Abstract
The epithelial to mesenchymal transition (EMT) is a developmental process enabling epithelial cells to gain a migratory mesenchymal phenotype. In cancer, this process contributes to metastases; however the regulatory signals and mechanistic details are not fully elucidated. Here, we sought to identify the subset of genes regulated in lung cancer by ZEB1, an E-box transcriptional repressor known to induce EMT. Using an Affymetrix-based expression database of 38 non-small cell lung cancer (NSCLC) cell lines, we identified 324 genes that correlated negatively with ZEB1 and 142 that were positively correlated. A mesenchymal gene pattern (low E-cadherin, high Vimentin or N-cadherin) was significantly associated with ZEB1 and ZEB2, but not with Snail, Slug, Twist1 or Twist2. Among 8 genes selected for validation, 7 were confirmed to correlate with ZEB1 by quantitative real-time RT-PCR in a series of 22 NSCLC cell lines, either negatively (CDS1, EpCAM, ESRP1, ESRP2, ST14) or positively (FGFR1, Vimentin). In addition, overexpression or knockdown of ZEB1 led to corresponding changes in gene expression, demonstrating that these genes are also regulated by ZEB1, either directly or indirectly. Of note, the combined knockdown of ZEB1 and ZEB2 led to apparent synergistic responses in gene expression. Furthermore, these responses were not restricted to artificial settings, since most genes were similarly regulated during a physiologic induction of EMT by TGF-β plus EGF. Finally, the absence of ST14 (matriptase) was linked to ZEB1 positivity in lung cancer tissue microarrays, implying that the regulation observed in vitro applies to the human disease. In summary, this study identifies a new set of ZEB-regulated genes in human lung cancer cells and supports the hypothesis that ZEB1 and ZEB2 are key regulators of the EMT process in this disease.
Keywords: lung cancer, ZEB1, ZEB2, EMT, ST14, NSCLC
1. INTRODUCTION
According to data from the World Health Organization, lung cancer accounts for 1.3 million deaths per year, or 18% of the total cancer deaths worldwide. The lung cancer mortality age-adjusted standardized rates increased rapidly from 1960 but then reached a plateau in 1990. Subsequently, lung cancer mortality has declined, although not in all countries and there has been little reduction in women [1]. Approximately half of patients undergoing complete surgical resection for low-stage lung cancer die from distant metastases. Therefore, targeting mechanisms that underlie the metastatic process might positively impact the outcome of patients with this disease.
The epithelial-to-mesenchymal transition (EMT), which converts epithelial cells into an elongated, motile and invasive phenotype, is thought to be a critical step in the dissemination of tumor cells [2–6]. Among hundreds of genes affected by EMT, the loss of E-cadherin and upregulation of Vimentin or N-cadherin have been most frequently described. Other frequent changes include the loss of cytokeratins, increased MMP activity and increased fibronectin. In epithelial cancers, EMT has been associated with resistance to therapy, the acquisition of tumor-initiating cell properties, and increased cell migration and invasion, all of which are associated with a poor prognosis [3; 5].
By immunostaining, severe E-cadherin loss occurs in approximately 10% of resected NSCLCs [7]. Using a low E-cadherin / high N-cadherin ratio and other markers including integrin αvβ6 and MMP-9, Prudkin et al. [8] concluded that a majority of lung adenocarcinomas and squamous cancers had evidence of EMT. These and other data suggest that EMT reflects a continuum with its assessment dependent on the markers used. Although E-cadherin loss is a major feature of EMT, this has often been considered a late event in tumor progression. E-cadherin loss can also be transient or occur preferentially at the tumor periphery [5]. Thus, a better understanding of the spectrum of EMT phenotypes and its regulation is indicated.
In part, EMT is mediated by transcription factors including ZEB [9], Snail [10], Slug, Twist [11] and E12/E47 that bind E-box elements (i.e., CANNTG) in genomic DNA [2]. In lung cancer, ZEB1 appears to be a major factor in the EMT process. In NSCLC cell lines, we previously found that loss of E-cadherin was inversely and specifically correlated with ZEB1 mRNA expression [12]. Takeyama et al. [13] recently confirmed these findings and demonstrated that ZEB1 positively influences anchorage-independent growth. In addition, we reported that the tumor suppressor gene, SEMA3F, was specifically repressed by ZEB1 in NSCLC cell lines [14], providing additional support for the importance of this factor in lung cancer.
In the present report, we aimed to identify genes that responded to ZEB1 in lung cancer cell lines. With Affymetrix data from 38 NSCLCs, we ranked genes depending on the correlation between their expression and that of ZEB1. A subset of genes was examined in more detail and their correlation and responsiveness to ZEB1 was validated. We also verified that the same variations in gene expression were obtained after treatment with TGF-β plus EGF to induce EMT. In addition, the expression of ST14, coding for matriptase, was compared to that of E-cadherin and ZEB1 in a tissue microarray (TMA) of lung cancers.
2. MATERIALS AND METHODS
2.1 Cell lines and primary tumor tissues
NSCLC cell lines were obtained from the Colorado Lung Cancer SPORE Cell Line Repository. They were grown in RPMI-1640 supplemented with 10% fetal calf serum and antibiotics (InVitrogen, Inc., Carlsbad, CA). Non-immortalized normal human broncho-epithelial cells (NHBE) were grown in BEBM supplemented with cytokines and growth factors according to manufacturer’s instructions (Lonza, Basel, Switzerland). The FC6625-2 3KT cell line, kindly provided by Dr. John Minna, was derived from human broncho-epithelial cells immortalized by telomerase. BEAS2B cells were derived from SV40-immortalized broncho-epithelia. FC6625-2 3KT and BEAS2B cells were also grown in BEBM supplemented with cytokines and growth factors. H358 FlpIn cells were engineered to express 6 Myc-tagged ZEB1 after induction with doxycycline [14]. TMA slides containing 109 lung tumor samples and 10 normal lung tissues were obtained from US Biomax (Rockville, MD, ref: BC041115).
2.2 RNA isolation and quantitative real-time RT-PCR analysis
Total RNA was extracted from cell lines using the RNeasy Plus Isolation kit (Qiagen, Inc., Valencia, CA). RT-PCR was performed with the Superscript III reverse transcriptase (InVitrogen, Inc.) using the procedure supplied by the manufacturer. mRNA levels were measured by quantitative real-time PCR using the GeneAmp 7500 quantitative PCR system with SYBR-Green chemistry (Applied Biosystems, Foster City, CA). Intron-spanning gene-specific PCR primers were designed to avoid genomic DNA amplification and the PCR products were confirmed by DNA sequencing (not shown). The primers are described in Supplementary Table S1. The results are displayed in terms of the relative expression (×100) compared to GAPDH or actin expression. RT-PCR was done in duplicate.
2.3 Western blot analysis
Protein lysates and SDS/PAGE were performed using standard techniques, as previously described [15]. Primary antibodies were incubated overnight at 4°C. These included rabbit anti-ZEB1 (1:1000; clone H-102; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), mouse anti E-cadherin (1:1000; Cell Signaling Technology, Inc., Danvers, MA), rabbit anti-ST14 (1:1000; Bethyl Laboratories, Inc., Montgomery, TX), mouse anti-EpCAM (1:1000; clone VU-1D9, ThermoFisher Scientific, Inc., Fremont, CA), rabbit anti-N-cadherin (1:1000; Abcam, Inc., Cambridge, MA), rabbit anti-Vimentin (1:1000; Cell Signaling Technology, Inc.), mouse anti-CDS1 (1:1000, clone 2D10; Novus Biologicals, Inc., Littleton, CO) and mouse anti-β-actin (1:20,000; Sigma-Aldrich, Inc., St. Louis, MO). Secondary antibodies were goat anti-mouse HRP or goat anti-rabbit HRP (1:5000; Perkin Elmer, Inc., Waltham, MA) with ECL (Perkin Elmer, Inc.) detection. For β-actin and E-cadherin, the filters were incubated at 4°C with Alexa 488-conjugated chicken anti-mouse secondary antibodies (1:1000; InVitrogen, Inc.) and detected using a Typhoon 9400 Image system (GE Healthcare Bio-sciences Corp., Piscataway, NJ).
2.4 Transfection of short-interfering RNA
Pre-validated siRNAs (InVitrogen, Inc., cat# HSS110549 and HSS190654) were used to inhibit endogenous ZEB1 and ZEB2, respectively, as previously reported [15]. Transfections were performed using HiPerFect according to manufacturer’s recommendations (Qiagen, Inc.). Cells were plated at 40–60% confluency and transfected 15 h later with final siRNA concentrations of 5 nM. Media were replaced at 24 h and cells harvested for analysis at 48 h or 96 h post-transfection.
2.5 TGF-β and EGF cell line treatment
Cells were treated for 48 h with 10 ng/ml TGF-β plus 50 ng/ml EGF. TGF-β (R&D Systems, Inc., Minneapolis, MN) was dissolved at 10 μg/ml in 4 mM HCl and 0.1% BSA. EGF (R&D Systems, Inc.) was dissolved at 500 μg/ml in 0.1% BSA. Control cells were treated with HCl and BSA at the corresponding final concentrations for 48 h; no difference was noticed with untreated cells.
2.6 Actin staining
Cells were fixed in 4% formaldehyde for 10 min at room temperature, permeabilized during 3 min using 0.5% Triton X-100 and stained for F-actin with Texas-red phalloidin (InVitrogen, Inc.). Representative images were captured by confocal microscopy (Leica DRME, Leica Microsystems GmbH, Wetzlar, Germany).
2.7 Lung tissue microarray analysis
TMA slides were deparaffinized by incubations (1 × 10 min; followed by 3 × 30 dips each) in Histoclear solution (National Diagnostics, Inc., Charlotte, NC) and rehydrated by sequential immersions in 100%, 95% and 70% ethanol. After washing 10 min in water, slides were incubated for 15 min in boiling Antigen Unmasking Solution (Vector Laboratories, Inc., Burlingame, CA) and left for an additional 15 min while the solution cooled. After washing in PBS (3 × 5 min each), endogenous peroxidases were inhibited with 0.3% H2O2 during 20 min at room temperature. Following washing in PBS (3 × 5 min each), tissues were permeabilized with 0.5% Triton in PBS for 20 min at room temperature. Blocking was performed with 3% BSA, 5% goat serum in PBS × 20 min (InVitrogen, Inc.). Slides were incubated for 2 h at room temperature with primary antibodies including rabbit anti-ZEB1 (1:50), mouse anti-E-cadherin (1:100) and rabbit anti-ST14 (1:100) antibodies. Following washing in PBS (3 × 5 min each), slides were incubated for 1 h with biotinylated anti-mouse or anti-rabbit antibodies and exposed to Vectastain ABC solution Standard, according to the manufacturer’s protocol (Vector Laboratories, Inc.). Slides were washed 3 times in PBS and incubated with DAB peroxidase substrate (ThermoFisher Scientific, Inc.). The development of the stain was followed by microscopy and stopped by placing the slides into water. Slides were mounted using Dako Fluorescent mounting medium (Dako, Carpinteria, CA). Slides were scanned with an Aperio system (Aperio Technologies, Inc., Vista, CA) and each sample was scored (from 0 to 300) by multiplying the percentage of positive cells (0 to 100%) by the average level of staining intensity (0 to 3). For ZEB1 nuclear staining, tumors with a score > 5 were considered positive (as only a low number of cells are positive for ZEB1). For E-cadherin and ST14, tumors with scores > 10 were considered positive.
2.8 Statistical analysis
Microarray data were preprocessed as previously described [16] yielding expression values for 44928 genes on 38 lung cancer cell lines. For each gene of interest (ZEB1, ZEB2, SNAIL, SLUG, TWIST1 and TWIST2), we calculated the Spearman correlation with each other gene. Due to the large false-discovery rate associated with p-values, a permutation approach was used to define a correlation threshold. Null distributions of correlations per gene were derived from permuted values from 5000 randomly chosen genes. As these null distributions were highly similar, they were combined to yield a threshold of +/− 0.525 to achieve a false discovery rate of 0.05. Gene lists were created based on the 0.525 threshold and sorted according to magnitude. In each case, <3% of genes were deemed to be correlated with the gene of interest.
TMA expression scores were log-transformed to symmetrize the data and correct for skewness. Associations between genes were measured using Pearson correlation; p-value testing identified correlations that were significantly different from 0. An alpha level of 0.05 was chosen as the threshold for statistical significance. Fisher’s Exact Test was used to measure association between expression categories.
3. RESULTS
3.1 Identification of ZEB1-correlated genes in NSCLC cell lines
To identify a set of ZEB1 correlated genes in lung cancer, we utilized Affymetrix data obtained from 38 NSCLC cell lines [16], which were examined using the Spearman rank correlation. Using a cut-off R value of −0.525 corresponding to a 5% false discovery rate, we identified 324 genes significantly negatively correlated with ZEB1, while 141 genes were significantly positively correlated. The top 50 genes from each group are shown in Tables 1 and 2, and the complete gene sets, including unidentified Affymetrix IDs, are provided in the corresponding Supplementary Tables S2 and S3.
Table 1.
Top 50 genes negatively correlated with ZEB1 in 38 NSCLC cell lines.
| Spearman R values1 | ||||||||
|---|---|---|---|---|---|---|---|---|
| RANK | SYMBOL | ZEB1 | ZEB2 | SNAIL | SLUG | TWIST1 | TWIST2 | GENE NAME |
| 1 | EPCAM | −0.85 | −0.79 | −0.19 | 0.00 | −0.20 | −0.14 | epithelial cell adhesion molecule |
| 2 | CDS1 | −0.85 | −0.69 | −0.30 | 0.19 | −0.18 | −0.19 | CDP-diacylglycerol synthase 1 |
| 3 | TSPAN1 | −0.83 | −0.56 | −0.22 | 0.16 | −0.16 | −0.04 | tetraspanin 1 |
| 4 | ESRP1 | −0.82 | −0.73 | −0.28 | 0.19 | 0.01 | −0.07 | epithelial splicing regulatory protein 1 |
| 5 | ST14 | −0.82 | −0.66 | −0.24 | 0.06 | −0.11 | −0.05 | suppression of tumorigenicity 14 (colon carcinoma) |
| 6 | SH3YL1 | −0.81 | −0.75 | −0.38 | 0.09 | −0.11 | −0.02 | SH3 domain containing, Ysc84-like 1 (S. cerevisiae) |
| 7 | IRF6 | −0.80 | −0.58 | −0.12 | 0.24 | −0.13 | −0.04 | interferon regulatory factor 6 |
| 8 | EPN3 | −0.80 | −0.46 | −0.24 | 0.26 | −0.05 | 0.03 | epsin 3 |
| 9 | ESRP2 | −0.79 | −0.59 | −0.33 | 0.04 | 0.04 | −0.02 | epithelial splicing regulatory protein 2 |
| 10 | TMEM125 | −0.79 | −0.60 | −0.11 | 0.02 | −0.30 | −0.16 | transmembrane protein 125 |
| 11 | TMEM30B | −0.79 | −0.60 | −0.12 | 0.11 | −0.17 | −0.23 | transmembrane protein 30B |
| 12 | BIK | −0.79 | −0.50 | −0.22 | 0.14 | 0.00 | −0.16 | BCL2-interacting killer (apoptosis-inducing) |
| 13 | F11R | −0.78 | −0.71 | −0.25 | −0.02 | −0.09 | −0.09 | F11 receptor |
| 14 | ITGB6 | −0.78 | −0.63 | −0.23 | 0.20 | −0.26 | −0.23 | integrin, beta 6 |
| 15 | PTK6 | −0.78 | −0.64 | −0.12 | 0.19 | −0.09 | −0.01 | PTK6 protein tyrosine kinase 6 |
| 16 | EPPK1 | −0.77 | −0.63 | −0.04 | 0.15 | −0.30 | −0.16 | epiplakin 1 |
| 17 | MAL2 | −0.77 | −0.60 | −0.26 | 0.26 | 0.02 | −0.16 | mal, T-cell differentiation protein 2 |
| 18 | PAK6 | −0.77 | −0.62 | −0.07 | 0.38 | −0.10 | −0.08 | p21 protein (Cdc42/Rac)-activated kinase 6 |
| 19 | VAMP8 | −0.77 | −0.44 | −0.30 | 0.19 | −0.16 | −0.08 | vesicle-associated membrane protein 8 (endobrevin) |
| 20 | FGFBP1 | −0.77 | −0.45 | −0.11 | 0.27 | −0.12 | −0.04 | fibroblast growth factor binding protein 1 |
| 21 | LAD1 | −0.77 | −0.54 | −0.10 | 0.21 | −0.10 | −0.16 | ladinin 1 |
| 22 | B3GNT5 | −0.76 | −0.66 | −0.28 | 0.28 | −0.07 | −0.03 | betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| 23 | FXYD3 | −0.76 | −0.54 | −0.26 | 0.28 | −0.10 | 0.01 | FXYD domain containing ion transport regulator 3 |
| 24 | GRHL1 | −0.75 | −0.68 | −0.12 | −0.08 | −0.10 | −0.19 | grainyhead-like 1 (Drosophila) |
| 25 | ADAP1 | −0.75 | −0.57 | −0.07 | 0.04 | −0.13 | −0.30 | ArfGAP with dual PH domains 1 |
| 26 | P2RY2 | −0.75 | −0.48 | 0.02 | 0.26 | −0.10 | −0.05 | purinergic receptor P2Y, G-protein coupled, 2 |
| 27 | S100A14 | −0.75 | −0.57 | −0.18 | 0.09 | −0.13 | −0.12 | S100 calcium binding protein A14 |
| 28 | MARVELD3 | −0.75 | −0.68 | −0.20 | 0.05 | −0.02 | −0.11 | MARVEL domain containing 3 |
| 29 | ANKRD22 | −0.75 | −0.55 | −0.16 | 0.13 | −0.19 | −0.23 | ankyrin repeat domain 22 |
| 30 | HOOK1 | −0.75 | −0.71 | −0.32 | −0.14 | −0.06 | −0.13 | hook homolog 1 (Drosophila) |
| 31 | CDH1 | −0.75 | −0.64 | −0.10 | −0.15 | −0.25 | −0.21 | cadherin 1, type 1, E-cadherin (epithelial) |
| 32 | MPZL2 | −0.74 | −0.63 | −0.13 | 0.11 | −0.24 | −0.04 | myelin protein zero-like 2 |
| 33 | MAPK13 | −0.74 | −0.66 | −0.11 | 0.18 | −0.06 | −0.17 | mitogen-activated protein kinase 13 |
| 34 | GPR87 | −0.74 | −0.45 | −0.12 | 0.42 | 0.04 | −0.08 | G protein-coupled receptor 87 |
| 35 | ZNF165 | −0.74 | −0.56 | −0.29 | 0.35 | 0.01 | 0.01 | zinc finger protein 165 |
| 36 | ELMO3 | −0.74 | −0.67 | −0.11 | 0.08 | −0.26 | −0.19 | engulfment and cell motility 3 |
| 37 | EXPH5 | −0.74 | −0.66 | −0.10 | −0.07 | −0.33 | 0.08 | exophilin 5 |
| 38 | RAB25 | −0.73 | −0.60 | −0.17 | 0.06 | −0.30 | −0.29 | RAB25, member RAS oncogene family |
| 39 | TACSTD2 | −0.73 | −0.55 | −0.21 | 0.15 | −0.13 | −0.22 | tumor-associated calcium signal transducer 2 |
| 40 | LIMA1 | −0.73 | −0.51 | −0.23 | 0.04 | −0.09 | −0.15 | LIM domain and actin binding 1 |
| 41 | CDH3 | −0.73 | −0.63 | 0.10 | 0.09 | −0.25 | −0.12 | cadherin 3, type 1, P-cadherin (placental) |
| 42 | TMPRSS4 | −0.72 | −0.60 | 0.00 | 0.00 | −0.24 | −0.22 | transmembrane protease, serine 4 |
| 43 | QSOX1 | −0.72 | −0.64 | −0.31 | 0.03 | −0.11 | −0.21 | quiescin Q6 sulfhydryl oxidase 1 |
| 44 | SPINT1 | −0.72 | −0.61 | −0.15 | 0.07 | −0.18 | −0.19 | serine peptidase inhibitor, Kunitz type 1 |
| 45 | CLDN7 | −0.72 | −0.57 | −0.12 | 0.02 | −0.37 | −0.22 | claudin 7 |
| 46 | LOC283278 | −0.72 | −0.51 | −0.21 | 0.09 | −0.09 | 0.01 | hypothetical protein LOC283278 |
| 47 | CRB3 | −0.71 | −0.71 | −0.43 | 0.03 | −0.09 | −0.07 | crumbs homolog 3 (Drosophila) |
| 48 | FGD4 | −0.71 | −0.30 | −0.23 | 0.10 | 0.08 | −0.23 | FYVE, RhoGEF and PH domain containing 4 |
| 49 | BSPRY | −0.71 | −0.63 | −0.06 | −0.06 | −0.30 | −0.30 | B-box and SPRY domain containing |
| 50 | PKP3 | −0.71 | −0.56 | −0.36 | 0.33 | 0.11 | 0.14 | plakophilin 3 |
Significant R values are bolded and italicized. Genes of interest are highlighted in gray.
Table 2.
Top 50 genes positively correlated with ZEB1 in 38 NSCLC cell lines.
| Spearman R values1 | ||||||||
|---|---|---|---|---|---|---|---|---|
| RANK | SYMBOL | ZEB1 | ZEB2 | SNAIL | SLUG | TWIST1 | TWIST2 | NAME |
| 1 | VIM | 0.76 | 0.62 | 0.11 | −0.06 | 0.15 | 0.00 | vimentin |
| 2 | MSRB3 | 0.75 | 0.67 | 0.13 | 0.01 | 0.19 | 0.10 | methionine sulfoxide reductase B3 |
| 3 | TXNDC15 | 0.74 | 0.40 | −0.05 | −0.33 | 0.16 | 0.16 | thioredoxin domain containing 15 |
| 4 | SACS | 0.73 | 0.66 | −0.01 | 0.12 | 0.18 | 0.25 | spastic ataxia of Charlevoix-Saguenay (sacsin) |
| 5 | ZEB2 | 0.71 | 1.00 | 0.17 | 0.11 | 0.15 | 0.04 | zinc finger E-box binding homeobox 2 |
| 6 | FGFR1 | 0.70 | 0.49 | 0.13 | −0.11 | 0.40 | 0.23 | fibroblast growth factor receptor 1 |
| 7 | DENND5A | 0.70 | 0.54 | −0.07 | −0.10 | 0.27 | 0.04 | DENN/MADD domain containing 5A |
| 8 | SLC2A14, A3 | 0.69 | 0.58 | 0.25 | −0.28 | −0.02 | 0.07 | solute carrier family 2, member 14 and 3 |
| 9 | LIX1L | 0.69 | 0.57 | 0.11 | −0.23 | 0.12 | 0.22 | Lix1 homolog (mouse)-like |
| 10 | SLC2A3 | 0.69 | 0.61 | 0.26 | −0.29 | −0.04 | 0.04 | solute carrier family 2 member 3 |
| 11 | ST3GAL2 | 0.69 | 0.69 | 0.39 | −0.02 | 0.29 | 0.09 | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| 12 | DENND5B | 0.68 | 0.66 | 0.21 | −0.09 | 0.17 | −0.06 | DENN/MADD domain containing 5B |
| 13 | JAM3 | 0.68 | 0.52 | 0.45 | −0.14 | 0.06 | 0.11 | junctional adhesion molecule 3 |
| 14 | SYDE1 | 0.68 | 0.69 | 0.35 | 0.06 | 0.19 | 0.23 | synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| 15 | PMP22 | 0.68 | 0.66 | 0.44 | −0.06 | −0.13 | 0.08 | peripheral myelin protein 22 |
| 16 | SLC16A6 | 0.68 | 0.48 | 0.01 | 0.10 | 0.14 | 0.09 | solute carrier family 16, member 6 |
| 17 | MAP1B | 0.68 | 0.51 | 0.10 | −0.08 | 0.22 | 0.06 | microtubule-associated protein 1B |
| 18 | TTC28 | 0.67 | 0.55 | 0.29 | −0.32 | 0.02 | 0.02 | tetratricopeptide repeat domain 28 |
| 19 | COL5A2 | 0.67 | 0.54 | 0.28 | −0.21 | −0.17 | −0.25 | collagen, type V, alpha 2 |
| 20 | NFX1 | 0.65 | 0.55 | 0.15 | −0.10 | −0.07 | 0.07 | nuclear transcription factor, X-box binding 1 |
| 21 | CNRIP1 | 0.65 | 0.75 | 0.27 | 0.26 | 0.30 | 0.11 | cannabinoid receptor interacting protein 1 |
| 22 | RECK | 0.65 | 0.75 | 0.09 | 0.07 | 0.28 | 0.24 | reversion-inducing-cysteine-rich protein with kazal motifs |
| 23 | EMP3 | 0.65 | 0.71 | 0.29 | 0.03 | 0.22 | −0.10 | epithelial membrane protein 3 |
| 24 | BATF3 | 0.65 | 0.59 | 0.30 | −0.21 | −0.07 | 0.02 | basic leucine zipper transcription factor, ATF-like 3 |
| 25 | TMEM158 | 0.65 | 0.41 | 0.11 | 0.18 | 0.38 | 0.21 | transmembrane protein 158 |
| 26 | OSBPL6 | 0.65 | 0.35 | 0.11 | −0.25 | −0.12 | −0.09 | oxysterol binding protein-like 6 |
| 27 | CAP2 | 0.64 | 0.60 | 0.15 | −0.03 | 0.36 | 0.14 | CAP, adenylate cyclase-associated protein, 2 |
| 28 | LEPRE1 | 0.64 | 0.53 | 0.02 | −0.16 | 0.18 | 0.13 | leucine proline-enriched proteoglycan (leprecan) 1 |
| 29 | AZI2 | 0.64 | 0.31 | 0.20 | −0.04 | 0.10 | 0.17 | 5-azacytidine induced 2 |
| 30 | TPK1 | 0.64 | 0.63 | 0.12 | −0.01 | −0.10 | −0.06 | thiamin pyrophosphokinase 1 |
| 31 | SIRT1 | 0.64 | 0.36 | 0.06 | −0.50 | −0.08 | −0.17 | sirtuin 1 |
| 32 | NBPF10, 11 | 0.64 | 0.40 | 0.06 | 0.10 | 0.29 | 0.14 | neuroblastoma breakpoint family, member 10 and 11 |
| 33 | STX2 | 0.64 | 0.60 | 0.19 | 0.03 | 0.24 | −0.06 | syntaxin 2 |
| 34 | DLX1 | 0.64 | 0.56 | 0.11 | 0.01 | 0.39 | 0.14 | distal-less homeobox 1 |
| 35 | SLC47A1 | 0.63 | 0.47 | 0.27 | −0.13 | 0.11 | 0.49 | solute carrier family 47, member 1 |
| 36 | FAM55C | 0.63 | 0.56 | −0.11 | 0.03 | 0.25 | 0.09 | family with sequence similarity 55, member C |
| 37 | C3orf19 | 0.63 | 0.37 | −0.04 | −0.32 | 0.20 | −0.06 | chromosome 3 open reading frame 19 |
| 38 | PHF17 | 0.63 | 0.40 | 0.12 | −0.21 | −0.01 | −0.28 | PHD finger protein 17 |
| 39 | STXBP4 | 0.63 | 0.49 | −0.11 | −0.21 | 0.18 | −0.14 | syntaxin binding protein 4 |
| 40 | TNPO1 | 0.63 | 0.49 | −0.02 | 0.07 | 0.21 | 0.06 | transportin 1 |
| 41 | ALPK2 | 0.63 | 0.58 | 0.24 | −0.03 | 0.22 | 0.22 | alpha-kinase 2 |
| 42 | LOC541471 | 0.62 | 0.47 | 0.08 | −0.19 | 0.00 | −0.14 | Hypothetical LOC541471 |
| 43 | FERMT2 | 0.62 | 0.41 | 0.32 | −0.11 | −0.09 | −0.13 | fermitin family homolog 2 |
| 44 | CSGALNACT2 | 0.61 | 0.37 | 0.07 | −0.04 | 0.16 | 0.17 | chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| 45 | EPB41L5 | 0.61 | 0.44 | 0.24 | −0.32 | −0.26 | −0.08 | erythrocyte membrane protein band 4.1 like 5 |
| 46 | CRTAP | 0.61 | 0.54 | −0.14 | 0.09 | 0.39 | 0.20 | cartilage associated protein |
| 47 | BCHE | 0.61 | 0.59 | 0.28 | −0.22 | 0.03 | −0.17 | butyrylcholinesterase |
| 48 | CEP170 | 0.61 | 0.46 | −0.02 | −0.22 | −0.07 | −0.15 | centrosomal protein 170kDa |
| 49 | CHN1 | 0.61 | 0.37 | 0.10 | −0.12 | 0.02 | 0.12 | chimerin 1 |
| 50 | DDAH1 | 0.61 | 0.56 | −0.23 | 0.12 | 0.51 | 0.33 | dimethylarginine dimethylaminohydrolase 1 |
Significant R values are bolded and italicized. Genes of interest are highlighted in gray.
EpCAM was the most significant negatively correlated gene (R = −0.85), followed closely by CDP-diacylglycerol synthetase (CDS1), tetraspanin 1 (TSPAN1), epithelial splicing regulatory protein 1 (ESRP1), and suppressor of tumorigenicity 14 (ST14). E-cadherin, a known ZEB1 target gene, ranked 31st with an R value of −0.75. Among the top positively correlated genes, Vimentin had the highest correlation (R = 0.76), while ZEB2 (R = 0.71) and FGFR1 (R = 0.70) ranked 5th and 6th, respectively. Interestingly, the correlation between ZEB1 and N-cadherin, another EMT marker, was considerably weaker (R = 0.395), well below the 5% false discovery rate cut-off. Many genes that were negatively or positively correlated with ZEB1 were also significantly correlated with ZEB2, but with lower R values. However, there was a notable absence of significant correlations for these specific genes with SNAIL, Slug, Twist1 and Twist 2, although each of the transcriptional factors have been reported to repress E-cadherin [4]. These results suggest that both ZEB1 and ZEB2 play a role in specific aspects of the EMT phenotype in lung cancer.
3.2 Validation of Affymetrix expression results by quantitative real-time RT-PCR and Western blot
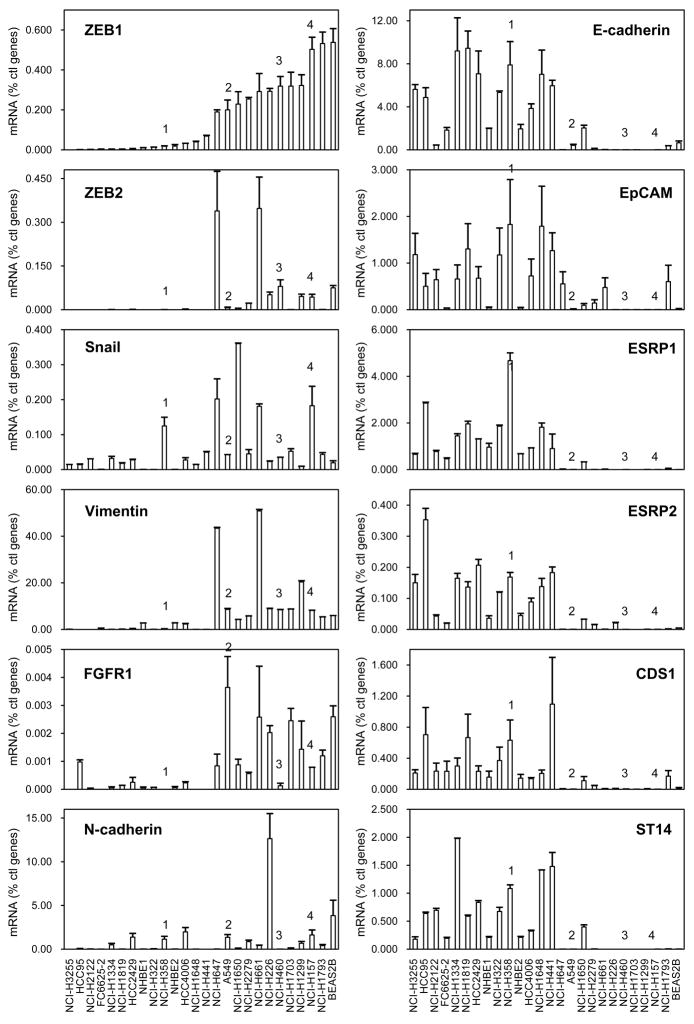
Among the most highly correlated genes from the Affymetrix results, we selected a subset to validate by quantitative real-time RT-PCR using a panel of 22 NSCLC cell lines, randomly chosen, and 4 controls. The selected genes were chosen because of their potential relevance to lung cancer or the EMT pathway (see Discussion) as well as the availability of commercial antibodies in most instances. The results for the negative correlates (EpCAM, ESRP1, ESRP2, CDS1, ST14), positive correlates (FGFR1 and Vimentin) and controls are presented in Fig. 1 with the cell lines arranged by increasing ZEB1 mRNA levels. Expression of the negatively correlated genes clustered with E-cadherin and was opposite to the expression of ZEB1 and ZEB2. Conversely, FGFR1 and Vimentin levels were predominant in cell lines expressing ZEB1 and ZEB2. Thus, the Affymetrix results for these genes, which demonstrated significant correlations with ZEB1, were confirmed by RT-PCR. As anticipated, Snail mRNA expression did not correlate with these genes, and the N-cadherin expression pattern showed considerably less clustering with ZEB1 than did Vimentin and FGFR1. We also tested SIRT1, which was positively correlated with ZEB1 (Table 2), but represented by a single Affymetrix ID. In this case, we could not confirm a correlation with ZEB1 expression (data not shown) and it was excluded from further analysis.
Figure 1. RNA screening in NSCLC cell lines.
RNA expression was measured by quantitative real-time RT-PCR in a series of 22 NSCLC cell lines, two NHBE cultures and two immortalized human airway primary cell lines (BEAS2B and FC6625-2 3KT). Cells were ranked from left to right according to ZEB1 mRNA expression. Values are expressed as percent of the geometric mean between GAPDH and actin. The experiment was done twice in duplicate. Bars = SD. Numbers 1–4 indicate H358, A549, H460 and H157 cell lines, respectively, discussed later in the text.
The two primary normal human broncho-epithelial (NHBE) cultures and the telomerase-immortalized cell line, FC6625-2 3KT, expressed E-cadherin and had low levels of ZEB1. In contrast, E-cadherin and ZEB1 expression in the SV40-TAg transformed BEAS2B cell line showed the opposite pattern. Indeed, while BEAS2B cells are often used as a model for normal lung epithelial cells, they present a mesenchymal pattern of gene expression.
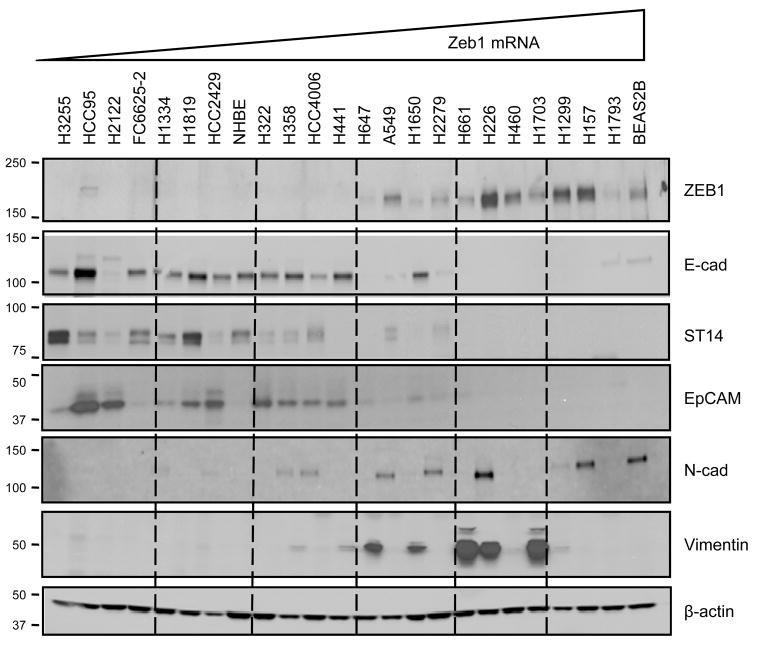
Gene expression at the protein level was confirmed by Western blots for ZEB1, ST14, EpCAM and Vimentin, along with E-cadherin and N-cadherin for comparison (Fig. 2). The overall patterns of ST14, EpCAM and E-cadherin expression were similar and opposite to those of ZEB1 and Vimentin. Therefore, both quantitative real-time PCR and Western blot analyses confirmed the observed correlations from the Affymetrix data.
Figure 2. Correlations of protein levels with ZEB1 in NSCLC.
For each cell line, 10 μg total protein were loaded on a 4–15% polyacrylamide gel. Cell lines were ranked on the gel according to ZEB1 mRNA from low to high levels (left to right). Western blots were performed with indicated antibodies. Molecular weights are provided in kDa. β-actin was used as a loading control. Among these cell lines, those with known mutations of EGFR and sensitivity to an EGFR inhibitor include H3255, HCC4006, H1650 and HCC2279 [48]. In addition, those with IC50s of 1 μM or less in a proliferation assay include H358 and H322 [48].
3.3 ZEB1-correlated genes are responsive to ZEB1 and ZEB2 overexpression as well as downregulation
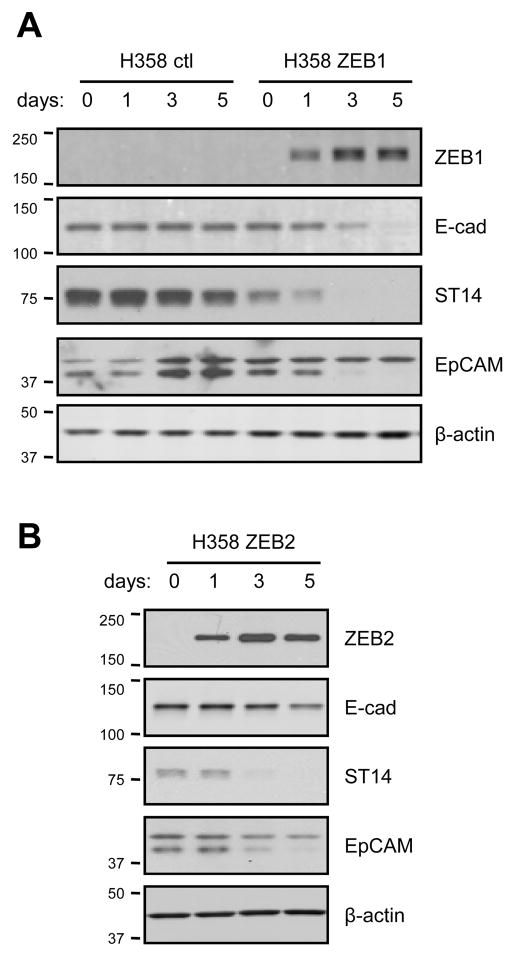
To determine whether ZEB1-correlated genes were responsive to changes in ZEB1 and ZEB2 expression, we separately overexpressed these genes in H358 cells using a dox-inducible system. These cells were chosen because of high endogenous E-cadherin expression, low ZEB1 and barely detectable ZEB2 (Fig. 1). H358 cells containing empty vector control (ctl), and Myc-tagged ZEB1 or ZEB2 were exposed to doxycycline for 0, 1, 3 and 5 days. Increased ZEB mRNA levels were detected after one day of induction and corresponding protein accumulated over time (Table 3, Fig. 3). The expression levels are shown in Table 3 as relative changes compared to baseline in the absence of doxycycline (day 0). ZEB1 induction led to a 55-fold upregulation of endogenous ZEB2 mRNA by day 5. Although we were unable to look for endogenous ZEB2 protein due to the lack of suitable antibodies, the ZEB2 RNA levels were still quite low even with this degree of upregulation. In contrast, ZEB2 induction had little to no effect on ZEB1 mRNA.
Table 3.
Changes in gene expression induced by exogenous ZEB1 or ZEB2 in H358 cells.
| day | ZEB1 | ZEB2 | E-cad | EpCAM | ESRP1 | ESRP2 | CDS1 | ST14 | FGFR1 | Vimentin | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| H358 ctl | 1 | 1.6 | 1.6 | 1.2 | 1.3 | 1.3 | 1.3 | 1.3 | 1.2 | 2.1 | 1.4 |
| 3 | 1.7 | 3.2 | 1.3 | 1.4 | 1.5 | 1.3 | 1.4 | 1.2 | 1.8 | 1.5 | |
| 5 | 1.4 | 2.2 | 1.2 | 1.3 | 1.2 | 1.3 | 1.3 | 1.1 | 1.5 | 1.3 | |
| H358 ZEB1 | 1 | 35 | 6.7 | 0.37 | 0.34 | 0.18 | 0.25 | 0.31 | 0.23 | 3.2 | 3.2 |
| 3 | 55 | 21 | 0.14 | 0.26 | 0.03 | 0.08 | 0.11 | 0.09 | 2.1 | 14 | |
| 5 | 27 | 55 | 0.03 | 0.20 | 0.01 | 0.08 | 0.17 | 0.04 | 3.6 | 19 | |
| H358 ZEB2 | 1 | 1.2 | 46 | 1.04 | 0.79 | 0.92 | 0.97 | 0.79 | 0.62 | 1.1 | 2.4 |
| 3 | 1.1 | 87 | 0.73 | 0.72 | 0.39 | 0.55 | 0.50 | 0.43 | 2.3 | 10 | |
| 5 | 2.9 | 47 | 0.49 | 0.58 | 0.20 | 0.58 | 0.51 | 0.27 | 2.1 | 11 | |
Figure 3. E-cadherin, ST14 and EpCAM proteins are decreased by exogenous ZEB1 or ZEB2 in H358 cells.
Cells stably transfected with inducible myc-tagged ZEB1 or ZEB2, or with vector (ctl) were grown for 5 days and concurrently induced with doxycycline (100 ng/ml) for 0, 1, 3 or 5 days. Protein lysates were analyzed by Western blot for induction of ZEB1 (A) or ZEB2 (B) using anti-myc antibodies. E-cadherin, ST14 and EpCAM were detected by corresponding antibodies. The asterisk indicates a variable background band. β-actin provided a loading control.
ZEB1 induction led to a substantial reduction in the expression of E-cadherin, which is a known target gene. As predicted by the Spearman correlations, EpCAM, ESRP1, ESRP2, CDS1 and ST14 mRNA levels were downregulated following ZEB1 induction, while Vimentin and FGFR1 levels were increased. ZEB2 induction led to similar, although more modest, changes in expression of the same genes (Table 3). Corresponding changes in protein expression were confirmed for E-cadherin, ST14 and EpCAM (Fig. 3). To explore the effects of lower levels of ZEB1 induction, we treated H358 cells with doxycycline (0–5 ng/ml) for 12 and 24 hr (Supplemental Fig. S1A). ZEB1 protein was already observed after 12 h and accumulated over time with increasing doxycycline concentrations. In these conditions, ESRP1 expression was affected more dramatically than the other tested genes (Supplemental Fig. S1B). Overall, these results demonstrate a dose and time-dependent response to ZEB1.
We next inhibited the expression of ZEB1, ZEB2, or both genes in four lung cancer cell lines using siRNA. As shown in Fig. 1, H157, H460 and H661 cells expressed elevated levels of ZEB1 and ZEB2 and had absent E-cadherin. A549 cells are intermediate in that they express ZEB1, have lower levels of ZEB2, and have reduced but detectable levels of E-cadherin, EpCAM and ST14. The results of transient ZEB silencing are shown in Table 4. In general, each of the ZEB negatively correlated genes showed some degree of upregulation, although this varied depending on the cell line. Also in general, the effects on gene expression were greater with knockdown of ZEB1 versus ZEB2, and the combined knockdown often resulted in the appearance of synergistic upregulation of these genes. Although we have not explored the mechanism of synergistic change, the loss of both factors appears necessary for substantial re-expression. In H157 cells, ZEB1 knockdown resulted in a 2-fold upregulation of ZEB2, which may compensate for the loss of ZEB1. However, this was not observed in the other cell lines. For ESRP2, we note that the levels, while generally paralleling ESRP1, were less responsive to ZEB1 and ZEB2 knockdown. Changes in Vimentin and FGFR1 mRNA were modest and variable. Protein levels for E-cadherin, CDS1 and EpCAM in H157 cells were increased by knockdown of ZEB (Fig. 4). As previously observed for E-cadherin [15], EpCAM levels were greatly increased by the combined knockdown of ZEB1 and ZEB2. Together, these results demonstrate that the ZEB negatively correlated genes are also responsive to ZEB overexpression and downregulation, whereas Vimentin and FGFR1 are predominantly responsive to ZEB overexpression.
Table 4.
Changes in gene expression induced by knockdown of ZEB1, ZEB2, or both in NSCLC cell lines.
| siRNA | ZEB1 | ZEB2 | E-cad | EpCAM | ESRP1 | ESRP2 | CDS1 | ST14 | FGFR1 | Vimentin | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| H157 | ZEB1 | 0.23 | 2.1 | 2.5 | 0.79 | 3.4 | 2.3 | 1.7 | 7.1 | 3.7 | 0.94 |
| ZEB2 | 0.72 | 0.10 | 0.20 | 1.4 | 2.2 | 4.8 | 0.29 | 1.4 | 1.5 | 1.2 | |
| ZEB1 + ZEB2 | 0.36 | 0.15 | 18 | 74 | 164 | 3.7 | 7.3 | 16 | 2.5 | 0.8 | |
| H460 | ZEB1 | 0.27 | 1.0 | 179 | 20 | 1.9 | 1.3 | 4.2 | 3.8 | 1.8 | 1.2 |
| ZEB2 | 1.0 | 0.21 | 0.65 | 1.7 | 0.85 | 1.8 | 1.3 | 1.2 | 1.2 | 1.5 | |
| ZEB1 + ZEB2 | 0.33 | 0.25 | 2280 | 139 | 4.5 | 2.5 | 10 | 12 | 2.7 | 1.9 | |
| H661 | ZEB1 | 0.25 | 1.1 | 3.7 | 1.6 | 1.1 | 0.86 | 2.1 | 1.5 | 1.0 | 0.85 |
| ZEB2 | 1.4 | 0.18 | 1.9 | 1.7 | 2.0 | 1.4 | 1.7 | 0.53 | 1.2 | 1.0 | |
| ZEB1 + ZEB2 | 0.41 | 0.33 | 32 | 3.4 | 6.2 | 1.4 | 3.8 | 0.38 | 1.7 | 1.3 | |
| A549 | ZEB1 | 0.27 | 0.16 | 1.6 | 4.0 | 2.5 | 0.86 | 2.4 | 2.4 | 0.29 | 0.36 |
| ZEB2 | 0.68 | 0.06 | 0.49 | 0.74 | 0.62 | 0.33 | 0.76 | 1.4 | 0.47 | 0.53 | |
| ZEB1 + ZEB2 | 0.26 | 0.11 | 2.5 | 2.6 | 4.2 | 0.32 | 2.5 | 1.4 | 0.69 | 0.59 | |
Figure 4. E-cadherin, CDS1 and EpCAM are increased by ZEB1 or ZEB2 knockdown in H157 cells.
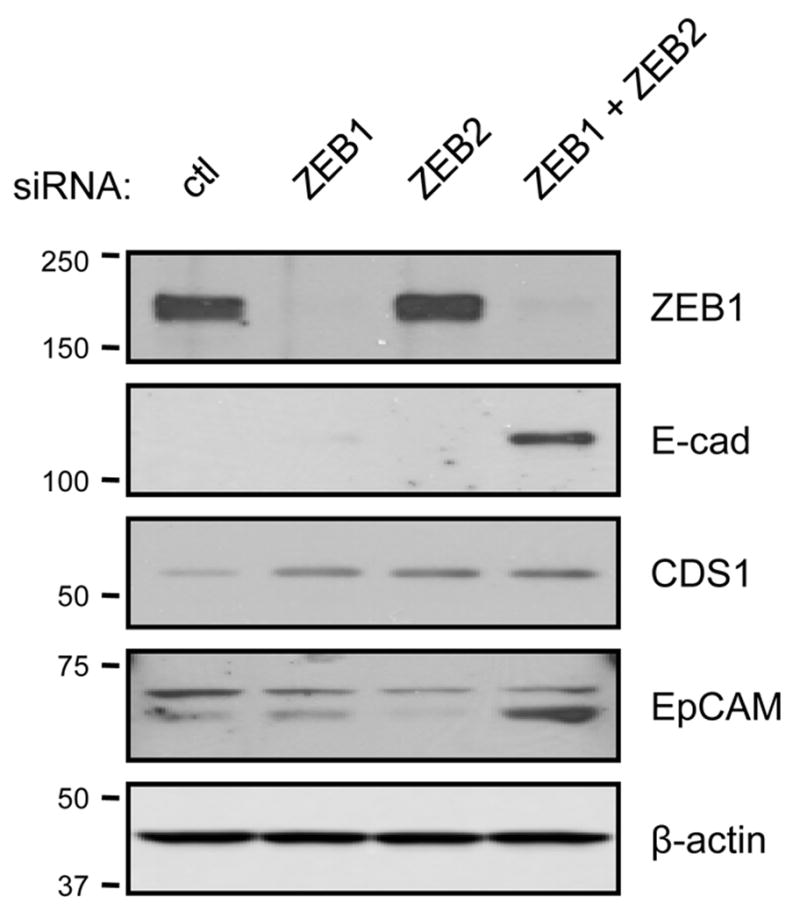
siRNAs targeting ZEB1, ZEB2 or a non-targeting control were transfected into H157 cells. Cells were harvested for analysis 5 days post-transfection. Protein lysates were examined by Western blot using the indicated antibodies. β-actin provided a loading control.
3.4 ZEB-responsive genes are also affected upon physiologic induction of EMT
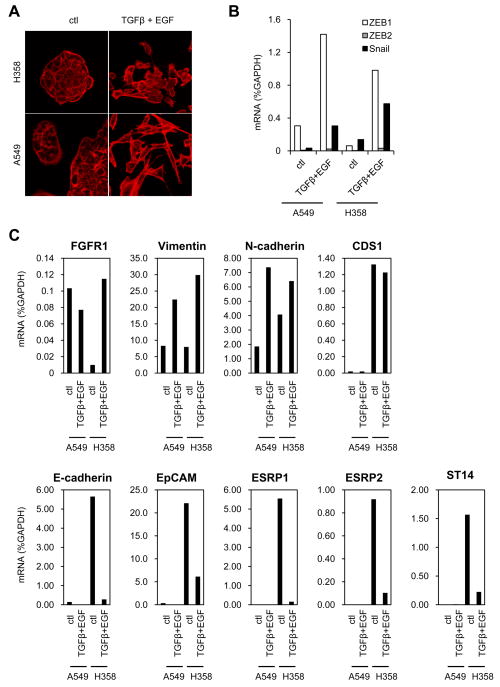
To determine whether the ZEB-responsive genes were also affected by a physiologic stimulator of EMT, we exposed A549 and H358 cells to TGF-β and EGF. Both untreated cell lines grew in typical epithelial-appearing clusters, but became more elongated and scattered in the presence of TGF-β plus EGF (Fig. 5A). This effect was more dramatic in A549 cells. The mRNA levels of ZEB1, Snail and, to a lesser extent ZEB2, were upregulated in both cell lines (Fig. 5B). In H358 cells, TGF-β plus EGF caused a marked reduction in the epithelial genes (E-cadherin, EpCAM, ESRP1, ESRP2 and ST14) and upregulation of mesenchymal genes (FGFR1, Vimentin, N-cadherin). However, CDS1 expression was unaffected (Fig. 5C). In A549 cells, the levels of epithelial genes (ESRP1, ESRP2 and ST14) were so low without treatment that further changes were negligible. Vimentin and N-cadherin were both upregulated. These results show that ZEB-responsive genes are also regulated in the same manner by a more physiologic induction of EMT.
Figure 5. ZEB1-responsive genes are similarly regulated by TGF-β plus EGF treatment.
(A) A549 and H358 cells were treated during 48 h with 10 ng/ml TGF-β plus 50 ng/ml EGF. Actin was stained by phalloidin. (B) Expression of ZEB1, ZEB2, Snail and (C) ZEB1 regulated genes was measured by quantitative real-time RT-PCR. Values are expressed as percent of GAPDH.
3.5 ZEB1 and ST14 staining in human lung cancers
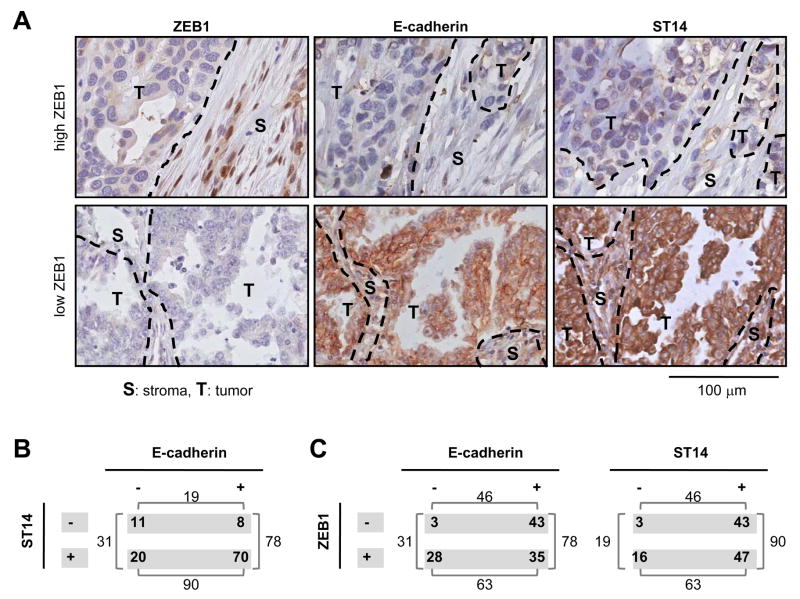
To determine if a correlation could be found in vivo between ZEB1 and one of the new ZEB1-responsive genes, we examined ST14 expression by immunohistochemistry on a TMA containing a series of 109 human lung tumors. ST14, coding for matriptase, was chosen because it was reported to have a tumor suppressor function in some contexts [17], and because of the availability of a highly specific commercial antibody. We examined E-cadherin as a control. As depicted in Supplemental Fig. S2, induced ZEB1 was detected in the nuclei, but not in the cytoplasm. In the majority of tumor samples from the TMA (Fig. 6A), ZEB1 positive nuclear staining was confined to elongated cells in the stromal compartment. In contrast, the vast majority of E-cadherin and ST14 positive cells were located in the tumor compartment but not in the stroma.
Figure 6. ST14 positively correlates with E-cadherin but negatively with ZEB1 in lung tumors.
(A) Examples of two tumors with either high (upper panel) or low (lower panel) ZEB1 level and corresponding immunostaining for E-cadherin and ST14. Dashed line: separation between tumor and stroma; T: Tumor compartment; S: Stromal compartment; scale bar = 100 μm. (B) Distribution of tumors for E-cadherin or ST14 staining. For ZEB1 nuclear staining, tumors with a score > 5 were considered positive (as only a low number of cells are positive for ZEB1). For E-cadherin and ST14, tumors with scores > 10 were considered positive. (C) Distribution of tumors stained for ZEB1 and compared with E-cadherin or ST14. (B, C) Results are presented as number of tumors.
Of these 109 tumors, 78 (72%) were E-cadherin positive and 90 (83%) were ST14 positive (Fig. 6B); the association between the two markers was significant (p = 0.004). However, among the 19 ST14-negative tumors, 8 (42%) were positive for E-cadherin. On the other hand, 20 of the 31 E-cadherin negative tumors (65%) retained ST14 staining. These two groups may represent examples of partial EMT, although additional markers would be required for confirmation. Other investigators have described partial EMT with retention of E-cadherin, for example in tumor cells expressing a CD44high/CD24low phenotype and increased N-cadherin (see Discussion).
An inverse pattern of expression was observed between the immunostaining for ZEB1 and E-cadherin or ST14 (Fig. 6A) and a statistically significant inverse association was found between E-cadherin and ZEB1 (p <0.001) and between ST14 and ZEB1 (p = 0.01). Of note, ZEB1 positive cells were present in 28/31 (90%) tumors that had lost E-cadherin, and similar results were observed for ST14 (16/19, 84%). These results indicate that when ZEB1 positive cells are found in the stroma, there is less probability to find E-cadherin or ST14 in the tumor compartment.
4. DISCUSSION
The epithelial to mesenchymal transition plays an important role in tumor progression, metastasis and treatment resistance [3–6; 18]. The changes associated with EMT are complex, progressive and dependent on the specific factors initiating the transition, not all of which are identified. EMT is reversible and often occurs in only a subset of cells or in specific locations. EMT can thus be difficult to visualize in tumors and may be underestimated. A set of EMT-related genes, in addition to the classical markers E-cadherin and Vimentin, could improve identification of this process in tumors and better define its features. Among EMT-inducing transcriptional repressors, ZEB1 appears to have an important role in lung cancer. To discover ZEB1 responsive genes, we used Affymetrix data sets from 38 NSCLC cell lines and identified 324 genes significantly inversely correlated with ZEB1 and 141 significantly positively correlated genes (Supplemental Tables S2 and S3). Among the positive correlates were the mesenchymal markers, Vimentin, ZEB2 and FGFR1. The negatively correlated genes included epithelial-specific adhesion molecules and cell surface antigens. This EMT phenotype was specific for ZEB1 (and ZEB2), but not associated with SNAIL, Slug or Twist. For E-cadherin, this confirms our previous findings [12] and those of Takeyama et al. [13]. In addition, seven of 45 published biomarkers reported to be prognostic in early stage adenocarcinoma were negatively correlated with ZEB1 (Supplemental Table S4), potentially implicating ZEB1 in this group of tumors.
EpCAM, ESRP1, ESRP2, CDS1, ST14, FGFR1 and Vimentin were chosen for validation and to serve as “proof of principle”, with E-cadherin included as a control. Correlation of these selected genes with ZEB1 was confirmed in 22 NSCLC cell lines at both the mRNA and protein levels (Fig. 1–2). Exogenous ZEB1 expression in H358 cells verified responsiveness of these genes (Table 3, Fig. 3). Although the genes were also responsive to ZEB2, the effects were generally more modest. However, the combined inhibition of ZEB1 and ZEB2 increased the expression of several ZEB downregulated genes in a synergistic manner (Table 4, Fig. 4), indicating they have complementary functions. Conversely, inhibiting ZEB1 or ZEB2, alone or together, had little to no effect on Vimentin, which was substantially upregulated by ZEB1 over-expression. This was surprising since Vimentin was the most positively correlated gene with ZEB1 (R = 0.76) and was also highly positively correlated with ZEB2 (R = 0.62). It is possible that the effects of ZEB1 on Vimentin are indirect and may require a longer time period to become manifest.
We further verified that corresponding variations in gene expression were obtained after treatment with TGF-β plus EGF in A549 and H358 cells (Fig. 5). TGF-β and EGF are known to induce EMT (for review see [19]). TGF-β acts via the Smad proteins, or through ERK and PI3K signaling pathways, to induce the expression of multiple E-box repressors including ZEB1, ZEB2, Snail and Slug [19]. The enhancing effect of EGF is thought to be through its phosphorylation and downregulation of E-cadherin, which can induce EMT over time [20]. Interestingly, we note that H358 cells were less sensitive than A549 cells to the morphologic changes induced by TGF-β plus EGF. This might result from the high endogenous expression of epithelial genes in H358 cells (Fig. 5C).
Several validated genes may be of particular interest for lung cancer or the EMT pathway including cell adhesion (EpCAM), epithelial-specific RNA splicing (ESRP1, ESRP2) and proteolytic activity (ST14). EpCAM had the highest negative correlation with ZEB1 expression (Table 1, R = −0.85). EpCAM is a homophilic adhesion molecule that connects to the actin cytoskeleton through β-actinin and inhibits cadherin-mediated adhesion [21]. In tumor cells, EpCAM undergoes regulated intramembrane proteolysis and subsequent nuclear translocation activates c-Myc expression [22]. Thus, EpCAM inhibition can have anti-proliferative effects. This suggests that some tumors may be driven, at least in part, by high EpCAM, which is associated with an epithelial phenotype. EpCAM was initially identified as a tumor antigen [23], and is overexpressed in ~ 25% of lung adenocarcinomas [24]. Our finding that EpCAM is ZEB-responsive is consistent with previous reports of low EpCAM and high ZEB1 associated with resistance to EGFR inhibitors [25; 26]. Of note, an anti-EpCAM antibody (catumaxomab) has been approved in the European Union for the treatment of malignant ascites [27].
ESRP1 and 2 are RNA binding proteins that regulate epithelial-specific splicing of several relevant genes including FGFR2, CD44 and p120-catenin [28]. ESRP1 was the fourth most negatively correlated gene with ZEB1, and while preliminary, we observed its levels to be more inhibited than E-cadherin by short-term, low dose ZEB1 induction (Supplemental Fig. S1). It is believed that the splicing patterns regulated by ESRP1 and 2 contribute to the epithelial phenotype. In agreement, induction of EMT in mammary epithelial cells by either Twist or E-cadherin knockdown resulted in ESRP1/2 inhibition [28; 29].
ST14 encodes a membrane-associated serine protease (matriptase) that co-localizes with E-cadherin [30], can process the hepatocyte growth factor precursor [31] and plays a role in epithelial differentiation and maintenance of tight junctions [30; 32]. In MDA-MB-231 cells, ST14 expression inhibited cell growth [17], although in other contexts its over-expression can facilitate transformation [33]. ST14 has also been reported to inhibit ZEB2-driven EMT [34]. In 109 lung cancers, we found that ST14 staining was highly significantly correlated with E-cadherin and negatively associated with ZEB1 (Fig. 6). Of note, 25% of tumors expressed either ST14 or E-cadherin, but not both, suggesting that ST14 may be a useful adjunct to E-cadherin for detection of EMT features. Among tumors that had lost either E-cadherin or ST14, there was a marked increase in ZEB1 positive stromal cells. Localization of ZEB1 in stromal cells has also been observed in both colon and breast cancers [35]. The nature of these cells is unknown. In lung cancer, the possibility that these represent tumor cells that have undergone EMT is supported by the recent work of Mink et al. [36]. In a xenograft model, these authors found that 24% of fibroblast-like cells in the stroma were tumor-derived, and when co-cultured, this stroma conferred resistance to gefitinib-sensitive cells. However, further studies such as FISH will be required to determine if these cells are tumor-derived. Two hypotheses could explain the loss of E-cadherin and ST14 in the tumor compartment. ZEB1-dependent paracrine signaling from the stroma compartment could repress both genes. Alternatively, an additional factor could repress E-cadherin in the tumor compartment and ZEB1 would act to maintain the repressed state in escaped tumor cells. Further investigations will be required to distinguish between these possibilities.
Our results suggest that both ZEB1 and ZEB2 may play major roles in lung cancer, in agreement with previous reports [12; 13; 37]. In addition to its predominance in E-cadherin suppression, ZEB1 knockdown has been shown to impair soft agar growth of H157, H460 and H1299 cells [13]. ZEB1 has also been implicated in EMT associated with loss of function of LKB1 [38], now recognized as the most common gene alteration in lung adenocarcinomas [39; 40]. However, ZEB1 does not act alone; multiple E-box binding factors are capable of inhibiting E-cadherin and inducing EMT. These include Snail, Slug, E12/E47 and Twist. EMT induction is complex and dependent on additional transcription and chromatin-modifying factors [41–44]. Snail as well as the T-box factor, Brachyury, have been shown to regulate EMT in NSCLC cell lines [45; 46]. Moreover, we observed a negative correlation of CD24 with ZEB1, while Twist1 was positively correlated with CD44, suggesting these two factors may cooperate to generate the CD44high/CD24low stem-like phenotype associated with EMT. In addition, Twist 1 was the only E-box factor examined that demonstrated a positive correlation with N-cadherin (data not shown).
In lung cancer cell lines, EMT features estimated by the loss of E-cadherin and upregulation of Vimentin is frequent. However, in resected lung cancers the frequency of EMT is less clear. Based on E-cadherin loss, the frequency ranges from 10–30% [7; 37; 47]. Using additional markers, including αVβ6 integrin, Prudkin et al. found evidence of EMT in the majority of adeno- and squamous cancers [8], emphasizing the importance of having additional markers and understanding their relationship to EMT.
In summary, this study has identified a set of over 400 genes whose expression is highly correlated with ZEB1 mRNA levels in 38 NSCLC cell lines. Based on our validation studies, we predict that a majority is likely to be both ZEB1 and EMT responsive. These genes should be useful in assessing the contribution of ZEB1 to the EMT phenotype in lung cancer, either through basal inhibition or following EMT induction by other factors (e.g., TGF-β). We hypothesize that a quantitative analysis of several selected ZEB1-responsive genes may provide an improved assessment of the EMT status in lung tumors. This gene collection should provide some excellent candidates in this regard. We also anticipate that several of these new ZEB-responsive genes may contribute to EMT in lung cancer.
Supplementary Material
Figure S1. Gene repression with low-level of induced ZEB1 in H358 cells. (A) ZEB1-inducible H358 cells at 50% confluency were treated with low doses of doxycycline (0 to 5 ng/ml, as indicated) for 12 and 24 h and harvested. Protein lysates were analyzed by Western blot for induced ZEB1 (anti-myc antibodies). β-actin provided a loading control. (B) RNA isolated from the same samples was analyzed by quantitative real-time RT-PCR for the indicated genes. Values were calculated as a percent of GAPDH and normalized to the untreated controls. Data are reported as the fold-change compared to cultures treated with vehicle alone.
Figure S2. Validation of ZEB1 nuclear staining on tumor xenografts. Control (H358 – ctl; left panel) or ZEB1-inducible H358 cells (H358 – ZEB1; right panel) were injected subcutaneously in NCrnu/nu mice. ZEB1 was induced by treating animals with doxycycline (1mg/ml) on a daily basis. Tumors were then excised, fixed and sectioned for immunohistochemical detection of ZEB1 using conditions identical to those used in Figure 6. Control ZEB1 tumors have very low ZEB1 levels and the faint cytoplasmic staining represents background. Animals were handled in accordance with established protocols approved by the institutional animal care committee.
Table S1. DNA primer sequences for quantitative real-time RT-PCR
Table S2. Spearman R values from Affymetrix data for all significant negative correlates with ZEB1 in 38 NSCLC cell lines.
Table S3. Spearman R values from Affymetrix data for all significant positive correlates with ZEB1 in 38 NSCLC cell lines.
Table S4. Spearman R values for 45 lung cancer biomarkers versus EMT factors from Affymetrix data of 38 NSCLC cell lines [1–8].
Acknowledgments
Supported by the University of Colorado Lung Cancer SPORE grant, NCI-CA58187 and by the Medical University of South Carolina – Cancer Center Support Grant, Biostatistics Core NCI-CA138313.
Footnotes
5. CONFLICT OF INTEREST
We have no conflict of interest to de disclosed.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Marugame T, Yoshimi I. Comparison of cancer mortality (lung cancer) in five countries: France, Italy, Japan, UK and USA from the WHO Mortality Database (1960–2000) Jpn J Clin Oncol. 2005;35:168–70. doi: 10.1093/jjco/hyi048. [DOI] [PubMed] [Google Scholar]
- 2.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 3.Sabbah M, Emami S, Redeuilh G, Julien S, Prevost G, Zimber A, Ouelaa R, Bracke M, De Wever O, Gespach C. Molecular signature and therapeutic perspective of the epithelial-to-mesenchymal transitions in epithelial cancers. Drug Resist Updat. 2008;11:123–51. doi: 10.1016/j.drup.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Voulgari A, Pintzas A. Epithelial-mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim Biophys Acta. 2009;1796:75–90. doi: 10.1016/j.bbcan.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Yilmaz M, Christofori G. Mechanisms of motility in metastasizing cells. Mol Cancer Res. 2010;8:629–42. doi: 10.1158/1541-7786.MCR-10-0139. [DOI] [PubMed] [Google Scholar]
- 7.Bremnes RM, Veve R, Gabrielson E, Hirsch FR, Baron A, Bemis L, Gemmill RM, Drabkin HA, Franklin WA. High-throughput tissue microarray analysis used to evaluate biology and prognostic significance of the E-cadherin pathway in non-small-cell lung cancer. J Clin Oncol. 2002;20:2417–28. doi: 10.1200/JCO.2002.08.159. [DOI] [PubMed] [Google Scholar]
- 8.Prudkin L, Liu DD, Ozburn NC, Sun M, Behrens C, Tang X, Brown KC, Bekele BN, Moran C, Wistuba Epithelial-to-mesenchymal transition in the development and progression of adenocarcinoma and squamous cell carcinoma of the lung. Mod Pathol. 2009;22:668–78. doi: 10.1038/modpathol.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Postigo AA, Dean DC. ZEB, a vertebrate homolog of Drosophila Zfh-1, is a negative regulator of muscle differentiation. Embo J. 1997;16:3935–43. doi: 10.1093/emboj/16.13.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peinado H, Quintanilla M, Cano A. Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: mechanisms for epithelial mesenchymal transitions. J Biol Chem. 2003;278:21113–23. doi: 10.1074/jbc.M211304200. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Ohira T, Gemmill RM, Ferguson K, Kusy S, Roche J, Brambilla E, Zeng C, Baron A, Bemis L, Erickson P, Wilder E, Rustgi A, Kitajewski J, Gabrielson E, Bremnes R, Franklin W, Drabkin HA. WNT7a induces E-cadherin in lung cancer cells. Proc Natl Acad Sci U S A. 2003;100:10429–34. doi: 10.1073/pnas.1734137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeyama Y, Sato M, Horio M, Hase T, Yoshida K, Yokoyama T, Nakashima H, Hashimoto N, Sekido Y, Gazdar AF, Minna JD, Kondo M, Hasegawa Y. Knockdown of ZEB1, a master epithelial-to-mesenchymal transition (EMT) gene, suppresses anchorage-independent cell growth of lung cancer cells. Cancer Lett. 2010 doi: 10.1016/j.canlet.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarhaut J, Gemmill RM, Potiron VA, Ait-Si-Ali S, Imbert J, Drabkin HA, Roche J. ZEB-1, a repressor of the semaphorin 3F tumor suppressor gene in lung cancer cells. Neoplasia. 2009;11:157–66. doi: 10.1593/neo.81074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kakihana M, Ohira T, Chan D, Webster RB, Kato H, Drabkin HA, Gemmill RM. Induction of E-cadherin in lung cancer and interaction with growth suppression by histone deacetylase inhibition. J Thorac Oncol. 2009;4:1455–65. doi: 10.1097/JTO.0b013e3181bc9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coldren CD, Helfrich BA, Witta SE, Sugita M, Lapadat R, Zeng C, Baron A, Franklin WA, Hirsch FR, Geraci MW, Bunn PA., Jr Baseline gene expression predicts sensitivity to gefitinib in non-small cell lung cancer cell lines. Mol Cancer Res. 2006;4:521–8. doi: 10.1158/1541-7786.MCR-06-0095. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Rathinam R, Walch A, Alahari SK. ST14 (suppression of tumorigenicity 14) gene is a target for miR-27b, and the inhibitory effect of ST14 on cell growth is independent of miR-27b regulation. J Biol Chem. 2009;284:23094–106. doi: 10.1074/jbc.M109.012617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 19.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–58. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Andersen H, Mejlvang J, Mahmood S, Gromova I, Gromov P, Lukanidin E, Kriajevska M, Mellon JK, Tulchinsky E. Immediate and delayed effects of E-cadherin inhibition on gene regulation and cell motility in human epidermoid carcinoma cells. Mol Cell Biol. 2005;25:9138–50. doi: 10.1128/MCB.25.20.9138-9150.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Litvinov SV, Balzar M, Winter MJ, Bakker HA, Briaire-de Bruijn IH, Prins F, Fleuren GJ, Warnaar SO. Epithelial cell adhesion molecule (Ep-CAM) modulates cell-cell interactions mediated by classic cadherins. J Cell Biol. 1997;139:1337–48. doi: 10.1083/jcb.139.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, Kieu C, Papior P, Baeuerle PA, Munz M, Gires O. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11:162–71. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 23.Koprowski H, Steplewski Z, Mitchell K, Herlyn M, Herlyn D, Fuhrer P. Colorectal carcinoma antigens detected by hybridoma antibodies. Somatic Cell Genet. 1979;5:957–71. doi: 10.1007/BF01542654. [DOI] [PubMed] [Google Scholar]
- 24.Kim Y, Kim HS, Cui ZY, Lee HS, Ahn JS, Park CK, Park K, Ahn MJ. Clinicopathological implications of EpCAM expression in adenocarcinoma of the lung. Anticancer Res. 2009;29:1817–22. [PubMed] [Google Scholar]
- 25.Frederick BA, Helfrich BA, Coldren CD, Zheng D, Chan D, Bunn PA, Jr, Raben D. Epithelial to mesenchymal transition predicts gefitinib resistance in cell lines of head and neck squamous cell carcinoma and non-small cell lung carcinoma. Mol Cancer Ther. 2007;6:1683–91. doi: 10.1158/1535-7163.MCT-07-0138. [DOI] [PubMed] [Google Scholar]
- 26.Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, Helfrich B, Dziadziuszko R, Chan DC, Sugita M, Chan Z, Baron A, Franklin W, Drabkin HA, Girard L, Gazdar AF, Minna JD, Bunn PA., Jr Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res. 2006;66:944–50. doi: 10.1158/0008-5472.CAN-05-1988. [DOI] [PubMed] [Google Scholar]
- 27.Seimetz D, Lindhofer H, Bokemeyer C. Development and approval of the trifunctional antibody catumaxomab (anti-EpCAMxanti-CD3) as a targeted cancer immunotherapy. Cancer Treat Rev. 2010 doi: 10.1016/j.ctrv.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–54. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 30.Buzza MS, Netzel-Arnett S, Shea-Donohue T, Zhao A, Lin CY, List K, Szabo R, Fasano A, Bugge TH, Antalis TM. Membrane-anchored serine protease matriptase regulates epithelial barrier formation and permeability in the intestine. Proc Natl Acad Sci U S A. 2010;107:4200–5. doi: 10.1073/pnas.0903923107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owen KA, Qiu D, Alves J, Schumacher AM, Kilpatrick LM, Li J, Harris JL, Ellis V. Pericellular activation of hepatocyte growth factor by the transmembrane serine proteases matriptase and hepsin, but not by the membrane-associated protease uPA. Biochem J. 2010;426:219–28. doi: 10.1042/BJ20091448. [DOI] [PubMed] [Google Scholar]
- 32.List K, Kosa P, Szabo R, Bey AL, Wang CB, Molinolo A, Bugge TH. Epithelial integrity is maintained by a matriptase-dependent proteolytic pathway. Am J Pathol. 2009;175:1453–63. doi: 10.2353/ajpath.2009.090240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.List K, Szabo R, Molinolo A, Sriuranpong V, Redeye V, Murdock T, Burke B, Nielsen BS, Gutkind JS, Bugge TH. Deregulated matriptase causes ras-independent multistage carcinogenesis and promotes ras-mediated malignant transformation. Genes Dev. 2005;19:1934–50. doi: 10.1101/gad.1300705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng H, Fukushima T, Takahashi N, Tanaka H, Kataoka H. Hepatocyte growth factor activator inhibitor type 1 regulates epithelial to mesenchymal transition through membrane-bound serine proteinases. Cancer Res. 2009;69:1828–35. doi: 10.1158/0008-5472.CAN-08-3728. [DOI] [PubMed] [Google Scholar]
- 35.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist P, Sommergruber W, Schweifer N, Wernitznig A, Beug H, Foisner R, Eger A. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–88. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mink SR, Vashistha S, Zhang W, Hodge A, Agus DB, Jain A. Cancer-associated fibroblasts derived from EGFR-TKI-resistant tumors reverse EGFR pathway inhibition by EGFR-TKIs. Mol Cancer Res. 2010;8:809–20. doi: 10.1158/1541-7786.MCR-09-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miura N, Yano T, Shoji F, Kawano D, Takenaka T, Ito K, Morodomi Y, Yoshino I, Maehara Y. Clinicopathological significance of Sip1-associated epithelial mesenchymal transition in non-small cell lung cancer progression. Anticancer Res. 2009;29:4099–106. [PubMed] [Google Scholar]
- 38.Roy BC, Kohno T, Iwakawa R, Moriguchi T, Kiyono T, Morishita K, Sanchez-Cespedes M, Akiyama T, Yokota J. Involvement of LKB1 in epithelial-mesenchymal transition (EMT) of human lung cancer cells. Lung Cancer. 2010 doi: 10.1016/j.lungcan.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C, Wu MC, Shimamura T, Perera SA, Liang MC, Cai D, Naumov GN, Bao L, Contreras CM, Li D, Chen L, Krishnamurthy J, Koivunen J, Chirieac LR, Padera RF, Bronson RT, Lindeman NI, Christiani DC, Lin X, Shapiro GI, Janne PA, Johnson BE, Meyerson M, Kwiatkowski DJ, Castrillon DH, Bardeesy N, Sharpless NE, Wong KK. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–10. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 40.Makowski L, Hayes DN. Role of LKB1 in lung cancer development. Br J Cancer. 2008;99:683–8. doi: 10.1038/sj.bjc.6604515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD, Rauscher FJ., 3rd The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198–207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez-Tillo E, Lazaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, Engel P, Postigo A. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29:3490–500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 43.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–95. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 44.Yori JL, Johnson E, Zhou G, Jain MK, Keri RA. Kruppel-like factor 4 inhibits epithelial-to-mesenchymal transition through regulation of E-cadherin gene expression. J Biol Chem. 2010;285:16854–63. doi: 10.1074/jbc.M110.114546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choudhary R, Li H, Winn RA, Sorenson AL, Weiser-Evans MC, Nemenoff RA. Peroxisome proliferator-activated receptor-gamma inhibits transformed growth of non-small cell lung cancer cells through selective suppression of Snail. Neoplasia. 2010;12:224–34. doi: 10.1593/neo.91638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernando RI, Litzinger M, Trono P, Hamilton DH, Schlom J, Palena C. The T-box transcription factor Brachyury promotes epithelial-mesenchymal transition in human tumor cells. J Clin Invest. 2010;120:533–44. doi: 10.1172/JCI38379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aokage K, Ishii G, Ohtaki Y, Yamaguchi Y, Hishida T, Yoshida J, Nishimura M, Nagai K, Ochiai A. Dynamic molecular changes associated with epithelial-mesenchymal transition and subsequent mesenchymal-epithelial transition in the early phase of metastatic tumor formation. Int J Cancer. 2010 doi: 10.1002/ijc.25500. ahead of publication. [DOI] [PubMed] [Google Scholar]
- 48.Helfrich BA, Raben D, Varella-Garcia M, Gustafson D, Chan DC, Bemis L, Coldren C, Baron A, Zeng C, Franklin WA, Hirsch FR, Gazdar A, Minna J, Bunn PA., Jr Antitumor activity of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor gefitinib (ZD1839, Iressa) in non-small cell lung cancer cell lines correlates with gene copy number and EGFR mutations but not EGFR protein levels. Clin Cancer Res. 2006;12:7117–25. doi: 10.1158/1078-0432.CCR-06-0760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Gene repression with low-level of induced ZEB1 in H358 cells. (A) ZEB1-inducible H358 cells at 50% confluency were treated with low doses of doxycycline (0 to 5 ng/ml, as indicated) for 12 and 24 h and harvested. Protein lysates were analyzed by Western blot for induced ZEB1 (anti-myc antibodies). β-actin provided a loading control. (B) RNA isolated from the same samples was analyzed by quantitative real-time RT-PCR for the indicated genes. Values were calculated as a percent of GAPDH and normalized to the untreated controls. Data are reported as the fold-change compared to cultures treated with vehicle alone.
Figure S2. Validation of ZEB1 nuclear staining on tumor xenografts. Control (H358 – ctl; left panel) or ZEB1-inducible H358 cells (H358 – ZEB1; right panel) were injected subcutaneously in NCrnu/nu mice. ZEB1 was induced by treating animals with doxycycline (1mg/ml) on a daily basis. Tumors were then excised, fixed and sectioned for immunohistochemical detection of ZEB1 using conditions identical to those used in Figure 6. Control ZEB1 tumors have very low ZEB1 levels and the faint cytoplasmic staining represents background. Animals were handled in accordance with established protocols approved by the institutional animal care committee.
Table S1. DNA primer sequences for quantitative real-time RT-PCR
Table S2. Spearman R values from Affymetrix data for all significant negative correlates with ZEB1 in 38 NSCLC cell lines.
Table S3. Spearman R values from Affymetrix data for all significant positive correlates with ZEB1 in 38 NSCLC cell lines.
Table S4. Spearman R values for 45 lung cancer biomarkers versus EMT factors from Affymetrix data of 38 NSCLC cell lines [1–8].