Plants sense shade (i.e., a proximity to their neighbors) as a decrease in the ratio of red to far-red light, which triggers a series of developmental responses. In Arabidopsis, phytochrome B (PHYB) is the major sensor of shade, but PHYB excitation has not been linked directly to a growth response. In this study, Chory and colleagues show that the bHLH transcription factor PIF7, an interactor with PHYB, accumulates in its dephosphorylated form in shade, allowing it to bind auxin biosynthetic genes and increase their expression. Thus, this study delineates a complete pathway from excitation of PHYB in response to shade to changes in plant architecture.
Keywords: auxin, phytochrome, shade avoidance
Abstract
Plants sense neighbor proximity as a decrease in the ratio of red to far-red light, which triggers a series of developmental responses. In Arabidopsis, phytochrome B (PHYB) is the major sensor of shade, but PHYB excitation has not been linked directly to a growth response. We show that the basic helix–loop–helix (bHLH) transcription factor PIF7 (phytochrome-interacting factor 7), an interactor of PHYB, accumulates in its dephosphorylated form in shade, allowing it to bind auxin biosynthetic genes and increase their expression. New auxin synthesized through a PIF7-regulated pathway is required for shade-induced growth, linking directly the perception of a light quality signal to a rapid growth response.
Plants sense self-shading or nearby plants as a change in the ratio of red (R) to far-red (FR) light. In full sunlight, the R:FR ratio is high (>1), but this ratio is reduced when plants are shaded, owing to an increase in far-red light that is reflected from cell walls and a drop in red light by chlorophyll absorption (R:FR <1). When a sun-loving plant such as Arabidopsis thaliana senses shade, a rapid response is triggered that alters plant architecture (stems and petioles elongate, leaf and root development is arrested, and leaf angle changes). If the plant remains under a canopy for prolonged periods of time, a shade avoidance syndrome (SAS) is elicited. During SAS, chlorophyll is reduced, flowering is initiated early with reduced seed set, and the plant becomes susceptible to herbivores (Izaguirre et al. 2006). Shade avoidance is thus an adaptive response to a change in light quality (Schmitt et al. 2003), which also contributes losses in yield (Ballare 1999; Franklin and Whitelam 2005).
The red/far-red-light-absorbing photoreceptor phytochrome B (PHYB) is the major sensor of shade light (Reed et al. 1993). When the R:FR is >1, the red-absorbing cytosolic form of PHYB (Pr, λmax = 660) is excited, photoconverts to a far-red-absorbing form (Pfr, λmax = 730), and moves to the nucleus, where it interacts with a subset of basic helix–loop–helix (bHLH) transcription factors called PIFs (phytochrome-interacting factors), leading to their rapid phosphorylation and degradation (Leivar and Quail 2011). In the stem, this causes a reduction of growth rate. In contrast, when the R:FR is low (<1), Pfr becomes Pr (an inactive form), allowing PIFs to accumulate and bind promoters of genes that promote cell elongation in the hypocotyl (Franklin 2008). Of the seven PIFs found in Arabidopsis, PIF4 and PIF5 have been implicated in SAS (Lorrain et al. 2008; Leivar and Quail 2011); however, a pif4pif5 double mutant retains a shade avoidance response to low R:FR, suggesting that there is redundancy with additional PIFs or that other mechanisms regulate this response. Moreover, although changes in the levels of multiple plant hormones are involved in SAS (Djakovic-Petrovic et al. 2007; Tao et al. 2008; Pierik et al. 2009; Kozuka et al. 2010), likely leading to the observed changes in growth rate and architecture, little is known about how light regulates plant hormone biosynthesis. Here we identify the bHLH transcription factor PIF7 as the missing link required for the early growth response to shade, thereby linking PHYB to the accumulation of new auxin in hypocotyls.
Results and Discussion
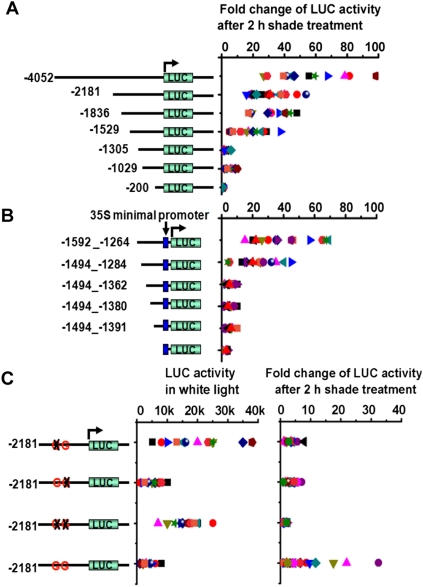
Previous studies have shown that >100 genes are up-regulated in Arabidopsis seedlings exposed to 1 h of shade (Sessa et al. 2005; Tao et al. 2008). One of these genes, PIL1 is induced >100-fold within 30 min of exposure to shade (Salter et al. 2003), making it an excellent marker for shade avoidance. To define the promoter elements that confer shade inducibility to PIL1's promoter, we made a series of 5′ deletions of the promoter, fused them to a luciferase (LUC) reporter with a 3′ untranslated region (UTR) of PIL1, and created stably transformed lines of Arabidopsis (Fig. 1A). At least 16 independent transgenic lines for each construct were grown in high R:FR (white light) and transferred for 2 h to low R:FR (simulated shade). These experiments indicated that a 210-base-pair (bp) region (from −1494 to −1284) was required for shade-induced expression of the PIL1 gene (Fig. 1A). This region was also sufficient for shade induction when fused to a minimal promoter (Fig. 1B).
Figure 1.
Identification of shade-responsive elements in the PIL1 promoter. (A) Induction of the PIL1 promoter after 2 h of shade treatment implicates a minimal region from −1529 to −1305 as required for shade responsiveness. A luciferase reporter was flanked with a series of 5′ deletions of PIL1 promoter and a full length of PIL1's 3′ UTR and transformed into Arabidopsis. Plotted is the fold change over white light of individual lines using a minimum of 16 independent lines. Each data point is a single measurement from an independent transformed line. (B) PIL1's 210-bp element (−1494 to −1284) confers shade responsiveness to a minimal 35S promoter. At least 16 independent lines were analyzed. (C) Two G-boxes contained within the 210-bp shade-responsive region are necessary for PIL1 induction by shade. LUC activities under the control of a wild-type or mutant G-box in the 2181-bp PIL1 native promoter are measured under the white light condition (left) or after 2-h low R:FR treatments (right). A minimum of 16 independent lines were analyzed.
A closer examination revealed that the 210-bp region contained two G-boxes (CACGTG), which are previously described light response elements involved in the regulation of many light-regulated genes (Martinez-Garcia et al. 2000), including PIL1 (Hornitschek et al. 2009). Sequential mutation of either G-box or mutation of both G-boxes indicated that both were required for shade induction of PIL1 (Fig. 1C); however, as reported for other light-regulated genes, multimerized G-boxes alone were not sufficient for shade-induced PIL1 expression (Supplemental Fig. S1; Puente et al. 1996).
To identify transcription factors that bind to the PIL1 210-bp shade-responsive region, wild type and a G-box mutant variant were used as baits in a yeast one-hybrid screen of ∼1600 Arabidopsis transcription factors (Pruneda-Paz et al. 2009). We identified several bHLH transcription factors that bound to the wild-type, but not a mutated, G-box (Supplemental Fig. S2). One of these is PIF7 (bHLH 72), previously described as a PHYB-interacting protein that plays a redundant role with other PIFs in seedling emergence (Leivar et al. 2008).
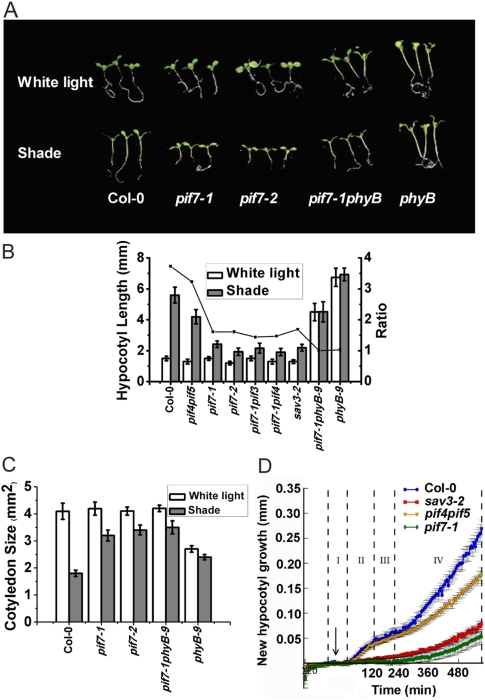
Two pif7 T-DNA insertion lines of Arabidopsis, pif7-1 (weak) and pif7-2 (strong), were obtained for functional studies (Leivar et al. 2008). In white light, both mutant lines appeared similar to wild type; however, in shade, pif7 mutants had short hypocotyls and expanded cotyledons (Fig. 2A–C), indicative of defects in SAS. pif7-1 partially rescued the constitutively shade response of a phyB-null allele in white light (Reed et al. 1993), indicating that PIF7 is a positive regulator of SAS that acts downstream from PHYB (Fig. 2A–C).
Figure 2.
pif7 mutants lack an early shade avoidance response. (A) Representative seedlings of Col-0 and pif mutants grown in white light and shade. Seedlings were grown in white light for 3 d and then moved to white light or simulated shade for 5 d. (B) Quantification of hypocotyl length of Col-0, pif4pif5 (pif4-101pif5-1), pif7 mutants and sav3-2. The left Y-axis is hypocotyl lengths, and the right Y-axis is the ratio of hypocotyl lengths in shade to that in white light. (C) Quantification of cotyledon area of Col-0, pif7 mutants, and phyB-9 in response to white light and shade. (D) pif7 hypocotyls have an early growth defect that is similar to sav3. New hypocotyl growth of pif7-1 after transfer to shade was quantified using HyDE (hypocotyl-determining engine) (Cole et al. 2010). The arrow indicates the start of shade treatment. Dotted lines and roman numerals indicate phases of growth.
During the first hour of shade avoidance, the levels of auxin in the Arabidopsis shoot increase by >50% over the levels found in white-light-grown wild type. New auxin is synthesized from tryptophan through TAA1, an aminotransferase encoded by the SAV3 gene (Tao et al. 2008). sav3 mutants have reduced auxin in white light and do not make new auxin in shade. In previous studies, we designed an imaging system and showed that the rapid, initial hypocotyl growth rate during SAS is biphasic and that sav3 mutants are lacking the first growth spurt (Cole et al. 2010). In the shade, pif7-1 hypocotyls elongated with a growth pattern similar to sav3-2 (Fig. 2D), suggesting that PIF7 is involved in regulating the auxin response to shade.
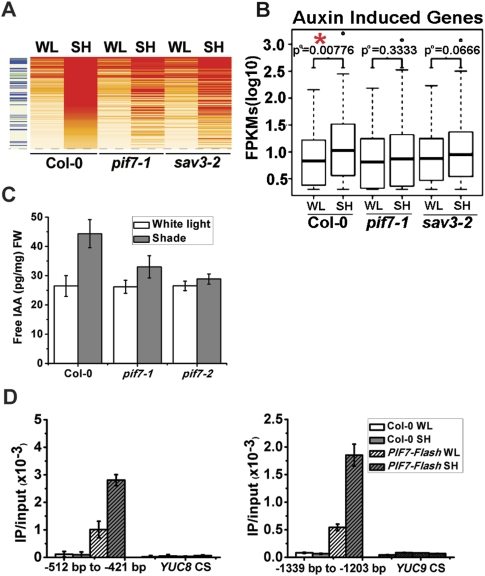
To determine possible targets of PIF7, we used RNA sequencing (Lister et al. 2008) to compare the genome-wide expression patterns of 5-d-old wild type, pif7-1, and sav3-2 in white light versus white light plus 1 h of shade. In wild type, shade treatment induced a total of 144 transcripts by >1.7 log2-fold (Supplemental Table S1). In pif7-1, 109 of these 144 transcripts did not respond to shade treatment, and 104 of 144 did not respond to shade in sav3-2 (Fig. 3A; Supplemental Table S1). Eighty-six genes that did not respond to shade treatment were shared between pif7-1 and sav3-2. Gene ontology (GO) functional prediction of these 86 genes showed an enrichment for auxin response genes (35%) (Supplemental Table S2). To determine the correlation between auxin and SAS pathways, we compared a list of 335 auxin-induced transcripts (Zhao et al. 2003; Nemhauser et al. 2004) against our shade-induced genes. In wild type, the distribution of the expression values of auxin-induced transcripts was significantly different between white light and shaded conditions (P-value = 0.00776), whereas these same transcripts were not differentially expressed between treatments in pif7-1 (P-value = 0.3333) and sav3-2 (P-value = 0.0666) (Fig. 3B; Supplemental Table S3). Together, these results suggest that PIF7 plays a role in auxin-related gene expression, a conclusion that was validated using quantitative real-time PCR (qPCR) (Supplemental Fig. S3). Among the genes whose expression did not change in shade in pif7 mutants were the auxin biosynthesis genes (YUCCA2, YUCCA5, YUCCA8, and YUCCA9), auxin transporters (PIN3 and PIN4), and auxin response genes (IAA29 and GH3.3), indicating a possible involvement of PIF7 in multiple aspects of auxin regulation in shade. PIF7 also regulates the expression of several transcription factors (PIL1 and ATHB2) in shade (Supplemental Fig. S3).
Figure 3.
pif7 mutants are defective in auxin biosynthesis. (A) Heat map representation of shade-regulated gene expression in Col-0 (wild type), pif7-1, and sav3-2. Red, orange, and white rows indicate RNA expression in high, medium, and low levels, respectively. The left column indicates auxin-regulated genes. Blue bars indicate auxin-induced genes, and the green bar indicates auxin-repressed genes. (B) Box plot representation of 335 auxin-induced genes between different genotypes under different light conditions. (FPKMs) Fragments per kilobase of exon per million fragments mapped. (C) Free IAA levels in Col-0 and pif7 mutants under white light and simulated shade. Col-0 and pif7 were grown in white light and moved to white light or shade for 1 h. The aerial parts were collected to measure free IAA level. (D) Direct binding of PIF7 with the G-box region of the promoters of YUC8 (−515 to −421, containing two G-boxes) and YUC9 (−1339 to −1203 bp, containing two G-boxes). After 1 h of shade treatment, ChIP assays were performed with Flag M2 antibody using 35S::PIF7-Flash transgenic plants. ChIP assays were quantified by real-time PCR after normalizing with the input DNA. PCR was performed with primers specific for the G-box region. The coding sequence (CS) region is used as a negative control.
We conducted functional studies of auxin biosynthesis and signaling pathways in pif7 mutants. An auxin signaling pathway appeared to be intact because (1) response genes (IAA5, IAA6, IAA29, and GH3.3) were induced by IAA (indole 3-acetic acid) treatment to the same or a greater degree in pif7 mutants compared with wild type (Supplemental Fig. S4), (2) pif7 hypocotyls elongated in white light in response to exogenous application of the auxin analog picloram (Supplemental Fig. S5; Sorin et al. 2005), and (3) 5 μM picloram rescued pif7's short hypocotyl phenotype in the shade (Supplemental Fig. S5). To investigate whether PIF7 controlled auxin availability, we measured the endogenous auxin levels in pif7 mutants in white light and in white light plus 1 h of shade. In white light, wild type and pif7 mutants had similar free IAA levels, while shade treatment resulted in a 69% increase in free IAA in wild type, a 26% increase in pif7-1, and a 7% increase in pif7-2 (Fig. 3C). Thus, PIF7 either directly or indirectly affects shade-induced auxin biosynthesis.
A revised pathway for IAA biosynthesis has recently been proposed (Mashiguchi et al. 2011; Phillips et al. 2011; Won et al. 2011) in which Trp is converted to indole-3-pyruvic acid (IPA) through TAA1 (encoded by SAV3), and IPA is converted to IAA through the action of a family of 11 spatially and dynamically expressed flavin mono-oxygenases encoded by the YUCCA gene family (Zhao et al. 2001). Whereas SAV3 and PIF7 transcriptional levels were slightly repressed by shade (Supplemental Fig. S6; Tao et al. 2008), the auxin biosynthesis genes YUCCA2, YUCCA5, YUCCA8, and YUCCA9 were induced by shade in wild type and sav3 (Tao et al. 2008; this study). In pif7, this induction was diminished (Supplemental Fig. S7; Supplemental Table 3). Since YUCCAs appear to encode a rate-limiting step in auxin biosynthesis (Zhao et al. 2001; Mashiguchi et al. 2011; Phillips et al. 2011; Won et al. 2011), this increase in YUCCA2, YUCCA5, YUCCA8, and YUCCA9 RNA might be the cause of increased auxin during shade avoidance. We found that multiple YUCCA genes contain at least one G-box (Supplemental Fig. S8). One-hybrid analysis in yeast showed that PIF7 can activate YUC8 and YUC9 promoters (Supplemental Fig. S8). PIF7 binds directly to these promoters in Arabidopsis, as shown by chromatin immunoprecipitation (ChIP)-PCR (Fig. 3D; Supplemental Fig. S8). We tested a yuc3,5,7,8,9 quintuple mutant for its hypocotyl elongation response in shade. Young seedlings (3 d old) had a small, but significant, defect in hypocotyl elongation in the shade (Supplemental Fig. S9.), suggesting that increased expression of YUCCA5, YUCCA8, and YUCCA9 genes is required for the early response to shade. It is possible that this weak phenotype is due to redundancy among YUCCA family members and the possible complication of the available pool of IPA in individual tissues (Tao et al. 2008; Won et al. 2011).
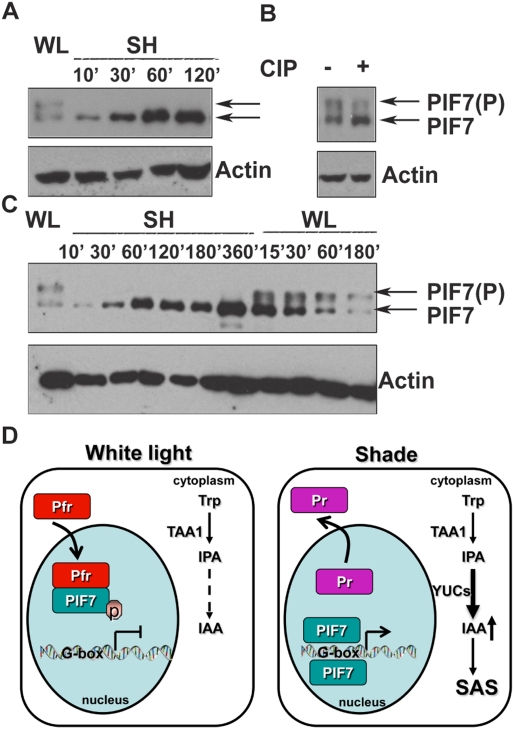
It has been shown that PIF7 can interact with PHYB during seedling emergence (Leivar et al. 2008), but the consequence of this interaction for shade avoidance is unknown. A previous study showed that unlike its close relatives, PIF7 is not rapidly degraded in the light (Leivar et al. 2008). To test whether shade regulates the activity of PIF7 by its post-translational modification, we stably overexpressed PIF7 tagged with a Flash tag (35S::PIF7-Flash; 9xMyc-6xHis-3xFlag) in the pif7-2 background. The transgenic lines grew taller than wild type under both white light and shade conditions (Supplemental Fig. S10). White-light-grown transgenic seedlings were treated with shade and used for Western analyses. As shown in Figure 4A, PIF7-Flash proteins from white-light-grown plants resolved as two different forms on SDS-PAGE gels. Treatment of extracted proteins with calf alkaline phosphatase (CIP) greatly attenuated the slowly migrating band (Fig. 4B), suggesting that the fast-migrating band is the dephosphorylated form and the slow band is the phosphorylated form of PIF7. Shade treatment rapidly decreased the amount of phosphorylated PIF7 while increasing the accumulation of dephosphorylated PIF7, which could be reversed when seedlings were returned to white light (Fig. 4C). This suggests that PIF7 phosphorylation is controlled by a phytochrome low-fluence response.
Figure 4.
PIF7 protein abundance and phosphorylation are regulated by shade. (A) Accumulation of a fast-migrating form of PIF7 within 10 min of transfer to shade. The Western blot shows reduction of a slow-migrating form of PIF7-Flash and accumulation of a faster-migrating form of PIF7-Flash in shade. PIF7-Flash protein was visualized by anti-Myc antibody, and loading was measured by anti-actin antibody. (WL) White light; (SH) shade. (B) PIF7's post-translational modification is a dephosphorylation. Treatment of lysates from white-light-grown transgenic plants with CIP attenuated the slowly migrating band. Loading was normalized to an actin control. (C) Dephosphorylation of PIF7 is rapid and photoreversible. Seedlings were grown in continuous white light for 5 d, treated with shade (R:FR = 0.7) for the times indicated, and then returned to white light. Note the reappearance of the slowly migrating band when seedlings were returned to white light. (D) A molecular model linking shade perception by PHYB to auxin-regulated gene expression. See the text for details.
These data, together with previous studies (Tao et al. 2008; Won et al. 2011), suggest a model for a series of events that link a change in light quality to modifications in plant architecture (Fig. 4D). High R:FR light is perceived by the photoreceptor PHYB, causing its translocation from the cytosol to the nucleus, where it interacts with phosphorylated PIF7. In early shade avoidance, the PHYB photoconverts to its inactive form (Pr) and is no longer associated with phospho-PIF7. PIF7 becomes rapidly dephosphorylated and binds G-boxes of auxin biosynthetic genes (as shown by ChIP-PCR in Fig. 4D; Supplemental Fig. S8) and other genes, resulting in a large increase in free IAA in the cotyledons (within 1 h in the shade) (Fig. 3D; Tao et al. 2008). This new IAA is transported out of the cotyledons into the stem and other tissues, where it initiates an elongation response. The phosphorylation and dephosphorylation of PIF7 is both fast and photoreversible (much faster than its modest degradation in white light). Thus, we favor a model in which dephosphorylation of PIF7 is the major regulatory mechanism for PIF7 by shade. Our model suggests the existence of a protein phosphatase and a protein kinase whose activities or availability are regulated by light quality changes.
PIF7 regulation of auxin biosynthesis is not the only mechanism by which the light environment controls growth. pif7 mutants have reduced expression of auxin signaling and transport genes, suggesting that these aspects of auxin response may also be altered by shade. In addition, during prolonged shade avoidance, when blue light becomes depleted by a thick canopy, we showed that the major transcription factors involved in elongation growth were PIF4 and PIF5 (Keller et al. 2011). More recently, PIF4 has been reported to control the expression of the genes for two auxin biosynthetic enzymes, SAV3 and CYP79B2, in response to high temperature (Franklin et al. 2011). This work and the current studies indicate that individual PIFs function to link discrete environmental stimuli to changes in plant architecture through similar transcriptional networks, a significant proportion of which is related to controlling the local levels and signaling of auxin (Leivar and Quail 2011).
Other plant hormones are known to have altered levels during shade avoidance. Whether similar mechanisms will account for the roles of gibberellins, cytokinins, brassinosteroids, and ethylene is a remaining piece of the puzzle. Nonetheless, these studies begin to explain how plants maintain phenotypic plasticity throughout their lives.
Materials and methods
Plant materials and growth conditions
All plant materials used in this study originated in the Columbia (Col-0) genetic background. The mutants used in this study have been described previously: pif7-1, pif7-2, pif7-1pif3-3, pif7-1pif4-2, pif7-1phyB-9 (Leivar et al. 2008), and pif4pif5 (pif4-101pif5-1) (Lorrain et al. 2008). Seeds were sterilized, stratified, and germinated on solid agar plates without sucrose. After stratification, plates were incubated in growth chambers under continuous white light (30–50 μE × m−2 × sec−1, R:FR ratio of 1.2) for 3 d, then the plates were either left in white light or transferred to simulated shade (LED light, red: 13 μE × m−2 × sec−1, and blue: 1.23 μE × m−2 × sec−1; far-red light: 20.2 μE × m−2 × sec−1, R:FR ratio of 0.7) for 5 d before hypocotyl measurements were made. Quantitative measurements of hypocotyl length and cotyledon area were performed on scanned images of seedlings using Scion Image software (http://www.scioncorp.com). For all measurements, at least 16 seedlings were used per treatment or genotype. In all figures, error bars represent standard error.
Hypocotyl growth measurements
New hypocotyl growth was measured using an image-based phenotyping platform as described previously (Cole et al. 2010).
RNA sequencing
Five-day-old seedlings were grown in duplicate under white light and then treated with shade for 1 h or an additional hour of white light. Roots were excised, and RNA was extracted from the aerial tissues using the Spectrum Plant Total RNA kit (Sigma-Aldrich). PolyA RNA was isolated from total RNA using the polyA Purist kit (Applied Biosystems/Ambion). One-hundred nanograms of polyA-selected RNA was used to construct a strand-specific RNA library construction following the manufacturer's protocol for the Whole Transcriptome kit (Ambion). Amplified libraries were size-selected using Ampure XP beads (Beckman) instead of the PAGE purification recommended in the protocol. RNA libraries were sequenced for 50 bp on the SOLiD4 platform (Life Technologies). Bioscope was used to align sequenced reads to the TAIR10 reference genome using the default parameters. Cufflinks version 0.9.3 (Trapnell et al. 2010) was used for determination of expression values. Any expression value <2 was replaced with a minimum expression value of 2. Data from each biological replicate were averaged for determination of shade induction. GO analysis of specific transcripts was determined using the DAVID Bioinformatics Resource 6.7 (Huang et al. 2009a,b; http://david.abcc.ncifcrf.gov/tools.jsp).
Quantification of IAA
For quantification of free IAA, Col-0 and pif7 mutants were grown under continuous white light for 5 d then treated with or without simulated shade for 1 h. Aerial parts of seedlings were weighed and collected. Measurements were performed as described previously (Ljung et al. 2005; Andersen et al. 2008) using four biological replicates.
Acknowledgments
We thank Dr. Peter H. Quail for the generous gifts of pif7-1, pif7-2, pif7-1phyB, pif7-1pif3, and pif7-1pif4 seeds; Dr. Bradley J.S.C. Olson (Kansas State University) for helpful comments on the manuscript; Dr. Christian Fankhauser (University of Lausanne) and Yunde Zhao (University of California at San Diego) for sharing unpublished data; and Roger Granbom for providing excellent technical assistance. R.J.S. was supported by an NIH NRSA post-doctoral fellowship (F32-HG004830). B.J.C. was supported by a National Science Foundation Integrative Graduate Education and Research Traineeship (0504645). K.L. was funded by the Swedish Governmental Agency for Innovation Systems (VINNOVA) and the Swedish Research Council (Vetenskapsrådet). J.R.E. was funded by the National Science Foundation (MCB-0929402 and MCB-1122246). These studies were funded by grants from the NIH (R01GM52413 to J.C., and 5RC2GM092412-02 and 5R01GM056006 to S.A.K.) and by the Howard Hughes Medical Institute to J.C. Sequencing data can be downloaded from NCBI SRA (SRA050699).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.187849.112.
References
- Andersen SU, Buechel S, Zhao Z, Ljung K, Novak O, Busch W, Schuster C, Lohmann JU 2008. Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 20: 88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballare CL 1999. Keeping up with the neighbours: Phytochrome sensing and other signalling mechanisms. Trends Plant Sci 4: 201. [DOI] [PubMed] [Google Scholar]
- Cole B, Kay SA, Chory J 2010. Automated analysis of hypocotyl growth dynamics during shade avoidance in Arabidopsis. Plant J 65: 991–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic-Petrovic T, de Wit M, Voesenek LA, Pierik R 2007. DELLA protein function in growth responses to canopy signals. Plant J 51: 117–126 [DOI] [PubMed] [Google Scholar]
- Franklin KA 2008. Shade avoidance. New Phytol 179: 930–944 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC 2005. Phytochromes and shade-avoidance responses in plants. Ann Bot (Lond) 96: 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. 2011. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C 2009. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009a. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Izaguirre MM, Mazza CA, Biondini M, Baldwin IT, Ballare CL 2006. Remote sensing of future competitors: Impacts on plant defenses. Proc Natl Acad Sci 103: 7170–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MM, Jaillais Y, Pedmale UV, Moreno JE, Chory J, Ballaré CL 2011. Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J 67: 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuka T, Kobayashi J, Horiguchi G, Demura T, Sakakibara H, Tsukaya H, Nagatani A 2010. Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol 153: 1608–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH 2011. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH 2008. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G 2005. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17: 1090–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C 2008. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH 2000. Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. 2011. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J 2004. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: e258 doi: 10.1371/journal.pbio.0020258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P 2011. vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23: 550–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, Djakovic-Petrovic T, Keuskamp DH, de Wit M, Voesenek LA 2009. Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and della proteins in Arabidopsis. Plant Physiol 149: 1701–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA 2009. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente P, Wei N, Deng XW 1996. Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J 15: 3732–3743 [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J 1993. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC 2003. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426: 680–683 [DOI] [PubMed] [Google Scholar]
- Schmitt J, Stinchcombe JR, Heschel MS, Huber H 2003. The adaptive evolution of plasticity: Phytochrome-mediated shade avoidance responses. Integr Comp Biol 43: 459–469 [DOI] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I 2005. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 19: 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C, Bussell JD, Camus I, Ljung K, Kowalczyk M, Geiss G, McKhann H, Garcion C, Vaucheret H, Sandberg G, et al. 2005. Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 17: 1343–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y 2011. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci 108: 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J 2001. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J 2003. SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301: 1107–1110 [DOI] [PubMed] [Google Scholar]