Chromatin post-translational modifications are widely studied, yet little is still known about the modification of linker histones, which play a key role in higher-order chromatin structure. Schneider and colleagues now provide the first characterization of an acetylation site on histone H1. Modified by Gcn5, H1.4K34ac is found enriched at active gene promoters and is able to regulate transcriptional activation by recruiting general transcription factors and promoting H1 mobility to create an open chromatin environment.
Keywords: linker histone H1, acetylation, chromatin, GCN5, transcription
Abstract
The linker histone H1 is a key player in chromatin organization, yet our understanding of the regulation of H1 functions by post-translational modifications is very limited. We provide here the first functional characterization of H1 acetylation. We show that H1.4K34 acetylation (H1.4K34ac) is mediated by GCN5 and is preferentially enriched at promoters of active genes, where it stimulates transcription by increasing H1 mobility and recruiting a general transcription factor. H1.4K34ac is dynamic during spermatogenesis and marks undifferentiated cells such as induced pluripotent stem (iPS) cells and testicular germ cell tumors. We propose a model for H1.4K34ac as a novel regulator of chromatin function with a dual role in transcriptional activation.
The linker histone H1 plays a key role in the formation of higher-order chromatin structure (Noll and Kornberg 1977; Allan et al. 1980; Zhou et al. 1998; Robinson and Rhodes 2006; Robinson et al. 2008), and early in vitro studies suggested that H1 acts as a global repressor of transcription (Schlissel and Brown 1984). However, in vivo H1 has been shown to be more dynamic than core histones (Lever et al. 2000; Misteli et al. 2000). Several knockout studies across a wide range of organisms revealed that H1 has multiple functions, including participation in the regulation of most nuclear processes (Izzo et al. 2008; Happel and Doenecke 2009), mediated at least in some cases by specific H1–protein interactions (McBryant et al. 2010). H1 depletion leads to perturbation in the expression of only a subset of genes, with both up-regulation and down-regulation (Shen and Gorovsky 1996; Hellauer et al. 2001; Fan et al. 2005). This suggests that in vivo, rather than being simply a global repressor, H1 can act as a fine tuner of gene expression. The activating role of the linker histone in gene expression is currently not understood. Post-translational modifications of proteins have been shown to regulate their function. In particular, the acetylation of lysine residues on core histone tails has been linked with transcription (Allfrey et al. 1964; Kurdistani and Grunstein 2003) by either a direct effect on chromatin structure (e.g., as shown for H4K16 acetylation) (Robinson et al. 2008) or the recruitment of effector bromodomain-containing proteins (Yang 2004). The linker histone H1 has been shown to be also acetylated in vivo (Garcia et al. 2004; Vaquero et al. 2004; Wisniewski et al. 2007), but this modification has not been functionally characterized.
Results and Discussion
The aim of this study was to provide for the first time functional insight into the acetylation of linker histone H1, a key player in chromatin function.
H1.4K34 is modified by acetylation
We chose the human linker histone subtype H1.4 as a model, as it is highly abundant, widely expressed (Lennox and Cohen 1983; Meergans et al. 1997), and the most conserved human linker histone subtype (Supplemental Fig. S1A). We focused on the N-terminal tail, a putative regulatory domain for H1 function (Allan et al. 1980). Within the H1.4 N-terminal tail, K34 is the most conserved lysine residue between both metazoan H1.4 homologs (Fig. 1A; Supplemental Fig. S1B, top) and the human H1 subtypes (Supplemental Fig. S1B, bottom). Interestingly, this residue is acetylated in vivo (Wisniewski et al. 2007). We therefore raised an antibody for H1.4K34 acetylation (H1.4K34ac). After affinity purification, this antibody specifically recognizes acetylated H1 (Fig. 1B; Supplemental Fig. S1C) and the H1.4K34ac peptide (Fig. 1C; Supplemental Fig. S1D). The H1 recognition is competed by the immunizing peptide, but not by any other peptide tested (Fig. 1D). The antibody has a preference for K34-acetylated H1.4 compared with other acetylated H1 subtypes (Supplemental Fig. S1E,F), and the signal is below the detection limit upon H1.4 knockdown (Supplemental Fig. S1G). Using this antibody, we confirmed the presence of acetylated H1.4K34 in vivo in a wide range of human cell lines (data not shown). In immunofluorescence (IF) assays, H1.4K34ac shows a distinct, dotted nuclear staining, depleted from DAPI-rich heterochromatic regions and nucleoli (Supplemental Fig. S2A), different from H1.4 distribution (Supplemental Fig. S2B). In contrast to the best-studied H1 modification, H1 phosphorylation, which is highly enriched during mitosis (Roth and Allis 1992), H1.4K34ac is stable throughout the cell cycle (Supplemental Fig. S2C–E).
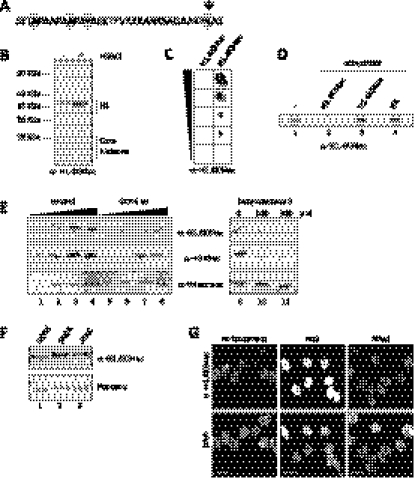
Figure 1.
H1.4K34 is acetylated in vivo and is a GCN5 target. (A) Sequence of the N-terminal tail of human H1.4. Residues showing 100% sequence identity between 10 metazoan species are highlighted. K34 is additionally indicated by an arrow (see also the alignment in Supplemental Fig. S1B). (B) Immunoblot analysis with anti-H1.4K34ac antibody of whole-cell lysates from MCF-7 cells not treated (−) or treated (+) with HDACi NaB and NAM. Corresponding Ponceau staining is shown in Supplemental Figure S1C. (C) Immuno-dot blot analysis of H1.4K34 acetylated (H1.4K34ac) and unmodified (H1.4K34un) peptides with anti-H1.4K34ac antibody. Triangle indicates fivefold peptide dilutions (starting from 3125 ng). (D) Competition in Western blot of anti-H1.4K34ac binding with no peptide (−) or H1.4K34 acetylated (H1.4K34ac) or unmodified (H1.4K34un) peptide and HA peptide (HA) as control. (E) Immunoblot analysis with anti-H1.4K34ac, anti-H3K9ac (positive control), and anti-tetra-ac-H4 (negative control) antibodies of control cells (lanes 1–4,9) and cells depleted for GCN5 activity by either shRNA-mediated knockdown (lanes 5–8) or chemical inhibition of GCN5 HAT activity with butyrolactone 3 (lanes 10,11). The triangles above lanes 1–4 and 5–8 indicate a twofold dilution series of loaded histones. For GCN5 knockdown control, see Supplemental Figure S3A. (F) Immunoblot analysis with anti-H1.4K34ac antibody of in vitro HAT assays on endogenous H1 with recombinant GCN5 and endogenous human GCN5-containing SAGA and ATAC complexes (see also control in Supplemental Fig. S3D). (G) IF with anti-H1.4K34ac antibody of MCF-7 cells untreated or treated with HDACi NaB or NAM.
H1.4K34 is a GCN5 target
In order to identify the enzymes that acetylate H1.4K34, we used a candidate approach and depleted key members of the three main HAT classes: GCN5 (GNAT family), MOF (MYST family), and p300/CBP (orphan class). Immunoblot analysis revealed that H1.4K34ac was clearly decreased upon GCN5 knockdown (Fig. 1E, left panel; Supplemental Fig. S3A), whereas we did not observe significant changes upon p300, CBP, or MOF depletion (Supplemental Fig. S3B,C). In line with this, we also observed a reduction of H1.4K34ac signal upon treatment of cells with an inhibitor of GCN5, butyrolactone 3 (Fig. 1E, right panel; Biel et al. 2004), and GCN5-containing SAGA and ATAC complexes purified from HeLa cells acetylated H1.4K34 in our in vitro HAT assays (Fig. 1F; Supplemental Fig. S3D). We also confirmed the activity of SAGA on H1.4K34 using mass spectrometry analysis (Supplemental Fig. S3E). Taken together, these results show that GCN5 is able to acetylate H1.4K34 both in vitro and in vivo. To gain insight into potential deacetylases targeting H1.4K34ac, we used histone deacetylase inhibitors (HDACi). HDACi targeting collectively class I and class II HDACs (sodium butyrate [NaB] and trichostatin A [TSA]) strongly enhanced H1.4K34ac levels, whereas class III (sirtuin) inhibitor (nicotinamide [NAM]) had no effect, as assayed by IF (Fig. 1G) and immunoblot (Fig. 1B; Supplemental Fig. S3F). This suggests that H1.4K34ac can be deacetylated by class I and class II HDACs, but not by sirtuins.
H1.4K34ac is enriched at transcription start sites (TSSs) of active genes as well as putative regulatory regions genome-wide
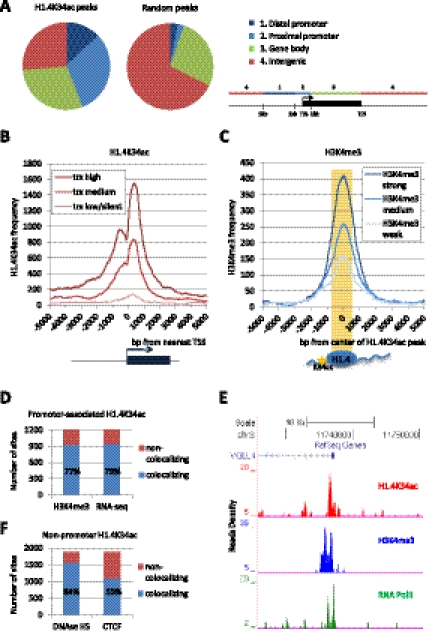
To reveal the genome-wide distribution of H1.4K34ac, we performed chromatin immunoprecipitation coupled to deep sequencing (ChIP-seq) in human MCF-7 cells and compared the distribution of H1.4K34ac with other chromatin features. We found that H1.4K34ac is highly enriched at promoters (Fig. 2A). Plotting the distribution of H1.4K34ac-binding regions around all human TSSs demonstrates that H1.4K34ac is enriched at the TSSs of active genes and is indicative of their level of transcription (Fig. 2B). A well-established mark for active genes is H3K4 trimethylation (H3K4me3) (Kouzarides 2007). The distribution of H3K4me3 around the center of H1.4K34ac peaks reveals that H1.4K34ac enrichment correlates with high H3K4me3 occupancy (Fig. 2C). In particular, 77% of promoter-associated H1.4K34ac-enriched regions colocalize with H3K4me3-enriched sites, and 79% colocalize with mRNA detected by RNA sequencing (RNA-seq), further strengthening the link of H1.4K34ac with active transcription (Fig. 2D). This can be exemplified by the VGLL4 locus, where H1.4K34ac co-occupies the promoter region together with H3K4me3 and RNA polymerase II (Fig. 2E). Furthermore, the H1.4K34ac-enriched sites outside of proximal promoter regions colocalize with DNase I-hypersensitive sites (84%) and CTCF-binding sites (59%), suggesting that H1.4K34ac marks regulatory regions (Fig. 2F). Additionally, we verified the presence of GCN5 at multiple H1.4K34ac sites by ChIP-qPCR (Supplemental Fig. S4C). Note also that H1.4K34ac-enriched regions tend to show a reduced linker histone occupancy (Supplemental Fig. S4D,E), consistent with nucleosome depletion from active regulatory regions (Lee et al. 2004; Heintzman et al. 2007). Taken together, our data show that H1.4K34ac is preferentially enriched at TSSs of active genes as well as putative regulatory regions. This also constitutes the first genome-wide mapping of a linker histone H1 modification.
Figure 2.
H1.4K34 acetylation is enriched at TSSs of active genes as well as regulatory regions. (A) Distribution of the H1.4K34ac-enriched sites (peaks) in MCF-7 cells relative to University of California at Santa Cruz RefGene coordinates. The distribution of random peaks obtained from control immunoprecipitation is shown as a control. Distal promoter has been defined as a region 1–5 kb upstream of the TSS, proximal promoter has been defined as the TSS flanked by 1 kb upstream and downstream, and the gene body has been defined as the region between the proximal promoter and the transcription end site (TES). The genome-wide H1.4K34ac occupancy was determined by ChIP-seq and bioinformatic analysis. For validation of H1.4K34ac-enriched regions, see Supplemental Figure S4, A and B. (B) H1.4K34ac occupancy profile 5 kb around the TSSs of three categories of transcripts: highly transcribed (top 33.3% of transcripts: trx high), medium transcribed (middle 33.3% of transcripts: trx medium), and lowly/not transcribed (bottom 33.3% of transcripts: trx low/silent). (C) H3K4me3 occupancy profile around the center of H1.4K34ac-enriched sites. Prior to plotting, H3K4me3 peaks have been divided into three equally sized categories according to their enrichment level (i.e., score): top 33.3%: H3K4me3 strong; middle 33.3%: H3K4me3 medium; bottom 33.3%: H3K4me3 weak. Note that strong H3K4me3 peaks are more frequently found at H1.4K34ac-binding regions than weak H3K4me3 peaks. (D) Bar graph showing the colocalization of proximal promoter-associated H1.4K34ac-enriched sites with H3K4me3-enriched regions and mRNA detected by RNA sequencing in MCF-7 cells (RNA-seq). (E) H1.4K34ac ChIP-seq tag profile at the VGLL4 locus. Shown are the ChIP-seq read densities of H1.4K34ac along with H3K4me3 and RNA polymerase II (Pol II) in a 20-kb region surrounding the VGLL4 TSS. (F) Bar graph showing the colocalization of H1.4K34ac-binding sites outside of proximal promoter regions (nonpromoter H1.4K34ac) with DNase I-hypersensitive sites (DNase HS) and CTCF-binding sites.
H1.4K34ac positively affects transcription
To complement this static snapshot of H1.4K34ac distribution, we followed the kinetics of H1.4K34ac upon transcriptional activation using TPA induction, since GCN5 had been implicated in this process (Nagy et al. 2010). As shown in Figure 3A (and Supplemental Fig. S5A,B), TPA induction results in recruitment of GCN5 as well as enrichment of H1.4K34ac and H3K4me3 at the promoters of immediate early-induced (IE) genes, demonstrating the dynamicity of H1.4K34ac.
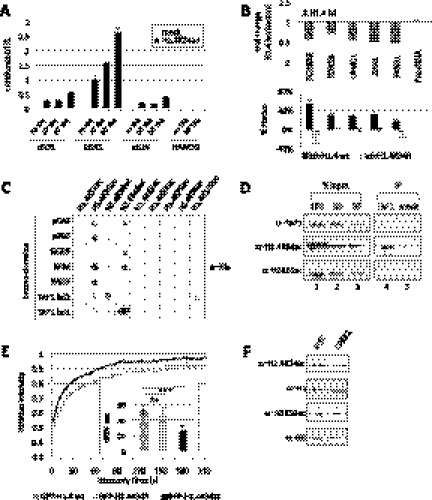
Figure 3.
H1.4K34ac affects transcription and is up-regulated in induced pluripotent stem (iPS) cells. (A) Time course of H1.4K34ac enrichment at the promoters of three immediate early (IE) genes (cFOS, EGR1, and cJUN) and NANOG (negative control) analyzed by ChIP before and after TPA induction for 10 or 30 min. Standard deviation was calculated from triplicates of qPCR for each time point. The enrichment of H1.4K34ac was normalized to H1 occupancy. See also Supplemental Figure S5, A and B, for gene induction and additional ChIP data. (B) H1.4 depletion-induced transcriptional defects can be partially rescued by H1.4 wild type but not H1.4K34R. Expression of representative H1.4K34ac target genes was analyzed by RT and real-time PCR in HeLa cells depleted of H1.4 (H1.4kd) and control cells. (Top panel) Fold changes in the expression of the indicated genes were calculated as a ratio between H1.4kd and control RNA levels in five biological replicates. (Bottom panel) Percentage of rescue of the H1.4kd-induced gene repression upon transfection of shRNA-resistant H1.4 wild type or H1.4K34R mutant, performed in two biological replicates. Error bars represent standard deviation. See also control in Supplemental Figure S5C. (C) H1.4K34ac can be bound by bromodomain 2 of TAF1 (TAF1 bd2). Screen of the indicated his-tagged bromodomains for binding to H1.4 acetylated peptides, as assayed using anti-His antibody. Specificity controls are shown in Supplemental Figure S6B. (D) H1.4K34ac coimmunoprecipitates with endogenous TAF1. Immunoblot analysis with anti-TAF1, anti-H1.4K34ac, and anti-H3K56ac antibodies of HeLa nuclear extract upon immunoprecipitation with anti-TAF1 antibody (TAF1-IP) or control immunoprecipitation (mock). (E) Mimicking H1.4K34 acetylation leads to increased linker histone mobility in living cells. FRAP analysis of HEK293 cell lines stably expressing GFP-H1.4 wild type, GFP-H1.4K34R mutant (unmodified protein mimic), or GFP-H1.4K34Q mutant (acetyl mimic). (Graph) FRAP recovery kinetics; (box graph) 80% recovery times; error bars indicate standard deviation. (F) H1.4K34ac is up-regulated in iPS cells. Immunoblot analysis using anti-H1.4K34ac, anti-H1, anti-H3K56ac, and anti-H3 antibodies of histones extracted from iPS cells (left) or the differentiated precursor CPRE2 cells (right).
In order to test the functional significance of H1.4K34ac in gene expression, we next examined the transcriptional phenotype of H1.4 depletion combined with H1.4 reintroduction. As shown in Figure 3B, we observed a down-regulation of multiple H1.4K34ac target genes upon H1.4 knockdown. Interestingly, we can partially rescue the effect of H1.4 depletion on transcription of these H1.4K34ac targets by reintroduction of wild-type H1.4, but not by reintroduction of the H1.4K34R mutant (which cannot be acetylated), strongly pointing toward a functional role of H1.4K34ac in transcriptional activation.
We hypothesized that there might be two—not mutually exclusive—mechanisms for how H1.4K34ac could activate transcription: H1.4K34ac could have a direct effect on transcription or could lower linker histone affinity to chromatin, making it more mobile and creating a more accessible chromatin environment. To test the first hypothesis and differentiate from effects on H1 mobility, we used Gal4 fusions of H1.4 wild type or mutants: H1.4K34Q mimicking K34 acetylation and H1.4K34R mimicking the unacetylated protein. These Gal4 fusions specifically recruit the linker histone isoform H1.4 to a promoter and allow testing for more direct effects of H1.4K34 on transcription. In our assays, both Gal4-H1.4 wild type and Gal4-H1.4K34R repressed transcription (Supplemental Fig. S6A). A similar effect for H1.4 wild type and H1.4K34R was expected, since the bulk of H1 is unacetylated (data not shown). Conversely, Gal4-H1.4K34Q could increase reporter activity. Although Gal4-H1.4 might not recapitulate normal linker histone–nucleosome interactions, these results suggest that specifically tethered H1.4K34ac is able to enhance gene expression and could turn a transcriptional repressor into an activator.
H1.4K34ac can recruit TAF1
In order to investigate whether there is an H1.4K34ac “binder” that might mediate transcriptional activation by H1.4K34ac, we performed a candidate screening for H1.4K34ac-binding partners. As shown in Figure 3C and Supplemental Fig. S6B, we found that the bromodomain 2 of the TAF1 subunit of the transcription factor TFIID can bind the H1.4K34ac peptide. This TAF1–H1.4K34ac binding is also relevant in vivo, as H1.4K34ac was enriched when endogenous TAF1 protein was immunoprecipitated (Fig. 3D). TAF1–H1.4K34ac binding is also reflected in their genomic distribution: Seventy-two percent of promoter-associated H1.4K34ac-enriched regions colocalize with TAF1-binding sites (Supplemental Fig. S6C). Note that only a fraction of TAF1 colocalizes with H1.4K34ac, consistent with the fact that this protein has been also shown to bind to acetylated core histones (Jacobson et al. 2000; Agalioti et al. 2002), and another TFIID subunit interacts with H3K4me3 (Vermeulen et al. 2007). Thus, the above data suggest that H1.4K34ac can help recruit a transcriptional activator to promoters.
H1.4K34ac increases H1 mobility and is up-regulated in induced pluripotent stem (iPS) cells
Next, we wanted to test whether H1.4K34ac can influence linker histone mobility in vivo. To this end, we created cell lines stably expressing GFP-H1.4 wild type and GFP-H1.4K34 mutants to use in fluorescence recovery after photobleaching (FRAP) assays (Fig. 3E). The wild-type H1.4 and the H1.4K34R mutant displayed comparable recovery kinetics. Importantly, the H1.4K34Q acetylation mimic showed an approximately twofold faster 80% recovery time (Fig. 3E). This strongly suggests that acetylation of H1.4K34 could lead to an increased linker histone mobility in living cells and hence a decreased H1.4 affinity to chromatin in vivo.
It has been previously proposed that the hyperdynamic binding of linker histone H1 is a functionally important hallmark of pluripotent stem cells, which allows generation of an active, “breathing” chromatin state (Meshorer et al. 2006). We hypothesized that H1 hyperacetylation could be involved in the increased H1 mobility. To test this, we checked the modification levels of histones from iPS cells as well as from the differentiated precursors (CPRE2). Interestingly, we found that H1.4K34ac levels increased in our iPS cells as compared with the CPRE2 cells (Fig. 3F).
H1.4K34ac shows a dynamic behavior during human spermatogenesis and marks seminomas
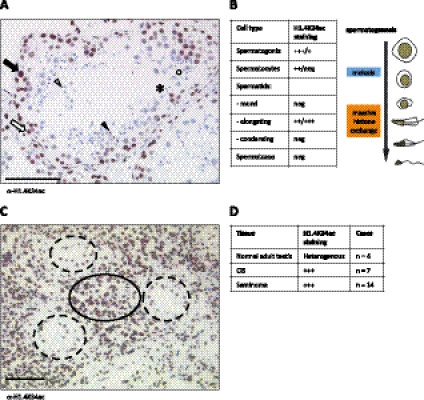
Dramatic changes in chromatin structure and in core histone modifications also occur during spermatogenesis: In maturating spermatids, histones are replaced by transition proteins and, later on, protamines (Gaucher et al. 2010). To study this differentiation process, we performed immunohistochemistry of biopsies from human testis and found that H1.4K34ac shows a dynamic behavior during spermatogenesis (Fig. 4A,B; Supplemental Fig. S7A). H1.4K34ac levels are high in the majority of immature germ cells (spermatogonia) and meiotic spermatocytes, but disappear at the post-meiotic round spermatid stage. Interestingly, we observed high levels of H1.4K34ac in the elongating spermatids (coinciding with chromatin reorganization) (Gaucher et al. 2010) and a loss of signal in condensing spermatids and spermatozoa (Fig. 4A,B). H1.4K34 acetylation could therefore be involved in the “opening” of chromatin necessary to exchange histones with their testis-specific variants and, later, protamines. This is also supported by our FRAP experiments, showing a decreased affinity of H1.4K34ac for chromatin.
Figure 4.
H1.4K34ac is dynamic during human spermatogenesis and strongly marks human seminomas. (A) Immunohistochemistry (IHC) of normal human testis stained for H1.4K34ac. (Black arrow) Spermatogonium; (white arrow) spermatocyte; (white arrowhead) round spermatid; (black arrowhead) elongated spermatid; (black circle) condensed spermatid; (*) spermatozoon. Bar, 100 μm. (B) Summary of H1.4K34ac staining patterns of consecutive stages of spermatogenesis. (neg) No staining detected; (+) faint staining; (++) apparent staining; (+++) strong staining; (+++/+) predominantly strong staining, some cells showing faint staining; (++/neg) predominantly apparent staining, some cells showing no staining; (++/+++) predominantly apparent staining, some cells showing strong staining. Schemata of spermatogenesis is shown on the right. (C) IHC with anti-H1.4K34ac antibody of a testicular biopsy showing both seminoma (black circle) and CIS lesions (dashed line circles). Bar, 100 μm. (D) Summary of H1.4K34ac staining patterns in normal testis, CIS, and invasive seminoma biopsies. Abbreviations as in B; (n) number of cases analyzed.
To follow up these findings in human testis and demonstrate the relevance of H1.4K34ac in human diseases, we went on to screen for alteration of H1.4K34ac levels in testicular germ cell tumors (TGCTs)—the most frequent malignancies in Caucasian males between 20 and 40 years of age (van de Geijn et al. 2009). We analyzed biopsies from two common types of testicular neoplastic lesions: carcinoma in situ (CIS) and seminomas (Fig. 4C,D). CIS is a common precursor of type II TGCTs, including seminomas. Seminomas constitute >50% of all invasive TGCTs. Relevant to our study, CIS and invasive TGCTs are totipotent, express stemness genes including OCT3/4 and NANOG, and can be considered stem cell tumors (Biermann et al. 2007; van de Geijn et al. 2009). We observed that in all patient samples tested (n = 21), H1.4K34ac strongly marks both CIS and invasive seminoma nuclei (Fig. 4C,D). Considering the increased levels of H1.4K34ac we observed in iPS cells, we believe that the marking of TGCTs by H1.4K34ac might reflect their undifferentiated stem cell-like phenotype. H1.4K34ac could therefore potentially serve as a novel diagnostic marker of those malignancies.
Conclusions
The data shown here allow us to propose the following model for H1.4K34ac functions in chromatin (Supplemental Fig. S8): In repressive chromatin, H1.4K34ac levels are low, and H1.4 has a higher affinity to chromatin, resulting in a longer residence time. When acetylated by GCN5 at K34, H1.4 binding to chromatin becomes more dynamic. This acetylated state is enriched at the promoters of transcriptionally active genes and at regulatory regions and can allow the local “opening up” of chromatin, facilitating the access for the transcriptional machinery and coactivators. Additionally, H1.4K34ac could help to recruit the general transcription factor TFIID to promoters via interaction with its TAF1 subunit. These events together facilitate transcription, thereby potentially turning H1.4 from a transcriptional repressor into an activator.
Importantly, TAF1 interactions with chromatin are an order of magnitude more transient than the residence time we observed for H1.4K34ac (de Graaf et al. 2010); therefore, H1.4K34ac mobility does not prohibit its ability to recruit TAF1. Note also that TAF1 has been shown to possess acetyltransferase activity toward the core histones, so its recruitment can lead to a positive feedback loop of acetylation.
We provided here the first systematic study of linker histone H1 acetylation and found that a single acetylation event on Lys 34 of H1.4 leads to significant effects on H1 mobility and transcription. However, H1 is also modified on other residues (Wisniewski et al. 2007; Zheng et al. 2010; and others), and those modifications might act synergistically. Further studies will be needed to unravel the cross-talk between different linker histone modifications as well as their interdependence with core histone modifications.
Materials and methods
All experiments were performed using human cell lines, including MCF-7, HeLa, HEK293, and 293T. The H1.4K34ac antibody used in this study was raised in rabbit against the AAKR(AcK)ASGPGC peptide. The affinity purification and antibody characterization procedures are described in the Supplemental Material. The expression and purification of hGCN5 using the baculovirus system was performed as published before (Demény et al. 2007). The purification of endogenous GCN5-containing SAGA and ATAC complexes from HeLa cells was as described previously (Nagy et al. 2009, 2010). Histone H1 extraction with perchloric acid (PCA) was performed as described previously (Wisniewski et al. 2007). The ChIP protocol and ChIP-seq data analysis are described in detail in the Supplemental Material. Supplemental Table S1 contains a list of peptides, Supplemental Table S2 contains a list of antibodies, and Supplemental Table S3 contains a list of primers used in this study. Further detailed Materials and Methods are also available in the Supplemental Material.
Acknowledgments
We thank L. Juan for H1.4 knockdown cells; W.L. Kraus and M. Gamble for sharing H1 chip–ChIP data; and A. Bannister, S. Robson, T. Manke, S. Diehl, A. Krebs, Z. Nagy, A. Riss, K. Karmodiya, T. Chelmicki, E. Hallacli, E. Wang, H. Kinyamu, and R.S. laboratory members for helpful discussions and reagents. Work in the R.S. laboratory is supported by the DFG (through SFB 746), an ERC starting grant, and the FRM (equipe labilise).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.182014.111.
References
- Agalioti T, Chen G, Thanos D 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111: 381–392 [DOI] [PubMed] [Google Scholar]
- Allan J, Hartman PG, Crane-Robinson C, Aviles FX 1980. The structure of histone H1 and its location in chromatin. Nature 288: 675–679 [DOI] [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE 1964. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci 51: 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biel M, Kretsovali A, Karatzali E, Papamatheakis J, Giannis A 2004. Design, synthesis, and biological evaluation of a small-molecule inhibitor of the histone acetyltransferase Gcn5. Angew Chem Int Ed Engl 43: 3974–3976 [DOI] [PubMed] [Google Scholar]
- Biermann K, Heukamp LC, Steger K, Zhou H, Franke FE, Sonnack V, Brehm R, Berg J, Bastian PJ, Muller SC, et al. 2007. Genome-wide expression profiling reveals new insights into pathogenesis and progression of testicular germ cell tumors. Cancer Genomics Proteomics 4: 359–367 [PubMed] [Google Scholar]
- de Graaf P, Mousson F, Geverts B, Scheer E, Tora L, Houtsmuller AB, Timmers HTM 2010. Chromatin interaction of TATA-binding protein is dynamically regulated in human cells. J Cell Sci 123: 2663–2671 [DOI] [PubMed] [Google Scholar]
- Demény MA, Soutoglou E, Nagy Z, Scheer E, Jànoshàzi A, Richardot M, Argentini M, Kessler P, Tora L 2007. Identification of a small TAF complex and its role in the assembly of TAF-containing complexes. PLoS ONE 2: e316 doi: 10.1371/journal.pone.0000316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI 2005. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123: 1199–1212 [DOI] [PubMed] [Google Scholar]
- Garcia BA, Busby SA, Barber CM, Shabanowitz J, Allis CD, Hunt DF 2004. Characterization of phosphorylation sites on histone H1 isoforms by tandem mass spectrometry. J Proteome Res 3: 1219–1227 [DOI] [PubMed] [Google Scholar]
- Gaucher J, Reynoird N, Montellier E, Boussouar F, Rousseaux S, Khochbin S 2010. From meiosis to postmeiotic events: The secrets of histone disappearance. FEBS J 277: 599–604 [DOI] [PubMed] [Google Scholar]
- Happel N, Doenecke D 2009. Histone H1 and its isoforms: Contribution to chromatin structure and function. Gene 431: 1–12 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Hellauer K, Sirard E, Turcotte B 2001. Decreased expression of specific genes in yeast cells lacking histone H1. J Biol Chem 276: 13587–13592 [DOI] [PubMed] [Google Scholar]
- Izzo A, Kamieniarz K, Schneider R 2008. The histone H1 family: Specific members, specific functions? Biol Chem 389: 333–343 [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288: 1422–1425 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Grunstein M 2003. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol 4: 276–284 [DOI] [PubMed] [Google Scholar]
- Lee C-K, Shibata Y, Rao B, Strahl BD, Lieb JD 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Lennox RW, Cohen LH 1983. The histone H1 complements of dividing and nondividing cells of the mouse. J Biol Chem 258: 262–268 [PubMed] [Google Scholar]
- Lever MA, Th'ng JP, Sun X, Hendzel MJ 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408: 873–876 [DOI] [PubMed] [Google Scholar]
- McBryant SJ, Lu X, Hansen JC 2010. Multifunctionality of the linker histones: An emerging role for protein-protein interactions. Cell Res 20: 519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meergans T, Albig W, Doenecke D 1997. Varied expression patterns of human H1 histone genes in different cell lines. DNA Cell Biol 16: 1041–1049 [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown David T, Misteli T 2006. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 10: 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Gunjan A, Hock R, Bustin M, Brown DT 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature 408: 877–881 [DOI] [PubMed] [Google Scholar]
- Nagy Z, Riss A, Romier C, le Guezennec X, Dongre AR, Orpinell M, Han J, Stunnenberg H, Tora L 2009. The human SPT20-containing SAGA complex plays a direct role in the regulation of endoplasmic reticulum stress-induced genes. Mol Cell Biol 29: 1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Riss A, Fujiyama S, Krebs A, Orpinell M, Jansen P, Cohen A, Stunnenberg HG, Kato S, Tora L 2010. The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell Mol Life Sci 67: 611–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M, Kornberg RD 1977. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol 109: 393–404 [DOI] [PubMed] [Google Scholar]
- Robinson PJJ, Rhodes D 2006. Structure of the ‘30 nm’ chromatin fibre: A key role for the linker histone. Curr Opin Struct Biol 16: 336–343 [DOI] [PubMed] [Google Scholar]
- Robinson PJJ, An W, Routh A, Martino F, Chapman L, Roeder RG, Rhodes D 2008. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol 381: 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth SY, Allis CD 1992. Chromatin condensation: Does histone H1 dephosphorylation play a role? Trends Biochem Sci 17: 93–98 [DOI] [PubMed] [Google Scholar]
- Schlissel MS, Brown DD 1984. The transcriptional regulation of Xenopus 5s RNA genes in chromatin: The roles of active stable transcription complexes and histone H1. Cell 37: 903–913 [DOI] [PubMed] [Google Scholar]
- Shen X, Gorovsky MA 1996. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell 86: 475–483 [DOI] [PubMed] [Google Scholar]
- van de Geijn G-JM, Hersmus R, Looijenga LHJ 2009. Recent developments in testicular germ cell tumor research. Birth Defects Res C Embryo Today 87: 96–113 [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D 2004. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell 16: 93–105 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WWMP, van Schaik FMA, Varier RA, Baltissen MPA, Stunnenberg HG, Mann M, Timmers HTM 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Krüger S, Mann M 2007. Mass spectrometric mapping of linker histone H1 variants reveals multiple acetylations, methylations, and phosphorylation as well as differences between cell culture and tissue. Mol Cell Proteomics 6: 72–87 [DOI] [PubMed] [Google Scholar]
- Yang X-J 2004. Lysine acetylation and the bromodomain: A new partnership for signaling. Bioessays 26: 1076–1087 [DOI] [PubMed] [Google Scholar]
- Zheng Y, John S, Pesavento JJ, Schultz-Norton JR, Schiltz RL, Baek S, Nardulli AM, Hager GL, Kelleher NL, Mizzen CA 2010. Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J Cell Biol 189: 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YB, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S 1998. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature 395: 402–405 [DOI] [PubMed] [Google Scholar]