Few, if any, proteins have been studied in as many organisms as have the hemoglobins. They are found in bacteria, fungi, plants, and animals, serving physiological roles ranging from oxygen transport in the blood of vertebrates to catalyzing the combination of oxygen and nitric oxide to form nitrate in bacteria, yeast, and worms (1, 2). Hence hemoglobins and the globin genes encoding them have been an important system for investigating many biochemical and evolutionary issues. Furthermore, the tissue-specific and developmental regulation of globin gene expression has been studied intensively in mammals and birds. This led to the discovery of many cis-regulatory sequences such as promoters, enhancers, locus control regions (LCRs) (3), and insulators (4). If any model system were well understood both in terms of gene evolution and regulation, this should be the one. However, by their discovery of an orphaned β-like globin gene in marsupials, Wheeler et al. (5) show in a recent issue of PNAS that previously inferred relationships between the β-globin gene clusters in eutherian mammals and birds were oversimplified. The new data force a reconsideration of the evolutionary course of this gene cluster, how these regulatory elements evolved, and how they may be acting today.
… the eutherian ɛ-γ-δ-β-globin gene cluster appears to be descended from a different gene cluster from that which led to the avian ρ-β-ɛ cluster.
In all jawed vertebrates (gnathostomes) studied, distinctive forms of hemoglobin are produced during primitive erythropoiesis in embryonic life, followed by different hemoglobins produced during definitive erythropoiesis in fetal and adult life. These hemoglobins are tetramers of two α-like globins and two β-like globins, each with an associated heme to which oxygen can bind reversibly. The genes encoding the globins are clustered, with the α-like globin gene cluster on a different chromosome from the β-like globin gene cluster in birds and mammals. The human β-like globin gene cluster has five active globin genes, encoding the embryonic ɛ-globin, two fetal γ-globins, the adult δ-globin produced at low levels, and the abundant adult β-globin (Fig. 1). Extensive comparisons with homologous genes from other eutherian mammals deduced a parsimonious pathway for the evolution of this gene cluster by repeated gene duplications (6, 7), including a predicted cluster containing the ancestor to ɛ-globin and β-globin genes (Fig. 1). This prediction was verified by the discovery of linked ɛ-globin and β-globin genes in two marsupial species, the American opossum, Didelphis virginiana (8), and the Australian dunnart, Sminthopsis crassicaudata (9).
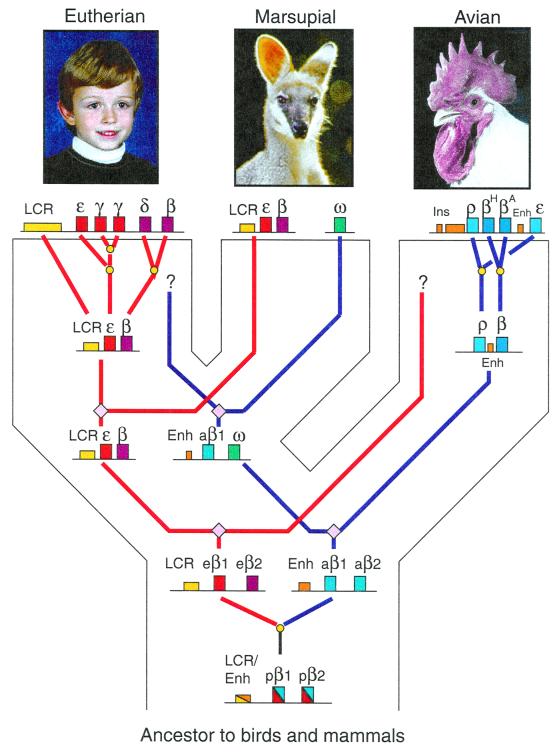
Figure 1.
Model for the independent evolution of β-like globin gene clusters in birds and mammals. The diagram shows major phyla as broad branches leading to eutherian mammals, marsupials, and birds, with the contemporary species at the top. Within the broad species branches are gene trees for the β-like globin gene clusters. The ancestral gene cluster (bottom of the diagram) has proto-β-globin genes (pβ1 and pβ2, each colored red and blue) and a regulatory element (yellow and orange rectangle for an LCR and/or enhancer). This gene cluster is proposed to have duplicated to generate the ancestor to the eutherian β-like globin genes (eβ1 and eβ2, colored in shades of red, with an LCR as a yellow rectangle) and the ancestor to the avian β-like globin genes (aβ1 and aβ2, colored in shades of blue, with an enhancer as an orange rectangle). The path of evolution to the genes related to the eutherian ɛ-γ-δ-β-globin gene cluster is drawn with red lines, and the path to the genes related to the avian ρ-β-ɛ-globin gene cluster is drawn with blue lines. Separations of genes by duplications are marked with yellow circles, and separations of genes by speciation events are marked with mauve diamonds. Note that the duplication to generate the ancestors to eutherian and avian β-like globin genes preceded the divergence of birds and mammals, hence both gene clusters were present in all three lineages depicted and still are found in contemporary marsupial mammals. The marsupial photographed is a pretty-faced wallaby.
Chickens also have a developmentally regulated β-like globin gene cluster, but it is rearranged relative to that in eutherians (Fig. 1), with two embryonic genes, encoding ρ-globin and ɛ-globin, at the ends of the cluster and two β-globin genes, one expressed at hatching (encoding βH-globin) and one expressed abundantly in adult red blood cells (encoding βA-globin). Given that the avian ρ-β-ɛ-globin gene cluster and the eutherian ɛ-γ-δ-β-globin gene clusters are the only β-like globin gene clusters known in these two taxa, it was natural to consider them to be orthologous, i.e., they were thought to be derived from the same gene cluster in the last common ancestor to birds and mammals. The discovery of the marsupial ω-globin (5) argues against this. Rather, the eutherian ɛ-γ-δ-β-globin gene cluster appears to be descended from a different gene cluster from that which led to the avian ρ-β-ɛ cluster.
Wallabies are medium-sized relatives of the kangaroos in Australia. The ω-globin was discovered as a component of a novel hemoglobin found in the blood of neonatal tammar wallabies (Macropus eugenii). The ω-globin gene is expressed just before and after the birth of the joey (5). A partial amino acid sequence of the tammar ω-globin indicated that it was more closely related to bird β-globin than to the eutherian, marsupial, or monotreme β-globins (10, 11). The new studies from Wheeler et al. (5) show that the ω-globin gene is widely distributed in marsupials and provide the DNA sequence of the ω-globin gene from two species of marsupials, the tammar and the dunnart. This allows a more rigorous and complete phylogenetic analysis. The marsupial ω-globin genes form a sister group with the avian β-, ɛ-, and ρ-globin genes, clearly separated from the group containing the eutherian, marsupial, and monotreme β-like globin genes. Finally, Wheeler et al. (5) show that the ω-globin gene is found on a separate chromosome from the ɛ- and β-globin gene cluster; hence it is an “orphaned” β-like gene.
These new data argue for a significant change in the view of hemoglobin gene evolution in the warm-blooded vertebrates (Fig. 1). Marsupial mammals have both a bird-like ω-globin gene and a β-like gene cluster similar to that of eutherian mammals. If they inherited both gene clusters from an ancestor common to birds and mammals, then the β-like globin gene clusters of eutherian mammals and avians are not orthologous. By this model, at a time preceding the divergence of birds and mammals, an ancestral gene (or more likely, gene cluster) duplicated. One gene cluster diverged to generate the ɛ-γ-δ-β-globin gene clusters found in eutherian mammals and the ɛ-β-globin gene clusters found in marsupials. The other gene cluster diverged to form the avian ρ-β-ɛ-globin gene cluster and the orphaned ω-globin gene found in marsupial species. The eutherian β-like globin gene cluster still shares a common ancestor with the chicken ρ-β-ɛ-globin gene cluster, but it is earlier than the bird-mammal divergence.
An alternative model in which these two gene clusters remain orthologous requires substantial evolutionary convergence between the marsupial ω-globin gene and the bird β-like globin genes. If there were a single gene cluster at the time of the bird-mammal split, then one would have to propose a gene duplication in the marsupial lineage to generate the contemporary ɛ-β-globin gene cluster and the ω-globin gene. The ω-globin gene should diverge to fulfill some function in marsupials, but the convergence model would hypothesize that all of the similarity between the ω-globin gene and the bird β-like globin genes arose because of some common function in the two species. Because the trees showing the sister-group relationship between these two clades are based on coding region comparisons (5), this alternative cannot be rigorously excluded at this time, but it seems unlikely. Further mapping of adjacent genes may show regions of conserved synteny that would resolve the issue definitively.
This revised view opens several new avenues for further investigation. For example, what happened to the descendants of the genes in the eutherian lineage that became the marsupial ω-globin gene and the avian β-like globin genes, and vice versa? None have been found in the working draft sequence or the extensive expressed sequence tags available by routine searches (5). Perhaps they have been lost, leaving only one β-like globin gene cluster in eutherians and birds, but two clusters in marsupials, the latter reflecting the ancestral state. Alternatively, more refined searches may uncover the remnants of the “lost” β-like globin genes, or possibly show that they have evolved to provide a different function. Certainly, all hemoglobins may not yet be known, as illustrated by the recent description of neuroglobin in human brain tissue (12). Further studies in intermediate taxa such as monotremes, in a greater variety of birds, and in reptiles will provide a more complete picture of the evolution of this gene cluster.
The interpretation that the eutherian and avian β-like globin gene clusters are paralogous, duplicating before the bird-mammal divergence, has implications for views of gene regulation and the extent to which regulatory elements are conserved in vertebrates. Genes within the β-like globin gene cluster are regulated by their promoters, proximal enhancers, and, in mammals, a distal, strong enhancer called the locus control region, or LCR. The LCR is needed for high-level expression of globin genes in erythroid cells, both at the normal chromosomal location (13) and at ectopic integration sites examined in transgenic mice (3). Sequences homologous to the eutherian β-globin LCR are found upstream of the marsupial ɛ-β-globin gene cluster (R. Hope, R. Baird, J. Kuliwaba, S. Cooper, J. Czelusniak, and M. Goodman, personal communication; Fig. 1). A major enhancer is located between the βA- and ɛ-globin genes of chickens (14). Although much smaller and in a different location than the eutherian LCR, it mimics the effects of the LCR in transgenic mice (15). Furthermore, an insulator at the 5′ end of the gene cluster in chickens blocks activation by an enhancer and prevents position effect variegation (4). The relationship between accessible, open chromatin and gene expression was first (16) and most thoroughly described for the chicken globin gene clusters of chickens. In the chicken ρ-β-ɛ-globin gene cluster, the region of DNase sensitivity also shows increased histone acetylation and is bounded precisely by DNase-hypersensitive sites, the 5′ most of which is the insulator (17, 18).
Because both gene clusters share several regulatory features, such as expression exclusively in erythroid cells and switches in expression during development, one could imagine that these control systems were in place well before the divergence of birds and mammals (Fig. 1). For instance, it was reasonable to suppose that regulatory functions such as enhancement, domain opening, and insulation would be found in both the eutherian and avian globin gene clusters. However, no clear counterpart to the avian insulator has been described in the eutherian β-like globin gene cluster (19). Further investigation of the sequences surrounding these globin gene clusters (18, 20, 21) may clarify whether the insulator is a unique adaptation for this locus in birds or if an equivalent function is found more distally in mammals. A discrete cis-regulatory element acting solely to open a chromatin domain for the β-like globin genes has yet to be mapped in either gene cluster. Such a regulator may be further away from the gene clusters, or perhaps no discrete cis element accounts for tissue-specific domain opening. Determining the extent of the open chromatin domain containing the eutherian β-globin gene cluster is of considerable interest, because some aspects of the domain described so far could be specific to the chicken ρ-β-ɛ-globin gene cluster.
Enhancers are located within the eutherian LCR and between two globin genes in chickens. If they had a common ancestor, as proposed in Fig. 1, then one might expect that their DNA sequences would have been conserved. Surprisingly, no significant sequence matches were seen in the regulatory regions in alignments between the chicken ρ-β-ɛ-globin gene cluster and the human β-like globin gene cluster (22, 23), even though the enhancers share common binding sites for transcriptional activators. Thus either the cis-regulatory elements do share a common ancestry but have diverged so much that sequence alignments cannot detect them above the background, or they have arisen independently (i.e., convergent evolution of cis-regulatory sequences) as suggested by Wheeler et al. (5). Study of the regulatory elements of the ω-globin gene in marsupials should provide important insights into this issue.
The absence of alignment between the human and chicken enhancer sequences had indicated that these two species were too far apart to see conserved regulatory elements. However, more recent studies show that some regulatory elements at the SCL/TAL1 locus are revealed by genomic sequence alignments between human and chicken (24). Thus the absence of sequence matches between regulatory elements in the β-like globin gene clusters can be explained, at least partially, by the longer phylogenetic distance between them.
Further investigation of the extent of conservation of known regulatory elements in mammalian and avian species should provide a better context for evaluating the effectiveness of sequence comparisons for finding regulatory elements. Given the differences in rate and mode of evolution at different loci, different species may be more useful for comparisons at various loci (25, 26). The intriguing new findings from the marsupials (5) remind us that this class, with a time of divergence from eutherians intermediate between the mammal-bird split (about 300 million yr ago) and the primate-rodent split (about 100 million yr ago), could provide a helpful comparison species with human for the identification of candidate regulatory elements at some loci.
Acknowledgments
This paper was supported by Grants DK27635 and HG02238 from the National Institutes of Health. The photo of the rooster is from Dr. G. Barbato.
Footnotes
See companion article on page 1101 in issue 3 of volume 98.
References
- 1.Gardner P R, Gardner A M, Martin L A, Salzman A L. Proc Natl Acad Sci USA. 1998;95:10378–10383. doi: 10.1073/pnas.95.18.10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minning D M, Gow A J, Bonaventura J, Braun R, Dewhirst M, Goldberg D E, Stamler J S. Nature (London) 1999;401:497–502. doi: 10.1038/46822. [DOI] [PubMed] [Google Scholar]
- 3.Grosveld F, van Assendelft G B, Greaves D, Kollias G. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 4.Chung J H, Whiteley M, Felsenfeld G. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 5.Wheeler D, Hope R, Cooper S J B, Dolman G, Webb G C, Bottema C D K, Gooley A A, Goodman M, Holland R A B. Proc Natl Acad Sci USA. 2001;98:1101–1106. doi: 10.1073/pnas.98.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman M, Koop B F, Czelusniak J, Weiss M L, Sightom J L. J Mol Biol. 1984;180:803–824. doi: 10.1016/0022-2836(84)90258-4. [DOI] [PubMed] [Google Scholar]
- 7.Hardison R C. Mol Biol Evol. 1984;1:390–410. doi: 10.1093/oxfordjournals.molbev.a040326. [DOI] [PubMed] [Google Scholar]
- 8.Koop B, Goodman M. Proc Natl Acad Sci USA. 1988;85:3893–3897. doi: 10.1073/pnas.85.11.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper S, Murphy R, Dolman G, Hussey D, Hope R. Mol Biol Evol. 1996;13:1012–1022. doi: 10.1093/oxfordjournals.molbev.a025651. [DOI] [PubMed] [Google Scholar]
- 10.Holland R A, Gooley A A., 2nd Eur J Biochem. 1997;248:864–871. doi: 10.1111/j.1432-1033.1997.00864.x. [DOI] [PubMed] [Google Scholar]
- 11.Holland R A, Gooley A A, Hope R M. Clin Exp Pharmacol Physiol. 1998;25:740–744. doi: 10.1111/j.1440-1681.1998.tb02288.x. [DOI] [PubMed] [Google Scholar]
- 12.Burmester T, Weich B, Reinhardt S, Hankeln T. Nature (London) 2000;407:520–523. doi: 10.1038/35035093. [DOI] [PubMed] [Google Scholar]
- 13.Bender M A, Bulger M, Close J, Groudine M. Mol Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- 14.Choi O-R B, Engel J D. Nature (London) 1986;323:731–734. doi: 10.1038/323731a0. [DOI] [PubMed] [Google Scholar]
- 15.Reitman M, Lee E, Westphal H, Felsenfeld G. Nature (London) 1990;348:749–752. doi: 10.1038/348749a0. [DOI] [PubMed] [Google Scholar]
- 16.Weintraub H, Groudine M. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- 17.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prioleau M N, Nony P, Simpson M, Felsenfeld G. EMBO J. 1999;18:4035–4048. doi: 10.1093/emboj/18.14.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bender M, Reik A, Close J, Telling A, Epner E, Fiering S, Hardison R, Groudine M. Blood. 1998;92:4394–4403. [PubMed] [Google Scholar]
- 20.Bulger M, von Doorninck J H, Saitoh N, Telling A, Farrell C, Bender M A, Felsenfeld G, Axel R, Groudine M. Proc Natl Acad Sci USA. 1999;96:5129–5134. doi: 10.1073/pnas.96.9.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulger M, Bender M A, von Doorninck J H, Wertman B, Farrell C, Felsenfeld G, Groudine M, Hardison R. Proc Natl Acad Sci USA. 2000;97:14560–14565. doi: 10.1073/pnas.97.26.14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reitman M, Grasso J A, Blumentahl R, Lewit P. Genomics. 1993;18:616–626. doi: 10.1016/s0888-7543(05)80364-7. [DOI] [PubMed] [Google Scholar]
- 23.Hardison R. J Exp Biol. 1998;201:1099–1117. doi: 10.1242/jeb.201.8.1099. [DOI] [PubMed] [Google Scholar]
- 24.Gottgens B, Barton L M, Gilbert J G, Bench A J, Sanchez M J, Bahn S, Mistry S, Grafham D, McMurray A, Vaudin M, et al. Nat Biotechnol. 2000;18:181–186. doi: 10.1038/72635. [DOI] [PubMed] [Google Scholar]
- 25.Miller W. Nat Biotechnol. 2000;18:148–149. doi: 10.1038/72588. [DOI] [PubMed] [Google Scholar]
- 26.Hardison R C. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]