, Sato S, Kataoka H, Moriyama T, Kawano H, Wakisaka S, Koono M 1997. Expression of c-Met correlates with grade of malignancy in human astrocytic tumours: An immunohistochemical study. Histopathology 31: 436–443 [DOI] [PubMed] [Google Scholar]
- Nagane M, Levitzki A, Gazit A, Cavenee WK, Huang HJ 1998. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc Natl Acad Sci 95: 5724–5729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME 2007. Molecular targets of glioma invasion. Cell Mol Life Sci 64: 458–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayana A, Kunnakkat SD, Medabalmi P, Golfinos J, Parker E, Knopp E, Zagzag D, Eagan P, Gruber D, Gruber ML 2012. Change in pattern of relapse after antiangiogenic therapy in high-grade glioma. Int J Radiat Oncol Biol Phys 82: 77–82 [DOI] [PubMed] [Google Scholar]
- Narita Y, Nagane M, Mishima K, Huang HJ, Furnari FB, Cavenee WK 2002. Mutant epidermal growth factor receptor signaling down-regulates p27 through activation of the phosphatidylinositol 3-kinase/Akt pathway in glioblastomas. Cancer Res 62: 6764–6769 [PubMed] [Google Scholar]
- Natsume A, Kato T, Kinjo S, Enomoto A, Toda H, Shimato S, Ohka F, Motomura K, Kondo Y, Miyata T, et al. 2011. Girdin maintains the stemness of glioblastoma stem cells. Oncogene doi: 10.1038/onc.2011.466 [DOI] [PubMed] [Google Scholar]
- Navin N, Hicks J 2011. Future medical applications of single-cell sequencing in cancer. Genome Med 3: 31 doi: 10.1186/gm247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. 2011. Tumour evolution inferred by single-cell sequencing. Nature 472: 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa R, Sugiyama T, Narita Y, Furnari F, Cavenee WK, Matsutani M 2004. Immunohistochemical analysis of the mutant epidermal growth factor, ΔEGFR, in glioblastoma. Brain Tumor Pathol 21: 53–56 [DOI] [PubMed] [Google Scholar]
- Norden AD, Drappatz J, Wen PY 2008a. Novel anti-angiogenic therapies for malignant gliomas. Lancet Neurol 7: 1152–1160 [DOI] [PubMed] [Google Scholar]
- Norden AD, Young GS, Setayesh K, Muzikansky A, Klufas R, Ross GL, Ciampa AS, Ebbeling LG, Levy B, Drappatz J, et al. 2008b. Bevacizumab for recurrent malignant gliomas: Efficacy, toxicity, and patterns of recurrence. Neurology 70: 779–787 [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. 2010. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt CL, Mani DR, Betensky RA, Tamayo P, Cairncross JG, Ladd C, Pohl U, Hartmann C, McLaughlin ME, Batchelor TT, et al. 2003. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res 63: 1602–1607 [PubMed] [Google Scholar]
- Nyberg P, Xie L, Kalluri R 2005. Endogenous inhibitors of angiogenesis. Cancer Res 65: 3967–3979 [DOI] [PubMed] [Google Scholar]
- Ogden AT, Waziri AE, Lochhead RA, Fusco D, Lopez K, Ellis JA, Kang J, Assanah M, McKhann GM, Sisti MB et al. 2008. Identification of A2B5+CD133− tumor-initiating cells in adult human gliomas. Neurosurgery 62: 505–515 [DOI] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P 2005. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol 64: 479–489 [DOI] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P 2009. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci 100: 2235–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Ichikawa T, Kurozumi K, Date I 2011. Angiogenesis and invasion in glioma. Brain Tumor Pathol 28: 13–24 [DOI] [PubMed] [Google Scholar]
- Ozawa T, Brennan CW, Wang L, Squatrito M, Sasayama T, Nakada M, Huse JT, Pedraza A, Utsuki S, Yasui Y, et al. 2010. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev 24: 2205–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Lazar DF, Saltiel AR, Dixit VM 1994. Activation of the Eck receptor protein tyrosine kinase stimulates phosphatidylinositol 3-kinase activity. J Biol Chem 269: 30154–30157 [PubMed] [Google Scholar]
- Panner A, Crane CA, Weng C, Feletti A, Parsa AT, Pieper RO 2009. A novel PTEN-dependent link to ubiquitination controls FLIPS stability and TRAIL sensitivity in glioblastoma multiforme. Cancer Res 69: 7911–7916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansuriya TC, van Eijk R, d'Adamo P, van Ruler MA, Kuijjer ML, Oosting J, Cleton-Jansen AM, van Oosterwijk JG, Verbeke SL, Meijer D, et al. 2011. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat Genet 43: 1256–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa AT, Wachhorst S, Lamborn KR, Prados MD, McDermott MW, Berger MS, Chang SM 2005. Prognostic significance of intracranial dissemination of glioblastoma multiforme in adults. J Neurosurg 102: 622–628 [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, Spath D, Kayser S, Zucknick M, Gotze K, et al. 2010. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol 28: 3636–3643 [DOI] [PubMed] [Google Scholar]
- Patenaude A, Parker J, Karsan A 2010. Involvement of endothelial progenitor cells in tumor vascularization. Microvasc Res 79: 217–223 [DOI] [PubMed] [Google Scholar]
- Paternot S, Roger PP 2009. Combined inhibition of MEK and mammalian target of rapamycin abolishes phosphorylation of cyclin-dependent kinase 4 in glioblastoma cell lines and prevents their proliferation. Cancer Res 69: 4577–4581 [DOI] [PubMed] [Google Scholar]
- Perren A, Komminoth P, Saremaslani P, Matter C, Feurer S, Lees JA, Heitz PU, Eng C 2000. Mutation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. Am J Pathol 157: 1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson AI, Petritsch C, Swartling FJ, Itsara M, Sim FJ, Auvergne R, Goldenberg DD, Vandenberg SR, Nguyen KN, Yakovenko S, et al. 2010. Non-stem cell origin for oligodendroglioma. Cancer Cell 18: 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister S, Janzarik WG, Remke M, Ernst A, Werft W, Becker N, Toedt G, Wittmann A, Kratz C, Olbrich H, et al. 2008. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest 118: 1739–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. 2006. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9: 157–173 [DOI] [PubMed] [Google Scholar]
- Phung TL, Ziv K, Dabydeen D, Eyiah-Mensah G, Riveros M, Perruzzi C, Sun J, Monahan-Earley RA, Shiojima I, Nagy JA, et al. 2006. Pathological angiogenesis is induced by sustained Akt signaling and inhibited by rapamycin. Cancer Cell 10: 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrak B, Zhao H, Qi H, Quinn C, Gao E, Boyer JG, Concha N, Brown K, Duraiswami C, Wooster R, et al. 2011. A tale of two subunits: How the neomorphic R132H IDH1 mutation enhances production of αHG. Biochemistry 50: 4804–4812 [DOI] [PubMed] [Google Scholar]
- Pillay V, Allaf L, Wilding AL, Donoghue JF, Court NW, Greenall SA, Scott AM, Johns TG 2009. The plasticity of oncogene addiction: Implications for targeted therapies directed to receptor tyrosine kinases. Neoplasia 11: 448–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, Bayani J, Head R, Lee M, Bernstein M, et al. 2009. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 4: 568–580 [DOI] [PubMed] [Google Scholar]
- Pope WB, Prins RM, Albert Thomas M, Nagarajan R, Yen KE, Bittinger MA, Salamon N, Chou AP, Yong WH, Soto H, et al. 2012. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol 107: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pore N, Liu S, Haas-Kogan DA, O'Rourke DM, Maity A 2003. PTEN mutation and epidermal growth factor receptor activation regulate vascular endothelial growth factor (VEGF) mRNA expression in human glioblastoma cells by transactivating the proximal VEGF promoter. Cancer Res 63: 236–241 [PubMed] [Google Scholar]
- Porter KR, McCarthy BJ, Freels S, Kim Y, Davis FG 2010. Prevalence estimates for primary brain tumors in the United States by age, gender, behavior, and histology. Neuro Oncol 12: 520–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quant EC, Norden AD, Drappatz J, Muzikansky A, Doherty L, Lafrankie D, Ciampa A, Kesari S, Wen PY 2009. Role of a second chemotherapy in recurrent malignant glioma patients who progress on bevacizumab. Neuro Oncol 11: 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimundo N, Baysal BE, Shadel GS 2011. Revisiting the TCA cycle: Signaling to tumor formation. Trends Mol Med 17: 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao JS 2003. Molecular mechanisms of glioma invasiveness: The role of proteases. Nat Rev Cancer 3: 489–501 [DOI] [PubMed] [Google Scholar]
- Rawlings CE III, Bullard DE, Burger PC, Friedman AH 1987. A case of Ollier's disease associated with two intracranial gliomas. Neurosurgery 21: 400–403 [DOI] [PubMed] [Google Scholar]
- Reardon DA, Neyns B, Weller M, Tonn JC, Nabors LB, Stupp R 2011a. Cilengitide: An RGD pentapeptide ανβ3 and ανβ5 integrin inhibitor in development for glioblastoma and other malignancies. Future Oncol 7: 339–354 [DOI] [PubMed] [Google Scholar]
- Reardon DA, Turner S, Peters KB, Desjardins A, Gururangan S, Sampson JH, McLendon RE, Herndon JE II, Jones LW, Kirkpatrick JP, et al. 2011b. A review of VEGF/VEGFR-targeted therapeutics for recurrent glioblastoma. J Natl Compr Canc Netw 9: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenberger G, Reifenberger J, Ichimura K, Meltzer PS, Collins VP 1994. Amplification of multiple genes from chromosomal region 12q13-14 in human malignant gliomas: Preliminary mapping of the amplicons shows preferential involvement of CDK4, SAS, and MDM2. Cancer Res 54: 4299–4303 [PubMed] [Google Scholar]
- Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, He Y, Bigner DD, Vogelstein B, Yan H 2011. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci 108: 3270–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Mugica M, Rieger-Christ K, Ohgaki H, Ekstrand BC, Helie M, Kleinman G, Yahanda A, Fearon ER, Kleihues P, Reale MA 1997. Loss of DCC expression and glioma progression. Cancer Res 57: 382–386 [PubMed] [Google Scholar]
- Reznik TE, Sang Y, Ma Y, Abounader R, Rosen EM, Xia S, Laterra J 2008. Transcription-dependent epidermal growth factor receptor activation by hepatocyte growth factor. Mol Cancer Res 6: 139–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T, Maira G, Parati EA, Stassi G, Larocca LM, et al. 2010. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature 468: 824–828 [DOI] [PubMed] [Google Scholar]
- Riddick G, Fine HA 2011. Integration and analysis of genome-scale data from gliomas. Nat Rev Neurol 7: 439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemenschneider MJ, Mueller W, Betensky RA, Mohapatra G, Louis DN 2005. In situ analysis of integrin and growth factor receptor signaling pathways in human glioblastomas suggests overlapping relationships with focal adhesion kinase activation. Am J Pathol 167: 1379–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin SN, Rodin AS 1998. Strand asymmetry of CpG transitions as indicator of G1 phase-dependent origin of multiple tumorigenic p53 mutations in stem cells. Proc Natl Acad Sci 95: 11927–11932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J 1994. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 370: 527–532 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Vanhaesebroeck B, Waterfield MD, Downward J 1996. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J 15: 2442–2451 [PMC free article] [PubMed] [Google Scholar]
- Rothenberg SM, Engelman JA, Le S, Riese DJ II, Haber DA, Settleman J 2008. Modeling oncogene addiction using RNA interference. Proc Natl Acad Sci 105: 12480–12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutka JT, Apodaca G, Stern R, Rosenblum M 1988. The extracellular matrix of the central and peripheral nervous systems: Structure and function. J Neurosurg 69: 155–170 [DOI] [PubMed] [Google Scholar]
- Rutka JT, Muller M, Hubbard SL, Forsdike J, Dirks PB, Jung S, Tsugu A, Ivanchuk S, Costello P, Mondal S, et al. 1999. Astrocytoma adhesion to extracellular matrix: Functional significance of integrin and focal adhesion kinase expression. J Neuropathol Exp Neurol 58: 198–209 [DOI] [PubMed] [Google Scholar]
- Sahm F, Capper D, Pusch S, Balss J, Koch A, Langhans CD, Okun JG, von Deimling A 2012. Detection of 2-hydroxyglutarate in formalin-fixed paraffin-embedded glioma specimens by gas chromatography/mass spectrometry. Brain Pathol 22: 26–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanai N, Nguyen T, Ihrie RA, Mirzadeh Z, Tsai HH, Wong M, Gupta N, Berger MS, Huang E, Garcia-Verdugo JM, et al. 2011. Corridors of migrating neurons in the human brain and their decline during infancy. Nature 478: 382–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandsmark DK, Zhang H, Hegedus B, Pelletier CL, Weber JD, Gutmann DH 2007. Nucleophosmin mediates mammalian target of rapamycin-dependent actin cytoskeleton dynamics and proliferation in neurofibromin-deficient astrocytes. Cancer Res 67: 4790–4799 [DOI] [PubMed] [Google Scholar]
- Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, El Hallani S, Boisselier B, Mokhtari K, Hoang-Xuan K, et al. 2009. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 27: 4150–4154 [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM 2005. Phosphorylation and regulation of Akt/PKB by the rictor–mTOR complex. Science 307: 1098–1101 [DOI] [PubMed] [Google Scholar]
- Sathornsumetee S, Rich JN 2007. Antiangiogenic therapy in malignant glioma: Promise and challenge. Curr Pharm Des 13: 3545–3558 [DOI] [PubMed] [Google Scholar]
- Scherer H 1940. The forms of growth in gliomas and their practical signficance. Brain 63: 1–35 [Google Scholar]
- Schindler G, Capper D, Meyer J, Janzarik W, Omran H, Herold-Mende C, Schmieder K, Wesseling P, Mawrin C, Hasselblatt M, et al. 2011. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol 121: 397–405 [DOI] [PubMed] [Google Scholar]
- Schlegel J, Stumm G, Brandle K, Merdes A, Mechtersheimer G, Hynes NE, Kiessling M 1994. Amplification and differential expression of members of the erbB-gene family in human glioblastoma. J Neurooncol 22: 201–207 [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Ichimura K, Reifenberger G, Collins VP 1994. CDKN2 (p16/MTS1) gene deletion or CDK4 amplification occurs in the majority of glioblastomas. Cancer Res 54: 6321–6324 [PubMed] [Google Scholar]
- Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, et al. 2008. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 14: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequist LV, Heist RS, Shaw AT, Fidias P, Rosovsky R, Temel JS, Lennes IT, Digumarthy S, Waltman BA, Bast E, et al. 2011. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 22: 2616–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Settleman J 2007. Oncogene addiction: Setting the stage for molecularly targeted cancer therapy. Genes Dev 21: 3214–3231 [DOI] [PubMed] [Google Scholar]
- Shervington A, Lu C 2008. Expression of multidrug resistance genes in normal and cancer stem cells. Cancer Invest 26: 535–542 [DOI] [PubMed] [Google Scholar]
- Shinojima N, Tada K, Shiraishi S, Kamiryo T, Kochi M, Nakamura H, Makino K, Saya H, Hirano H, Kuratsu J, et al. 2003. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res 63: 6962–6970 [PubMed] [Google Scholar]
- Shweiki D, Itin A, Soffer D, Keshet E 1992. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature 359: 843–845 [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB 2003. Identification of a cancer stem cell in human brain tumors. Cancer Res 63: 5821–5828 [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB 2004. Identification of human brain tumour initiating cells. Nature 432: 396–401 [DOI] [PubMed] [Google Scholar]
- Sirko S, von Holst A, Wizenmann A, Gotz M, Faissner A 2007. Chondroitin sulfate glycosaminoglycans control proliferation, radial glia cell differentiation and neurogenesis in neural stem/progenitor cells. Development 134: 2727–2738 [DOI] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274 [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, et al. 2001. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 344: 783–792 [DOI] [PubMed] [Google Scholar]
- Smith JS, Wang XY, Qian J, Hosek SM, Scheithauer BW, Jenkins RB, James CD 2000. Amplification of the platelet-derived growth factor receptor-A (PDGFRA) gene occurs in oligodendrogliomas with grade IV anaplastic features. J Neuropathol Exp Neurol 59: 495–503 [DOI] [PubMed] [Google Scholar]
- Snuderl M, Eichler AF, Ligon KL, Vu QU, Silver M, Betensky RA, Ligon AH, Wen PY, Louis DN, Iafrate AJ 2009. Polysomy for chromosomes 1 and 19 predicts earlier recurrence in anaplastic oligodendrogliomas with concurrent 1p/19q loss. Clin Cancer Res 15: 6430–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, et al. 2011. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 20: 810–817 [DOI] [PubMed] [Google Scholar]
- Sohr S, Engeland K 2008. RHAMM is differentially expressed in the cell cycle and downregulated by the tumor suppressor p53. Cell Cycle 7: 3448–3460 [DOI] [PubMed] [Google Scholar]
- Song H, Li Y, Lee J, Schwartz AL, Bu G 2009. Low-density lipoprotein receptor-related protein 1 promotes cancer cell migration and invasion by inducing the expression of matrix metalloproteinases 2 and 9. Cancer Res 69: 879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Carracedo A, Salmena L, Song SJ, Egia A, Malumbres M, Pandolfi PP 2011. Nuclear PTEN regulates the APC–CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell 144: 187–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroceanu L, Kharbanda S, Chen R, Soriano RH, Aldape K, Misra A, Zha J, Forrest WF, Nigro JM, Modrusan Z, et al. 2007. Identification of IGF2 signaling through phosphoinositide-3-kinase regulatory subunit 3 as a growth-promoting axis in glioblastoma. Proc Natl Acad Sci 104: 3466–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettner MR, Wang W, Nabors LB, Bharara S, Flynn DC, Grammer JR, Gillespie GY, Gladson CL 2005. Lyn kinase activity is the predominant cellular SRC kinase activity in glioblastoma tumor cells. Cancer Res 65: 5535–5543 [DOI] [PubMed] [Google Scholar]
- Stockhausen MT, Broholm H, Villingshoj M, Kirchhoff M, Gerdes T, Kristoffersen K, Kosteljanetz M, Spang-Thomsen M, Poulsen HS 2011. Maintenance of EGFR and EGFRvIII expressions in an in vivo and in vitro model of human glioblastoma multiforme. Exp Cell Res 317: 1513–1526 [DOI] [PubMed] [Google Scholar]
- Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, et al. 2007. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 318: 287–290 [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA 2009. The cancer genome. Nature 458: 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. 2005. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352: 987–996 [DOI] [PubMed] [Google Scholar]
- Sugiarto S, Persson AI, Munoz EG, Waldhuber M, Lamagna C, Andor N, Hanecker P, Ayers-Ringler J, Phillips J, Siu J, et al. 2011. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell 20: 328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Hillmann P, Hofmann BT, Hart JR, Vogt PK 2010. Cancer-derived mutations in the regulatory subunit p85α of phosphoinositide 3-kinase function through the catalytic subunit p110α. Proc Natl Acad Sci 107: 15547–15552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunayama J, Sato A, Matsuda K, Tachibana K, Suzuki K, Narita Y, Shibui S, Sakurada K, Kayama T, Tomiyama A, et al. 2010. Dual blocking of mTor and PI3K elicits a prodifferentiation effect on glioblastoma stem-like cells. Neuro-oncol 12: 1205–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S, et al. 2012. Intratumoral heterogeneity of EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci 109: 3041–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate MC, Aghi MK 2009. Biology of angiogenesis and invasion in glioma. Neurotherapeutics 6: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CM, Sauvageot CM, Wen PYC, Stiles CD 2010. Cellular origins of malignant glioma: The cancer stem cell polemic. In Primary central nervous system tumors: Pathogenesis and therapy (ed. AD Norden et al.), pp. 45–54. Springer, New York [Google Scholar]
- Teodorczyk M, Martin-Villalba A 2010. Sensing invasion: Cell surface receptors driving spreading of glioblastoma. J Cell Physiol 222: 1–10 [DOI] [PubMed] [Google Scholar]
- Tian Y, Rich BE, Vena N, Craig JM, Macconaill LE, Rajaram V, Goldman S, Taha H, Mahmoud M, Ozek M, et al. 2011. Detection of KIAA1549–BRAF fusion transcripts in formalin-fixed paraffin-embedded pediatric low-grade gliomas. J Mol Diagn 13: 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso CL, Freije WA, Day A, Chen Z, Merriman B, Perlina A, Lee Y, Dia EQ, Yoshimoto K, Mischel PS, et al. 2006. Distinct transcription profiles of primary and secondary glioblastoma subgroups. Cancer Res 66: 159–167 [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AWM, Lu C, Ward PS, et al. 2012. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature doi: 10.1038/nature10866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki K, Ono Y, Henson JW, Efird JT, von Deimling A, Louis DN 1996. CDKN2/p16 or RB alterations occur in the majority of glioblastomas and are inversely correlated. Cancer Res 56: 150–153 [PubMed] [Google Scholar]
- Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, Hennessy BT, Tseng H, Pochanard P, Kim SY, et al. 2009. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell 16: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeff JJ, van Tellingen O, Claes A, Stalpers LJ, van Linde ME, Richel DJ, Leenders WP, van Furth WR 2009. Concerns about anti-angiogenic treatment in patients with glioblastoma multiforme. BMC Cancer 9: 444 doi: 10.1186/1471-2407-9-444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE 2011. Cells of origin in cancer. Nature 469: 314–322 [DOI] [PubMed] [Google Scholar]
- Vivanco I, Rohle D, Versele M, Iwanami A, Kuga D, Oldrini B, Tanaka K, Dang J, Kubek S, Palaskas N, et al. 2010. The phosphatase and tensin homolog regulates epidermal growth factor receptor (EGFR) inhibitor response by targeting EGFR for degradation. Proc Natl Acad Sci 107: 6459–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW 2004. Cancer genes and the pathways they control. Nat Med 10: 789–799 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL 1988. Genetic alterations during colorectal-tumor development. N Engl J Med 319: 525–532 [DOI] [PubMed] [Google Scholar]
- von Deimling A, Korshunov A, Hartmann C 2011. The next generation of glioma biomarkers: MGMT methylation, BRAF fusions and IDH1 mutations. Brain Pathol 21: 74–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vredenburgh JJ, Desjardins A, Herndon JE II, Dowell JM, Reardon DA, Quinn JA, Rich JN, Sathornsumetee S, Gururangan S, Wagner M, et al. 2007a. Phase II trial of bevacizumab and irinotecan in recurrent malignant glioma. Clin Cancer Res 13: 1253–1259 [DOI] [PubMed] [Google Scholar]
- Vredenburgh JJ, Desjardins A, Herndon JE II, Marcello J, Reardon DA, Quinn JA, Rich JN, Sathornsumetee S, Gururangan S, Sampson J, et al. 2007b. Bevacizumab plus irinotecan in recurrent glioblastoma multiforme. J Clin Oncol 25: 4722–4729 [DOI] [PubMed] [Google Scholar]
- Wakimoto H, Mohapatra G, Kanai R, Curry WT Jr, Yip S, Nitta M, Patel AP, Barnard ZR, Stemmer-Rachamimov AO, Louis DN, et al. 2012. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro-oncol 14: 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, Hung MC 2009. Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin Cancer Res 15: 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Shen W, Huang H, Hu L, Ramdas L, Zhou YH, Liao WS, Fuller GN, Zhang W 2003. Insulin-like growth factor binding protein 2 enhances glioblastoma invasion by activating invasion-enhancing genes. Cancer Res 63: 4315–4321 [PubMed] [Google Scholar]
- Wang H, Zhang W, Huang HJ, Liao WS, Fuller GN 2004. Analysis of the activation status of Akt, NFκB, and Stat3 in human diffuse gliomas. Lab Invest 84: 941–951 [DOI] [PubMed] [Google Scholar]
- Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C, et al. 2007. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang J, Zheng H, Tomasek GJ, Zhang P, McKeever PE, Lee EY, Zhu Y 2009. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell 15: 514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Chadalavada K, Wilshire J, Kowalik U, Hovinga KE, Geber A, Fligelman B, Leversha M, Brennan C, Tabar V 2010. Glioblastoma stem-like cells give rise to tumour endothelium. Nature 468: 829–833 [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. 2010. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17: 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrington NM, Gianino SM, Jackson E, Goldhoff P, Garbow JR, Piwnica-Worms D, Gutmann DH, Rubin JB 2010. Cyclic AMP suppression is sufficient to induce gliomagenesis in a mouse model of neurofibromatosis-1. Cancer Res 70: 5717–5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick RS, Cheng YN, Rangarajan A, Ince TA, Weinberg RA 2003. Ras modulates Myc activity to repress thrombospondin-1 expression and increase tumor angiogenesis. Cancer Cell 3: 219–231 [DOI] [PubMed] [Google Scholar]
- Wee B, Charles N, Holland EC 2012. Animal models to study cancer-initiating cells from glioblastoma. Front Biosci 17: 2243–2258 [DOI] [PubMed] [Google Scholar]
- Weir B, Zhao X, Meyerson M 2004. Somatic alterations in the human cancer genome. Cancer Cell 6: 433–438 [DOI] [PubMed] [Google Scholar]
- Weis SM, Cheresh DA 2011. Tumor angiogenesis: Molecular pathways and therapeutic targets. Nat Med 17: 1359–1370 [DOI] [PubMed] [Google Scholar]
- Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, Degroot J, Wick W, Gilbert MR, Lassman AB, et al. 2010. Updated response assessment criteria for high-grade gliomas: Response assessment in neuro-oncology working group. J Clin Oncol 28: 1963–1972 [DOI] [PubMed] [Google Scholar]
- Whiteman DC, Zhou XP, Cummings MC, Pavey S, Hayward NK, Eng C 2002. Nuclear PTEN expression and clinicopathologic features in a population-based series of primary cutaneous melanoma. Int J Cancer 99: 63–67 [DOI] [PubMed] [Google Scholar]
- Wiedemeyer R, Brennan C, Heffernan TP, Xiao Y, Mahoney J, Protopopov A, Zheng H, Bignell G, Furnari F, Cavenee WK, et al. 2008. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell 13: 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemeyer WR, Dunn IF, Quayle SN, Zhang J, Chheda MG, Dunn GP, Zhuang L, Rosenbluh J, Chen S, Xiao Y, et al. 2010. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci 107: 11501–11506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, Young GS, Makale M, Hu X, Yildirim N, Cui K, Wong ST, Kesari S 2011. Characterization of a human tumorsphere glioma orthotopic model using magnetic resonance imaging. J Neurooncol 104: 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. 2012. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44: 251–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykosky J, Gibo DM, Stanton C, Debinski W 2005. EphA2 as a novel molecular marker and target in glioblastoma multiforme. Mol Cancer Res 3: 541–551 [DOI] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. 2009. IDH1 and IDH2 mutations in gliomas. N Engl J Med 360: 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, Schlederer M, Johns C, Altorki N, Mittal V, et al. 2010. TGF-β IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci 107: 15535–15540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip S, Butterfield YS, Morozova O, Chittaranjan S, Blough MD, An J, Birol I, Chesnelong C, Chiu R, Chuah E, et al. 2012. Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J Pathol 226: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordanova YN, Moritz-Gasser S, Duffau H 2011. Awake surgery for WHO Grade II gliomas within ‘noneloquent' areas in the left dominant hemisphere: Toward a ‘supratotal' resection. Clinical article. J Neurosurg 115: 232–239 [DOI] [PubMed] [Google Scholar]
- Yuan TL, Cantley LC 2008. PI3K pathway alterations in cancer: Variations on a theme. Oncogene 27: 5497–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. 2009. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science 324: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, et al. 2008. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 455: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Wiedemeyer R, Yan H, Quayle SN, Ivanova EV, Paik JH, Zhang H, Xiao Y, Perry SR, et al. 2010. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell 17: 497–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XP, Loukola A, Salovaara R, Nystrom-Lahti M, Peltomaki P, de la Chapelle A, Aaltonen LA, Eng C 2002. PTEN mutational spectra, expression levels, and subcellular localization in microsatellite stable and unstable colorectal cancers. Am J Pathol 161: 439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Guignard F, Zhao D, Liu L, Burns DK, Mason RP, Messing A, Parada LF 2005. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell 8: 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Acquaviva J, Ramachandran P, Boskovitz A, Woolfenden S, Pfannl R, Bronson RT, Chen JW, Weissleder R, Housman DE, et al. 2009. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci 106: 2712–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler DS, Wright RD, Kesari S, Lemieux ME, Tran MA, Jain M, Zawel L, Kung AL 2008. Resistance of human glioblastoma multiforme cells to growth factor inhibitors is overcome by blockade of inhibitor of apoptosis proteins. J Clin Invest 118: 3109–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L 2005. Mosaic analysis with double markers in mice. Cell 121: 479–492 [DOI] [PubMed] [Google Scholar]