Abstract
The cranial neural crest (CNC) is a transient cell population that originates at the crest of the neural fold and gives rise to multiple cell types during craniofacial development. Traditionally, researchers have used tissue explants, such as the neural tube, to obtain primary neural crest cells for their studies. However, this approach has inevitably resulted in simultaneous isolation of neural and non-neural crest cells as both of these cells migrate away from tissue explants. Using the Wnt1-Cre/R26R mouse model, we have obtained a pure population of neural crest cells and established a primary CNC cell culture system in which the cell culture medium best supports the proliferation of E10.5 first branchial arch CNC cells and maintains these cells in their undifferentiated state. Differentiation of CNC cells can be initiated by switching to a differentiation medium. In this model, cultured CNC cells can give rise to neurons, glial cells, osteoblasts, and other cell types, faithfully mimicking the differentiation process of the post-migratory CNC cells in vivo. Taken together, our study shows that the Wnt1-Cre/R26R mouse first branchial arch provides an excellent model for obtaining post-migratory neural crest cells free of any mesodermal contaminants. The cultured neural crest cells are under sustained proliferative, undifferentiated, or lineage-enhanced conditions, hence, serving as a tool for the investigation of the regulatory mechanism of CNC cell fate determination in normal and abnormal craniofacial development.
Keywords: cranial neural crest (CNC) cell culture, proliferation, differentiation, apoptosis
Introduction
Neural crest cells are critical for the development and evolution of the vertebrates. They originate at the crest of the closing neural folds in vertebrate embryos and migrate into the periphery along stereotypical paths to a number of sites where these cells restrict their potential and differentiate into a variety of derivatives (LaBonne and Bronner-Fraser, 1999). The development of facial structures depends upon the successful contribution of cranial neural crest (CNC) cells. Although both intrinsic and extrinsic regulatory signals are critical for the proper migration and expansion of the CNC cell population, recent studies have suggested that the fate specification of CNC cells may be instructed by surrounding tissues during or after migration, as this group of progenitor cells contributes to the formation of various craniofacial structures (Trainor et al., 2002; Couly et al., 2002).
Derivatives of neural crest cells include sensory neurons, autonomic neurons, glia, melanocytes, adrenal medulla cells, and smooth muscle cells. In the craniofacial region, additional neural crest derivatives include cartilage, bone, and odontoblasts. Through in vitro clonal analysis, it has been suggested that prior to migration the neural crest cells are a multipotent population and can give rise to multiple types of derivatives from a single progenitor cell (Stemple and Anderson, 1992). This finding has also been confirmed by in vivo study, in which individual premigratory or migratory neural crest cells are labeled with dye and the fates of their descendants are then followed (Bronner-Fraser and Fraser 1988, 1989; Fraser and Bonner-Fraser, 1991). However, some individual neural crest cells give rise to only one type of derivative in both in vivo and in vitro studies. Thus, it has been proposed that even at the onset of migration, the neural crest is composed of a heterogeneous population of cells with different proliferation and differentiation potentials (Le Douarin et al., 2004). Interestingly, multipotent neural crest cells have also been identified in sites where post-migratory neural crest cells reside (Lo and Anderson 1995; Lo et al., 1997; Sieber-Blum et al., 1993). This implies that some of the premigratory neural crest cells maintain their multipotent differentiation ability and undergo self-renewal or simply keep quiescence after migration. Others may restrict their differentiation potential progressively and commit to one specific lineage (Le Douarin and Dupin, 2003).
Cell labeling experiments show that branchial arches are composed of neural crest cells that have emigrated from posterior midbrain and anterior hindbrain segments (Graham et al., 1993; Kulesa and Fraser, 1998). Although previous studies on neural crest and other multipotential populations have shown that restricted and pluripotent cells can co-exist during embryonic development (Smith, 1990), we do not have a clear understanding of the differentiation status of post-migratory CNC cells within the first branchial arch. In order to investigate the differentiation of CNC cell, we need an in vitro neural crest cell culture model because of its accessibility and the absence of interference from other cells and tissues. However, because of difficulties in obtaining a pure population of these cells for analysis, it has not been easy to characterize the properties of neural crest (NC) cells in vitro.
In this study, we have used the Wnt1-Cre/R26R first branchial arch as a model to investigate the proliferation and differentiation potential of post-migratory cranial neural crest cells. In vitro cell culture was used to study the effects of several different types of medium on CNC cell proliferation and differentiation. Based on our analysis, an optimal culture system was established for sustaining proliferative activity and maintaining undifferentiated status of CNC cells. Overall, the establishment of this primary CNC cell culture model will greatly facilitate the investigation of the regulatory mechanism for post-migratory CNC cell fate determination during craniofacial morphogenesis.
Results and Discussion
Establishing a Pure Population of CNC Cells for the Primary Cell Culture Model
In order to characterize the post-migratory CNC cells and their differentiation status within the first branchial arch, we took advantage of the Wnt1-Cre/R26R system to obtain a pure population of CNC cells for the primary cell culture. As shown in our previous study, the lacZ staining pattern indicated the distribution of CNC cells (Chai et al., 2000). At E9.5 (21–29 somites), X-gal-positive CNC-derived cells densely populate both the first and second branchial arches. Very few non-CNC cells, which are believed to be mesoderm derived, appear within the branchial arch intermingled with the CNC-derived cells (Chai et al., 2000). At E10.5 (35–39 somites), the CNC-derived cells account for 90–95% of the cells in the first branchial arch (Fig. 1A, and FACS analysis). By E11.5, the CNC-derived cells still account for a majority of cells in the first branchial arch (Fig. 1B).
Fig. 1.
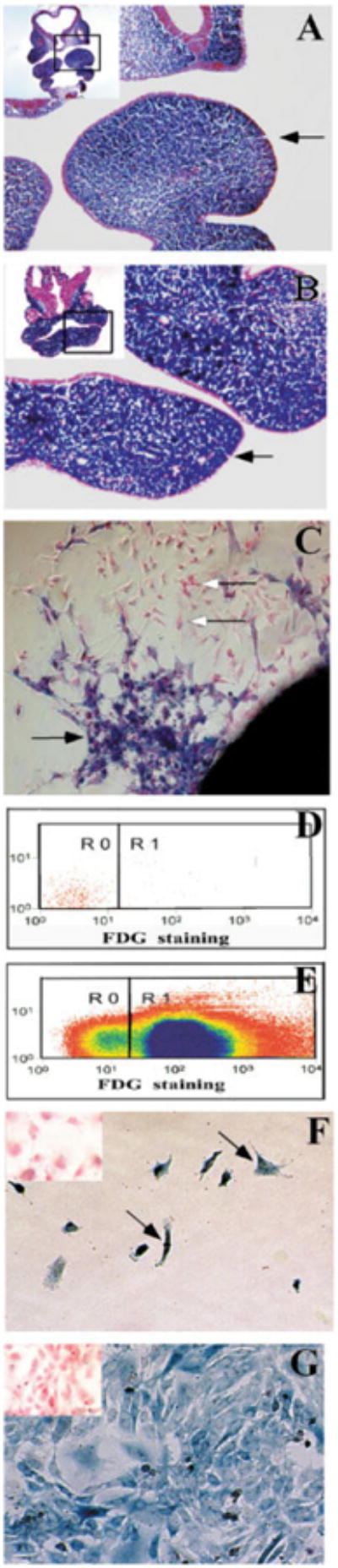
Establishing a pure population of post-migratory CNC cells in vitro. CNC-specific β-galactosidase activity is present in transverse sections of the first branchial arch from Wnt-cre/R26R mouse embryos at E10.5 (A) and E11.5 (B). The arrows indicate the first branchial arch. The position of each section in the embryo is framed out in the inlet. C: Cultured E9.0 neural tube explant (for one day) shows neural crest (blue, black arrow) and non-neural crest (pink, white arrow) cells migrate away from the explant. D: Fluorescence activated cell sorting (FACS) was used to separate CNC from non-CNC cells. Cells without FDG staining are used to establish a base line (R0) prior to cell sorting. E: Following FDG staining, FDG positive CNC cells (R1) are collected for cell culture study, while FDG negative cells are treated as non-CNC cells (R0). F: One day following cell sorting, CNC cells (arrow) have attached to the plate in culture. G: Five days following cell sorting, CNC cells have expanded and become confluent in the dish. All cells show CNC-specific β-galactosidase activity (blue). There is no non-CNC-derived cell in the culture dish. The cell cultures are counterstained with fast red. Insets (E,F): The non-CNC derived cells are pink in color following β-gal and fast red staining.
Traditionally, neural crest cells are obtained by harvesting cells that have migrated away from the neural tube explant. We show here that both neural crest (blue) and non-neural crest (pink) cells migrate away from the E9 Wnt1-Cre/R26R neural tube plant following one day of culture (Fig. 1C). However, these migratory cells have classically been used as a source of neural crest cells. Clearly, caution needs to be taken when dealing with this mixed population of neural crest and non-neural-crest-derived cells without cell sorting. In other studies, neural crest cells are obtained by using NGF receptor p75 (NGFR p75) surface marker-activated cell sorting, which is suitable for premigratory neural crest cell studies, but may not be suitable for post-migratory neural crest cell studies because the expression specificity of p75 within post-migratory neural crest cells has not been well studied. We do not know how long the expression of p75 lasts in neural crest cells. As a matter of fact, some in vitro studies have indicated that neural crest cells lose p75 expression after differentiating into smooth muscle cells (Stemple and Anderson, 1992; Shah et al., 1996; Morrison et al., 1999).
In this study, following two days of primary cell culture, CNC cells were separated from non-CNC cells by cell sorting based on β-gal expression in the CNC-derived cells. FACS analysis revealed that we were able to clearly segregate the CNC from non-CNC cells (Fig. 1D,E). The CNC cells were lacZ positive (blue staining), attached to the cell culture plate, and showed robust proliferative capability one day following sorting (Fig. 1F). There were no lacZ-positive cells in the sorted non-CNC cells (Fig. 1F, inset), while there were no lacZ-negative cells in the sorted CNC cells. This suggested that we were able to obtain a pure population of CNC cells for the primary cell culture. At five days after sorting, these CNC cells had expanded by active proliferation while remaining undifferentiated (Fig. 1G; also see Fig. 3). The non-CNC cells cultured in a separate dish did not show any lacZ activity, indicating that there was no contamination of CNC cells (Fig. 1G, inset). Clearly, the Wnt1-Cre/R26R model can provide an extremely pure population of CNC cells for cell culture.
Fig. 3.
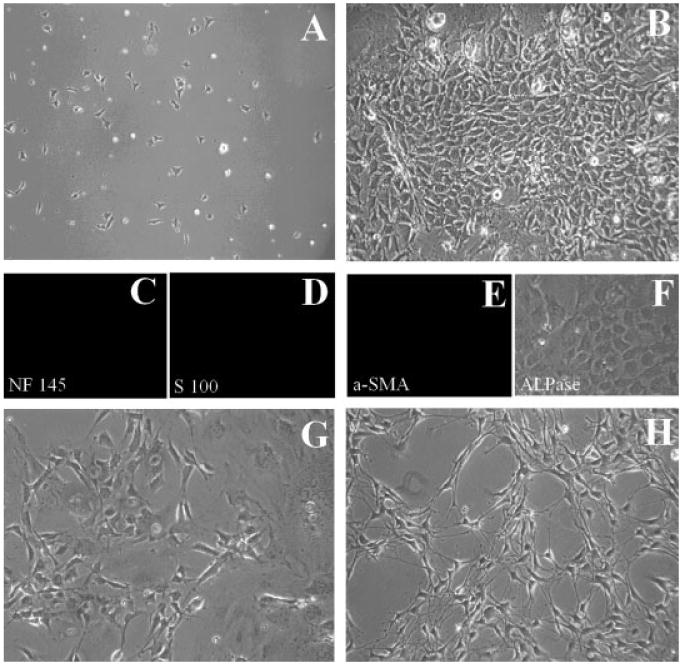
MCDM medium maintains the first branchial arch post-migratory CNC cells in an undifferentiated state. A: Sorted CNC cells 4 hr after plating on FN-coated dish in medium II (MCDM medium). These cells show attachment to the plate. B: The undifferentiated cells, which are featured by their small body size, intense nuclei, and deep color, have reached confluence on day 4. Following immunofluorescent staining with antibodies against NF145 (C), S100 (D), or α-SMA (E), it is clear that these are undifferentiated CNC cells following 4 days of culture in medium II. Alkaline phosphatase staining with BCIP/NBT reagent is also negative (F). When MCDM is switched to DMCDM medium on day 5, morphological changes begin to take place, suggesting differentiation of CNC cells following 2 (G) or 4 (H) additional days of culture.
The E10.5 First Branchial Arch Provides a Suitable Population of Cells for Investigating the Differentiation Potential of Post-Migratory CNC
In order to investigate the differentiation potential of post-migratory CNC cells, we designed experiments to determine a suitable embryonic stage for obtaining undifferentiated CNC cells from the first branchial arch. Neuron, glial cell, smooth muscle cell, and osteocyte are examples of terminally differentiated cell fates of the CNC-derived cells residing within the first branchial arch. To evaluate the differentiation status of the post-migratory neural crest cells within the first branchial arch, we examined the expression of cellular markers characteristic of neurons (NF145), glial cells (GFAP, S100), and smooth muscle cells (α-SMA) by immunohistochemistry. We were unable to detect the expression of any of the three differentiation markers in the first branchial arch at E9.5 (data not shown) or at E10.5 (Fig. 2A–C). At E11.5, NF145 expression was present in the proximal part of the first branchial arch, which obviously marked branches of the trigeminal nerve (Fig. 2D). GFAP and S100 expressions were yet to be detected at this stage (Fig. 2E and data not shown). The α-SMA expression was seen widely dispersed in the first branchial arch (Fig. 2F). In addition, Runx2, one of the earliest markers of osteoblast differentiation, was not seen until E13.5 in the first arch derivatives (our unpublished data), inferring an undifferentiated CNC population within the first branchial arch at E10.5 or earlier. The absence of differentiation marker staining at E10.5, and the presence of differentiation markers following cell culture (presented below), fulfill the criteria used to define a CNC progenitor cell population suitable for cell fate determination analysis. Taken together, we concluded that E9.5 and E10.5 embryo first branchial arch provided a suitable source of undifferentiated CNC cells for our study. We decided to use E10.5 first branchial arch as our starting material because it provided more undifferentiated CNC cells for us to work with.
Fig. 2.
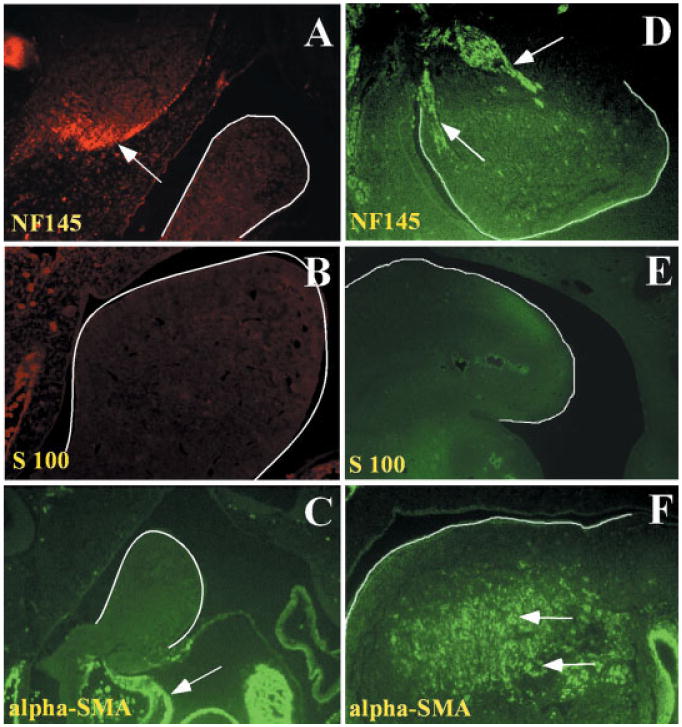
Differentiation marker analysis of post-migratory CNC cells in the first branchial arch at E10.5 and E11.5. Cell marker analysis shows the differentiation status of CNC cells within the first branchial arch of E10.5 (A–C) or E11.5 (D–F) embryos. The first branchial arch is highlighted by the solid white line. A: NF145 immunofluorescent staining (red). Arrow points to positive staining in the brain. B: There is no S100 immunofluorescent staining (red) in the first branchial arch. C: There is no α-SMA immunofluorescent staining (green) in the first arch. Positive staining (arrow) is detected in the developing heart. D: The trigeminal nerve is positive for NF145 immunofluorescent staining (green). E: There is no S100 immunofluorescent staining (green) in the first branchial arch. F: Positive α-SMA immunofluorescent staining (green) is detected throughout the first branchial arch. The white arrows indicate the positive staining of α-SMA. The red signals in the peripheral region of B are non-specific staining of red blood cells.
Maintaining the Survival, Proliferation, and Undifferentiation of CNC Cells in Different Cell Culture Mediums
In general, an ideal cell culture system for investigating the pluripotent properties of CNC cells should fulfill the following requirements: (1) it can support the survival and proliferation of CNC progenitor cells; (2) it can maintain the CNC progenitor cells in undifferentiated state for a relatively long time before they undergo differentiation; (3) the differentiation of CNC cells can be initiated in a controlled way; (4) it can support the clonal culture; (5) it is desirable to have a chemically-defined medium; and (6) it should be easy to work with.
In order to develop the ideal cell culture condition, we tested five different types of cell culture media for their ability to support CNC cell proliferation and undifferentiation. Following cell sorting, different media showed different abilities to support dissociated CNC cell cultures. The survival of CNC cells in the culture depended on the plating density. When CNC cells were plated at high density (104 cells/cm2), medium II and medium V supported cell proliferation, while the other media did not. Furthermore, CNC cells cultured in medium V showed early signs of differentiation, such as positive staining of NF145 and S100 following five days in culture, suggesting that this medium is not ideal for maintaining cultured cells in an undifferentiated status.
When CNC cells were cultured in MCDM (medium II), they attached to the plates 4 hr after plating, with an attaching efficiency of 80–90% (Fig. 3A). On day 4, the undifferentiated progenitor cells, characterized by small, deep colored cell bodies and intense nuclei, grew up in clusters in the culture (Fig. 3B). Their undifferentiated status was confirmed by immunofluorescent staining on the 14th day following plating. There was no indication of CNC differentiation [NF145(-), S100(-), α-SMA(-), and ALPase(-)] (Fig. 3C–F).
In order to fulfill the criteria of being a useful cell culture system for cell fate determination analysis, the cultured cells should also be easily manipulated to initiate terminal differentiation. In this study, the differentiation of cultured CNC cells could be facilitated by switching the medium to differentiation DMCDM with a lower content of CEE (from 10 to 1%) after 5 days in culture. The dramatic morphological changes from undifferentiated CNC cells to differentiated cells were seen 2 or 4 days after the switch (Fig. 3G,H, also see Fig. 4). Taken together, we have established a primary CNC cell culture model in which these progenitor cells can actively proliferate without undergoing differentiation. These CNC cells remain sensitive to changes in their environment, which can be manipulated to induce differentiation. This model fulfills the requirement for CNC fate determination analysis during normal and abnormal development.
Fig. 4.
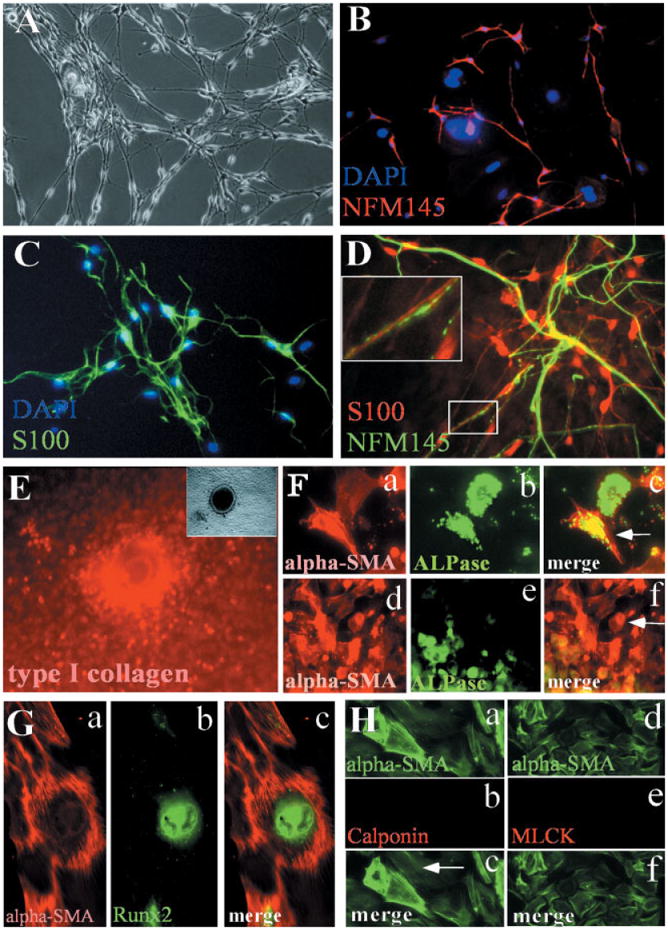
Differentiation potentials of CNC cells in culture. Sorted CNC cells are maintained in MCDM for 5 days and are switched to DMCDM medium for additional 4 days. The differentiation status of cultured CNC cells is evaluated by using different markers. A: Some of the neural crest–derived cells display typical neural cell morphology following the switch of cell culture medium from MCDM to DMCDM. B: Double staining with antibodies against NF145 (red) and DAPI (blue). C: Double staining with antibodies against S100 (green) and DAPI (blue). D: Double staining with antibodies against NF145 (green) and S100 (red). Part of the axon is enlarged to show the parallel alignment of neuron and glial cell (inset), faithfully mimicking the relationship in vivo. E: Calcified nodule is stained with antibody against type I collagen (red). Inset at top left corner is the same calcified nodule under light microscope and is positive for osteopontin expression, therefore, validating the differentiation of CNC cells into osteoblasts. F: Double staining with ALPase (green) and α-SMA (red) antibodies indicates that some CNC cells express both alpha-SMA and ALPase (F, c: arrow). While some CNC cells only express alpha-SMA (F, f: arrow), most of ALPase positive cells are also alpha-SMA positive (F, f: green and red staining merged as yellow). G: Double staining with α-SMA (G, a, red) and RUNX2 (G, b, green) antibodies indicates that some CNC cells express both alpha-SMA and RUNX2 (G, c, merged a and b). H: Double staining with α-SMA + calponin antibodies (a–c) or †-SMA + MLCK (d–f) show that these α-SMA positive cells do not express calponin or MLCK.
Differentiation Potentials of Post-Migratory CNC Cells In Vitro
Using this in vitro culture model, we investigated the differentiation of CNC cells into neurons, glial cells, smooth muscle cells, and osteoblasts. The differentiation of CNC cells into neurons and glia cells was obtained by changing the medium into DMCDM on day 5 after plating and continuing the culture for an additional 4 days. Under a light microscope, it is difficult to distinguish between neurons and Schwann cells in the culture by their morphological appearance (Fig. 4A). NF145 or S100 were used as the differentiation markers for neurons or glial cells, respectively (Iwanaga et al., 1989). Some CNC cells differentiated into neurons (NF145+), while others differentiated into Schwann cells (S100+) (Fig. 4B,C). Most of these NF145- or S100-positive cells exhibited small and slim cell bodies with long projections. Our differentiation marker analysis provided definitive identity for cultured CNC cells. Overall, Schwann cells outnumbered neurons dramatically (3:1) in the culture and they usually co-localized in the same areas, as they do in vivo (Fig. 4D).
CNC cells are known to give rise to craniofacial bone structures in vivo. To facilitate differentiation towards osteoblasts, dissociated CNC cells were cultured in a DMCDM medium. Supplement reagents (10 mM β-glycerophosphate, 50 μg/ml ascorbic acid and 10-7M dexamethasone) were added 3 days following cell sorting and plating. The CNC culture was maintained for an additional 3 to 7 days and then examined for the presence of calcified nodules (Fig. 4E), alkaline phosphatase (ALPase) activity, and/or Runx2 expression (Fig. 4F,G), which validated the differentiation of CNC cells into osteoblasts. A fairly high percentage of osteoblast progenitor cells in the bone marrow express α-SMA (Bianco et al., 2001; Kinner and Spector, 2002). Given the fact that a large number of post-migratory CNC cells expresses α-SMA (Fig. 2F) but only a small percentage of them will become smooth muscle cells in adult, we decided to test whether some of the α-SMA positive cells may differentiate into osteoprogenitors. Compared with α-SMA expression, which was seen in some cells 2 to 3 days after plating, the expression of ALPase did not appear until 7 days after plating. Double staining with α-SMA and ALPase antibodies revealed that some CNC cells in the culture expressed both markers (Fig. 4F, a–c). Although there were CNC cells expressing only ALPase or only a-SMA (Fig. 4F, d–f), we found that most ALPase-positive cells also expressed α-SMA (Fig. 4F, f). To further confirm differentiation of CNC cells into an osteoblast fate, we double stained cultured crest cells with α-SMA and RUNX2 antibodies and verified that some α-SMA-positive cells were indeed osteoblast progenitors (Fig. 4G, a–c)
Previous studies used α-SMA as a marker for differentiated smooth muscle cells. The co-expression of AL-Pase/RUNX2 and α-SMA in many CNC-derived cells raises the question of whether these α-SMA positive cells will differentiate into true smooth muscle cells. To investigate this issue further, we cultured the CNC cells in MCDM for 3 days, and then switched to DMCDM for an additional 7 days. Calponin and myosin light chain kinase (MLCK) are two other markers for differentiated smooth muscle cells (Birukov et al., 1991). However, double staining with α-SMA plus either calponin or MLCK showed that the majority (85%) of α-SMA-positive cells did not express these two markers (Fig. 4H, a–f), while only 10–15% of α-SMA-positive cells also showed positive staining with MLCK (data not shown). Therefore, our data suggest that it may not be appropriate to define all α-SMA expressing cells as smooth muscle cells. Instead, we define them as “myofibroblast” cells, which clearly have the potential to differentiate into osteoblasts as well as smooth muscle cells.
Multipotent post-migratory CNC cells must have some self-renewal capacity and be able to generate various differentiated cell types in an organized fashion during craniofacial morphogenesis. Although the migration of CNC into the first branchial arch occurs between E8 and E9, the post-migratory CNC cells go through self-renewal and remain undifferentiated until E11.5. Here, we show that the post-migratory CNC cells are able to give rise to neurons, Schwann cells, myofibroblasts, and osteoblast cells as validated by differentiation markers. The study clearly demonstrates that post-migratory CNC cells are a multipotent cell population and have the potential to differentiate into an array of cell types in vitro. It is difficult to conclude, however, whether the post-migratory CNC population consists of only multipotent progenitors or a mixed population of uncommitted and committed cells without performing the clone culture.
Smooth muscle cells represent one of the fates of CNC differentiation. They populate the walls of blood vessel in the craniofacial region and contribute to the development of cardiac outflow tract (Chai et al., 2000; Jiang et al., 2000; Mann et al., 2004; Berg-werff et al., 1998; Topouzis and Majesky, 1996). Trunk neural crest cells, however, do not become smooth muscle cells during embryogenesis (our unpublished data). α-SMA as a cytoskeleton element is expressed by all the differentiated smooth muscle cells both in vivo and in vitro and, therefore, has long been employed as a marker for validating differentiated smooth muscle cells in many studies. However, based on the information presented here, α-SMA must be used in conjunction with other smooth muscle differentiation markers in order to validate the terminal differentiation into smooth muscle cell.
In summary, we have developed a new primary neural crest cell culture model, which can be used to investigate how extrinsic factors can alter the fate of post-migratory CNC cells. Taking advantage of both wild type and mutant animal models, we can now use this CNC cell culture model and begin to investigate the functional significance of a signaling network in regulating the cell fate determination.
Experimental Procedures
CNC Cell Cultures
Wnt1-Cre/R26R heterozygous embryos were removed at embryonic day 9.5 (E9.5) or 10.5 (E10.5) and placed in a Petri dish containing Hank's buffer solution (Invitrogen, La Jolla, CA). The first branchial arches of embryos were dissected and digested for 4 min at 37°C, with a collagenase mixture, which contained 0.1% collagenase (Sigma, St. Louis, MO) and 0.025% trypsin (Sigma) in Ca2+, Mg2+ free DPBS (Invitrogen). The tissue blocks were pipetted up and down to break them into single cell suspension. The dissociated tissue was then washed with a fresh culture medium to stop digestion. Cells were counted and plated on a fibronectin coated 6-well plate (Becton Dickinson Labware). Cells were maintained in the indicated mediums at 37°C in 5% CO2/ 95% air. After 2 days in culture, CNC cells were segregated from non-CNC cells by using Fluorescence Activated Cell Sorting (see below). Then, CNC cells were plated and cultured for additional days as indicated in the results.
FDG (Fluorescence di-β-D-Galactopyranoside) Staining for β-Galactosidase
Exponentially growing cells were treated with trypsin (GIBCO no. 610-5400, Grant Island, NY; diluted to 1× solution) in phosphated-buffered saline until they could be removed from the culture dish with mild agitation and were then centrifuged to a pellet. Next, the cell mass was re-suspended into 107 per ml of fresh culture medium (MCDM, medium II), incubated in 37°C for 5 min, and mixed gently with equal volume of pre-warmed FDG working solution [2 mM FDG (fluorescence di-β-D-galactopyranoside)] in H2O. Then cells in mixed medium were incubated at 37°C for 1 min. We then added regular cell culture medium (10 times the volume of the mixed medium) to stop the reaction on ice.
Fluorescence Activated Cell Sorting (FACS)
FACS was set up at the flow cytometry core facility at USC Keck School of Medicine. The auto-fluorescence of cultured cells was compensated by control cells (the neural crest cultures were stained by the staining medium with PBS instead of FDG). Both the FDG-positive and -negative cells were collected. The FDG-positive cells were used for primary CNC cell culture.
X-Gal Staining
The cultured cells were rinsed by a rinse solution (150 mM NaCl, 15 mM Na phosphate, in PBS pH 7.3), fixed for 10 min in a fixative solution (0.2% gluteraldehyde in PBS) and washed by PBS. Then, the cells were overlaid with a histochemical reaction mixture (1 mg/ml 4-CL-5-Br-3indolyl-β-galactosidase [X-Gal], 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 2 mM MgCl2 in PBS), and were incubated for 14–18 hr at 37°C.
Culture Mediums
Five mediums were tested in this study. Medium I is a chemically-defined medium consisting of L-15 CO2, supplemented with 10% chicken embryo extract (CEE) prepared as previously described (Stemple and Anderson, 1992), and the additives are as described by Sieber-Blum and Chokshi (1985). Specifically, the medium contained L15 CO2 with 10% CEE; 100 μg/ml insulin, 16 μg/ml putrescin, 20 nM progesterone, 30 nM selenious acid, 1 mg/ml bovine serum albumin, 39 pg/ml dexamethasone, 35 ng/ml retinoic acid, 5 μg/ml α-d-1-tocopherol, 63 μg/ml β-hydroxybutyrate, 25 ng/ml cobalt chloride, 1 μg/ml biotin, 10 ng/ml oleic acid, 3.6 mg/ml glycerol, 100 ng/ml α-MSH, 10 ng/ml prostaglandin E1, 67.5 ng/ml triiodothyronine, and 4 ng/ml bFGF (all from Sigma); 100 ng/ml EGF and 20 ng/ml 2.5S NGF (Upstate Biotechnology, Lake Placid, NY); 25 U/ml penicillin and 25 μg/ml streptomycin sulphate (Penicillin/Streptomycin, Invitrogen).
Medium II (MCDM) is modified from a chemically-defined medium described by Morrison et al. (1999). The culture medium contained DMEM-low glucose (Invitrogen) with 10% CEE, 20 ng/ml bFGF (Sigma), N2 supplement (Invitrogen), B-27 Supplement minus Vitamin A (Invitrogen), 50 μM 2-mercaptomethanol (Sigma), 35 ng/ml retinoic acid (Sigma), and 25 U/ml penicillin and 25 μg/ml streptomycin sulphate (Invitrogen). Differentiation MCDM (DMCDM) was used in the cell culture to initiate CNC cell differentiation. It consisted of the same components as the MCDM medium with the exception of a lower concentration of CEE (1%) and bFGF (10 ng/ml).
Medium III consists of DMEM-F12 (Invitrogen), 10% CEE, 10 ng/ml bFGF (Sigma), and 25 U/ml penicillin and 25 μg/ml streptomycin sulphate (Invitrogen).
Medium IV is composed of DMEM-F12 (Invitrogen), 5% CEE, 5% FBS (Invitrogen), 10 ng/ml bFGF (Sigma), and 25 U/ml penicillin and 25 μg/ml streptomycin sulphate (Invitrogen).
Medium V is based on the NEP medium as described by Kalyani et al. (1997). The medium contained DMEM-F12 (Invitrogen) with 100 μg/ml transferrin (Calbiochem, San Diego, CA), 5 μg/ml insulin (Sigma), 16 μg/ml putrescin (Sigma), 20 nM progesterone (Sigma), 30 nM selenious acid (Sigma), 1 mg/ml bovine serum albumin (Invitrogen), plus B27 additives (Invitrogen), 25 ng/ml bFGF (Sigma), 10% CEE, and 25 U/ml penicillin and 25 μg/ml streptomycin sulphate (Invitrogen).
As a control, a regular FBS medium was used, which was composed of high glucose DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen), and 25 U/ml penicillin and 25 μg/ml streptomycin sulphate (Invitrogen).
Immunocytochemistry
For routine analysis of cell differentiation, cultures were fixed in 4% paraformaldehyde for 10 min. Primary antibodies against NGFR p75 1:400 (Chemicon, Temecula, CA), NFM145 1:400 (Chemicon), GFAP 1:400 (Sigma), S100 1:400 (Sigma), α-SMA 1:400 (Sigma), Cy3 conjugated α-SMA (Sigma), type I collagen 1:500 (Sigma), RUNX2 1:400 (R&D, Minneapolis, MN), and β-galactosidase 1:400 (Abcam) were used. Secondary antibodies conjugated with either FITC or rhodamine red against rabbit IgG or mouse IgG were all from Molecular Probes, Eugene, OR. Detailed immunofluorescent staining protocols are available upon request. VECTASHIELD Hard set (Vector, Burlingame, CA) was used for DAPI staining.
Acknowledgments
Grant sponsor: National Institute of Dental and Craniofacial Research; Grant sponsor: NIH; Grant numbers: DE012711; DE014078; Grant sponsor: March of Dimes; Grant number: 6-FY02-137.
This study was supported by grants from the National Institute of Dental and Craniofacial Research, NIH (DE012711 and DE014078), and the March of Dimes (6-FY02-137) to Yang Chai.
References
- Bergwerff M, Verberne ME, DeRuiter MC, Poelmann RE, Gittenberger-de Groot AC. Neural crest cell contribution to the developing circulatory system: implications for vascular morphology? Circ Res. 1998;82:221–231. doi: 10.1161/01.res.82.2.221. [DOI] [PubMed] [Google Scholar]
- Bianco P, Riminucci M, Gronthos S, Robey PG. Bone marrow stromal stem cells: nature, biology, and potential applications. Stem Cells. 2001;19:180–192. doi: 10.1634/stemcells.19-3-180. [DOI] [PubMed] [Google Scholar]
- Birukov KG, Stepanova OV, Nanaev AK, Shirinsky VP. Expression of calponin in rabbit and human aortic smooth muscle cells. Cell Tissue Res. 1991;266:579–584. doi: 10.1007/BF00318599. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M, Fraser SE. Cell lineage analysis reveals multipotency of some avian neural crest cells. Nature. 1988;335:161–164. doi: 10.1038/335161a0. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M, Fraser SE. Developmental potential of avian trunk neural crest cells in situ. Neuron. 1989;3:755–766. doi: 10.1016/0896-6273(89)90244-4. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Couly G, Creuzet S, Bennaceur S, Vincent C, Le Douarin NM. Interactions between Hox-negative cephalic neural crest cells and the foregut endoderm in patterning the facial skeleton in the vertebrate head. Development. 2002;129:1061–73. doi: 10.1242/dev.129.4.1061. [DOI] [PubMed] [Google Scholar]
- Fraser S, Bronner-Fraser M. Migrating neural crest cells in the trunk of the avian embryo are multipotent. Development. 1991;112:913–920. doi: 10.1242/dev.112.4.913. [DOI] [PubMed] [Google Scholar]
- Graham A, Heyman I, Lumsden A. Even-numbered rhombomeres control the apoptotic elimination of neural crest cells from odd-numbered rhombomeres in the chick hindbrain. Development. 1993;119:233–245. doi: 10.1242/dev.119.1.233. [DOI] [PubMed] [Google Scholar]
- Iwanaga T, Takahashi Y, Fujita T. Immunohistochemistry of neuron-specific and glia-specific proteins. Arch Histol Cytol. 1989;52(Suppl):13–24. doi: 10.1679/aohc.52.suppl_13. [DOI] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Kalyani A, Hobson K, Rao MS. Neuroepithelial stem cells from the embryonic spinal cord: isolation, characterization, and clonal analysis. Dev Biol. 1997;186:202–223. doi: 10.1006/dbio.1997.8592. [DOI] [PubMed] [Google Scholar]
- Kinner B, Spector M. Expression of smooth muscle actin in osteoblasts in human bone. J Orthop Res. 2002;20:622–632. doi: 10.1016/S0736-0266(01)00145-0. [DOI] [PubMed] [Google Scholar]
- Kulesa PM, Fraser SE. Neural crest cell dynamics revealed by time-lapse video microscopy of whole embryo chick explant cultures. Dev Biol. 1998;204:327–344. doi: 10.1006/dbio.1998.9082. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Molecular mechanisms of neural crest formation. Annu Rev Cell Dev Biol. 1999;15:81–112. doi: 10.1146/annurev.cellbio.15.1.81. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Dupin E. Multipotentiality of the neural crest. Curr Opin Genet Dev. 2003;13:529–536. doi: 10.1016/j.gde.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Creuzet S, Couly G, Dupin E. Neural crest cell plasticity and its limits. Development. 2004;131:4637–4650. doi: 10.1242/dev.01350. [DOI] [PubMed] [Google Scholar]
- Lo L, Anderson DJ. Postmigratory neural crest cells expressing c-RET display restricted developmental and proliferative capacities. Neuron. 1995;15:527–539. doi: 10.1016/0896-6273(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Lo L, Sommer L, Anderson DJ. MASH1 maintains competence for BMP2-induced neuronal differentiation in post-migratory neural crest cells. Curr Biol. 1997;7:440–450. doi: 10.1016/s0960-9822(06)00191-6. [DOI] [PubMed] [Google Scholar]
- Mann KM, Ray JL, Moon ES, Sass KM, Benson MR. Calcineurin initiates smooth muscle differentiation in neural crest stem cells. J Cell Biol. 2004;165:483–491. doi: 10.1083/jcb.200402105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, White PM, Zock C, Anderson DJ. Prospective identification, isolation by flow cytometry, and in vivo self-renewal of multipotent mammalian neural crest stem cells. Cell. 1999;96:737–749. doi: 10.1016/s0092-8674(00)80583-8. [DOI] [PubMed] [Google Scholar]
- Shah NM, Groves AK, Anderson DJ. Alternative neural crest cell fates are instructively promoted by TGFbeta super-family members. Cell. 1996;85:331–343. doi: 10.1016/s0092-8674(00)81112-5. [DOI] [PubMed] [Google Scholar]
- Sieber-Blum M, Chokshi HR. In vitro proliferation and terminal differentiation of quail neural crest cells in a defined culture medium. Exp Cell Res. 1985;158:267–272. doi: 10.1016/0014-4827(85)90450-1. [DOI] [PubMed] [Google Scholar]
- Sieber-Blum M, Ito K, Richardson MK, Langtimm CJ, Duff RS. Distribution of pluripotent neural crest cells in the embryo and the role of brain-derived neurotrophic factor in the commitment to the primary sensory neuron lineage. J Neurobiol. 1993;24:173–184. doi: 10.1002/neu.480240205. [DOI] [PubMed] [Google Scholar]
- Smith J. The avian neural crest as a model system for the study of cell lineages. Int J Dev Biol. 1990;34:157–162. [PubMed] [Google Scholar]
- Stemple DL, Anderson DJ. Isolation of a stem cell for neurons and glia from the mammalian neural crest. Cell. 1992;71:973–985. doi: 10.1016/0092-8674(92)90393-q. [DOI] [PubMed] [Google Scholar]
- Topouzis S, Majesky MW. Smooth muscle lineage diversity in the chick embryo. Two types of aortic smooth muscle cell differ in growth and receptor-mediated transcriptional responses to transforming growth factor-beta. Dev Biol. 1996;178:430–445. doi: 10.1006/dbio.1996.0229. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Sobieszczuk D, Wilkinson D, Krumlauf R. Signalling between the hindbrain and paraxial tissues dictates neural crest migration pathways. Development. 2002;129:433–442. doi: 10.1242/dev.129.2.433. [DOI] [PubMed] [Google Scholar]


