Abstract
Molecular imaging is a newly emerged multiple disciplinary field that aims to visualize, characterize and quantitatively measure biological processes at cellular and molecular levels in humans and other living systems. A reporter gene is a piece of DNA encoding reporter protein, which presents as a readily measurable phenotype that can be distinguished easily from the background of endogenous protein. After being transferred into cells of organ systems (transgenes), the reporter gene can be utilized to visualize transcriptional and posttranscriptional regulation of gene expression, protein-protein interactions, or trafficking of proteins or cells in living subjects. Herein, we review previous classification of reporter genes and regroup the reporter gene based imaging as basic, inducible and activatable, based on the regulation of reporter gene transcription and post-translational modification of reporter proteins. We then focus on activatable reporters, in which the signal can be activated at the posttranslational level for visualizing protein-protein interactions, protein phosphorylation or tertiary structure changes. The applications of several types of activatable reporters will also be summarized. We conclude that activatable reporter imaging can benefit both basic biomedical research and drug development.
Keywords: Molecular imaging, reporter gene, activatable, optical imaging, PET
Introduction
Recently, molecular imaging has been evolved to be an important tool for both preclinical research and clinical applications. By converging multiple disciplinary subjects, the new imaging paradigm aims to visualize, characterize and measure quantitatively biological processes at the cellular and molecular levels in humans and other living systems 1. Advanced multiple molecular imaging modalities are of great value in a wide array of biomedical applications ranging from reflecting specific cellular and molecular processes including gene expression or protein-protein interactions to optimizing drug and gene therapy 2.
The reporter genes form the basis of reporter gene expression imaging in living subjects, a subfield of molecular imaging 3. A reporter gene is a piece of DNA encoding reporter protein molecules, which represents a readily measurable phenotype that can be distinguished easily from the background of endogenous proteins 3, 4. Different from the methods including tissue biopsy, sampling and in situ hybridization, reporter gene imaging provides a noninvasive way to evaluate the location, magnitude, and extent of gene expression in a living subject 5. After being externally transferred into cells of organ systems (transgenes) or endogenous genes, molecular imaging with reporter genes have been utilized to visualize transcriptional and posttranscriptional regulation of gene expression, protein-protein interactions, or trafficking of proteins or cells in living subjects 3.
Classification of reporter genes
Intracellular vs. cell membrane reporter genes
In fact, long before the concept of molecular imaging was coined, reporter genes have been invented and adopted by scientists to follow transgenic expression either macroscopically or microscopically 6, 7. As part of the evolving concept of molecular medicine, molecular imaging technologies have been developed to examine the integrative functions of molecules, cells, organ systems, and whole organisms 8. Consequently, reporter genes have been involved with many different imaging technologies as an important subfield of molecular imaging. There are several ways to categorize reporter genes. A broad classification of the reporter genes could be made based on the cellular localization of the gene products, either being intracellular or associated with the cell membrane 9. Examples of the former include green fluorescent protein (GFP), luciferases, cytosine deaminase, and thymidine kinase. Examples of reporter proteins on or in the cell surface include the receptors for somatostatin or transferrin and the sodium iodide symporter 9, 10.
Imaging modality specific reporter genes
An application-oriented classification strategy is based on the different imaging modalities used for visualization of a particular reporter gene. Almost every imaging modality used for molecular imaging has the corresponding reporter genes available. Positron emission tomography (PET) is the most sensitive and specific technique for imaging molecular pathways in vivo in humans 11. So far, the most popular PET reporter genes are herpes simplex virus type 1 thymidine kinase (HSV-tk) 12, 13 and human or rat sodium iodide symporter (hNIS or rNIS) 14, 15.
Non-radionuclide-based imaging techniques include magnetic resonance imaging (MRI), ultrasound and optical imaging. MRI is a noninvasive diagnostic technique based on the interaction of protons (or other nuclei) with each other and with surrounding molecules in a tissue of interest 16. Different tissues have different relaxation times which can result in endogenous contrast. The major advantage of MRI over radionuclide-based imaging is the absence of radiation and higher spatial resolution (usually sub-millimeter level). The major disadvantage of MRI is its inherent low sensitivity, which makes it very difficult for reporter gene MRI to get decent target/background signal ratio in vivo 17. Native ferritin has been used as MRI reporter gene since it can concentrate the body's natural iron to the amount detectable by MRI 18, 19. There is no reporter gene suitable for ultrasound imaging so far.
Two main branches of optical imaging are fluorescence imaging and bioluminescence imaging. In fluorescence imaging, excitation light illuminates the subject, and a charge-coupled device (CCD) camera collects the emission light at a shifted wavelength 2. A series of fluorescent proteins, such as the green fluorescent protein, have also enabled sophisticated studies of protein function and wide-ranging processes from gene expression to second-messenger cascades and intercellular signaling, typically through fusion protein approach rather than through direct labeling 20. The major limitation of optical imaging is tissue light scattering and absorption that affect both image resolution and depth of light penetration of tissues 21. Bioluminescence imaging (BLI) is based on the expression of luciferases in target cells and tissues 22. In the presence of its substrate (such as D-luciferin), an energy-dependent reaction releases photons that can be detected using sensitive detection systems. BLI has been used to study gene-expression patterns 22, to measure gene transfer efficiency 23, to monitor tumor growth and response to therapy 24, to investigate protein-protein interactions in vivo 25, 26, and to determine the location and proliferation of stem cells 27. The most commonly used bioluminescence reporter gene for research purposes has been the luciferase from the North American firefly (Photinus Pyralis, Fluc) 28.
Basic, inducible and activatable reporter imaging
Based on the regulation of reporter gene transcription and post-translational modification of the proteins encoded by the different reporter genes, we categorized the reporter systems into three subgroups: basic, inducible and activatable. Most commonly used reporter gene system consists of a constitutive promoter and a coding sequence for a reporter protein. For example, Fluc expressing vector driven by CMV promoter has been intensively used as a reporter system for tumor growth monitoring, especially for orthotropic tumor models 29. Vectors encoding HSV-tk has been used to transduce T cells for in vivo cell trafficking study by SPECT and PET 30. The proportional correlation between the imaging signal and the constant expression of the reporter proteins guarantees accurate quantification of the images.
With an inducible reporter gene system, the expression of the reporter genes is regulated or manipulated at the transcriptional or post-transcriptional level. The most common strategy is to introduce specific promoter/enhancer elements before the reporter gene. Thus the transcription and expression of the reporter gene is regulated by corresponding transcription factors. One example is HIF-1, which involves one of the most intensively studied oxygen response pathways in molecular biology. Under hypoxic conditions, accumulated HIF-1 binds to the hypoxia responsive element (HRE) and triggers the recruitment of co-activators essential for transcriptional activation 31. Several elegant methods have been developed to directly measure HIF-1 activity by the introduction of transgenes with HREs as promoter sequences coupled to reporter genes such as luciferase 32, 33 or GFP 34. Another example is that heat-shock protein promoters, particularly HSP70 promoters, have been commonly used for gene therapy strategies because they are both heat-inducible and efficient 35. GFP has been utilized extensively as the reporter gene to evaluate the spatial and temporal control of gene expression driven by HSP70 promoters 36-39. With PET imaging, spontaneous in vivo activation of the HSV1-tk suicide gene driven by the Grp78 promoter, a member of HSP70 family, in growing tumors and its activation by photodynamic therapy (PDT) in a controlled manner has been observed 40. Noguchi et al. 41 have established a dual-color transgenic mouse that carries transgenes encoding green-emitting luciferase (ELuc, λmax = 538 nm) expressed under the control of the Bmal1 promoter and red-emitting luciferase (SLR2, λmax=630 nm) expressed under the control of the Per2 promoter. Consequently, the antiphasic oscillations of Bmal1 and Per2 expression were able to be monitored simultaneously in a single tissue of the dual-color luciferase mouse by splitting their emissions.
MicroRNAs (miRNAs) are approximately 21-nucleotide (nt) single-stranded, small noncoding RNAs acting as post-transcriptional regulators of gene expression in animals and plants 42. To non-invasively image and quantify endogenous miRNAs in vivo, Kim et al. 43 developed a Gaussia princeps luciferase (Gluc) imaging system in which three perfectly matched complementary sequences of miR-221 was fused immediately after the stop codon of Gluc reporter vector. Then the transcript of this vector was expected to be bound by endogenous miR-221, thus the transcript of Gluc would be degraded. Consequently, the translation of Gluc would be suppressed, resulting in a decrease of bioluminescent signal. This study provides an example of reporter genes which can be induced at the post-transcriptional level.
The ability to noninvasively image protein-protein interactions has important implications for a wide variety of biological research endeavors, including drug discovery and molecular medicine 44. There are three popular strategies to visualize protein-protein interaction by molecular imaging including modified mammalian two-hybrid system, bioluminescence resonance energy transfer (BRET) technology and split reporter genes. The two-hybrid system is fairly simple, rapid and the strategies developed should be generally applicable for many protein partners, as long as the interaction complex can lead to transactivation of the reporters of choice. The main limitation is that it can only be used to detect interacting proteins in the nucleus 44. Through non-radioactive transfer of energy between donor and acceptor molecules by the Förster mechanism, both FRET and BRET are capable of studying protein-protein interactions. However, in vivo application is limited because of the high requirement for separating emissions between the donor and the acceptor 26.
Activatable reporter gene imaging
With the advance of molecular imaging techniques, more sophisticated strategies have been adapted for the design of reporter gene system to broaden their biomedical applications. One category of reporter genes can be activated at the post-translational level with protein-protein interaction, enzymatic reaction, phosphorylation or tertiary structure changes 45. In order to distinguish these reporter systems from those inducible reporter gene imaging, we named them here as “activatable reporter gene imaging”. Most of split reporter genes designed to image protein-protein interactions fall into this category.
Imaging protein-protein interaction
The principle mechanism of split reporter gene lies in that splitting a specific reporter protein into two distinct fragments abolishes its function and bringing the two fragments back together in a controlled manner restores the functional activity 46. To date, several reporter proteins (e.g. β-lactamase, β-galactosidase, ubiquitin, dihydrofolate reductase, luciferases and GFP) have been adapted for split protein strategies by finding various split sites for each reporter protein 47-50. For in vivo imaging, luciferase is the most popular reporter protein to be split apart. Firefly luciferase has been cleaved into two fragments as Nfluc (residues 1-437) and Cfluc (residues 438-550) and fused to Id and MyoD respectively as test proteins 48.
The rapamycin-induced interaction between FKBP12 (FK506 binding protein) and FRB (FKBP-Rapamycin binding domain) can be considered a textbook example of a re-usable protein interaction device 51. The crystal structure of the ternary complex of FKBP12 and FRB revealed extensive interactions between rapamycin and both proteins, but fewer interactions between the proteins 52. With alternative complementary N- and C-terminal fragments of Fluc, Luker et al. 53 monitored rapamycin-mediated interaction of rapamycin binding proteins FRB and FKBP12 using two fragments of Fluc (residues 2-416 and 398-550).
Hida et al. 54 fused fragments from firefly luciferase, click beetle luciferase in green and click beetle luciferase in red with FRB and FKBP12. By mutation of the C-terminal fragment from the click beetle luciferase, they developed an imaging method for real-time analysis of protein-protein interactions using multicolor luciferases with different spectral characteristics. They applied this method to visualize protein-protein interactions of Smad proteins and demonstrated Smad1-Smad4 and Smad2-Smad4 interactions in early developing stages of a single living Xenopus laevis embryo. This technique supports quantitative analysis and imaging of versatile protein-protein interactions with a selective luminescence wavelength in opaque or strongly autofluorescent living subjects.
Mammalian target of rapamycin (mTOR) plays critical roles in regulating protein synthesis and growth, ribosomal protein synthesis, cell cycle, autophagy and apoptosis of eukaryotic cells 55. As a potent immunosuppressant, Rapamycin was approved by the Food and Drug Administration (FDA) in 1999 for use in kidney transplant recipients 56. Later on, rapamycin has been found to be a potential anticancer drug, since it can act as a cytostatic, arresting cells in G1 phase or potentially inducing apoptosis, in many malignant cells in culture 57. Rluc fragmentation generated by splitting between residues 229 and 230 has been used to study rapamycin-induced interaction of the human proteins FRB and FKBP12 58. In the range from 0.2 nM to 10 nM, bioluminescence signal intensity increased along with the rapamycin concentration, implying that the reporter gene system could be applied to monitor drug delivery.
Imaging protein trafficking
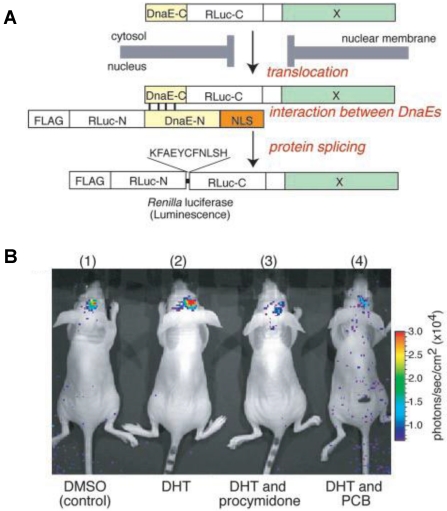
Nucleocytoplasmic trafficking of functional proteins plays a key role in regulating gene expressions in response to extracellular signals 59. Kim et al. 60 developed an activatable BLI reporter system for monitoring and imaging the nuclear trafficking of target proteins in vitro and in vivo. The assay is based on the reconstitution of split fragments of Renilla reniformis luciferase (Rluc) by protein splicing with a DnaE intein. A target cytosolic protein fused to the N-terminal half of Rluc is expressed in mammalian cells. If the protein translocates into the nucleus, the Rluc moiety meets the C-terminal half of Rluc and the full-length Rluc is reconstituted by protein splicing (Figure 1). The authors demonstrated quantitative cell-based in vitro sensing and in vivo imaging of ligand induced translocation of the androgen receptor, which allowed high-throughput screening of exogenous and endogenous agonists and antagonists. Furthermore, the reporter gene enabled noninvasive in vivo imaging of the androgen receptor translocation in the brains of living mice, justifying the potential of this strategy for drug screening.
Figure 1.
Basic strategy for the detection of AR translocation. A. Principle for monitoring translocation of a particular protein (X) into the nucleus using protein splicing of split-Rluc. RLuc-N (1-229 aa) is connected with DnaE-N (1-123 aa) and NLS [(DPKKKRKV)3], which is predominantly localized in the nucleus. DnaE-C (1-36 aa) is connected with RLuc-C (230-311 aa) and a protein X, which is localized in the cytosol. When the tandem fusion protein consisting of DnaE-C, Rluc-C, and protein X translocates into the nucleus, the DnaE-C interacts with DnaE-N, and protein splicing results. Rluc-N and -C are linked by a peptide bond, and the reconstituted Rluc recovers its bioluminescence activity. FLAG means epitope (DYKDDDDK). B. The inhibitory effect of chemicals on AR translocation into the nucleus in the mouse brain. The COS-7 cells transiently cotransfected with pcRDn-NLS and pcDRc-AR were implanted in the forebrain of the nude mice at a depth of 3 mm through a 1-mm burrhole. Of mouse groups 1-4, groups 1 and 2 were stimulated with1%DMSO, whereas groups 3 and 4 were stimulated with procymidone (10 mg/kg body weight) and PCB (10 mg/kg of body weight), respectively. Two hours after the stimulation, mouse groups 2-4 were then stimulated with DHT (10µg/kg of body weight). Two hours after DHT stimulation, the mice were imaged in 2-min intervals until reaching the maximum photon counts after intercerebral injection of coelenterazine (1.4 mg/kg of body weight). Reproduced with permission from ref. 60.
Imaging physiopathological pathways
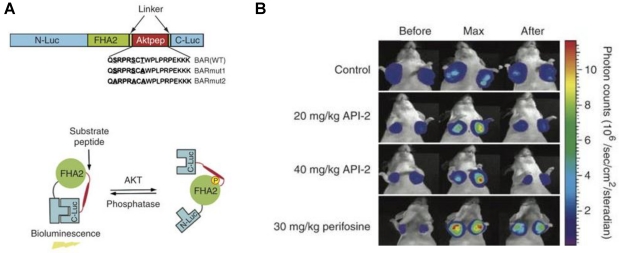
One advantage of activatable reporter genes lies in that the design is complicated enough to be used for visualization and quantification of particular physiological or pathological pathways directly. One example is the bioluminescent Akt reporter for Akt activity imaging 61. The serine/threonine kinase Akt functions as a signaling hub where many upstream signaling pathways converge 62. Because Akt and its upstream regulators are dysregulated in some forms of cancer, they are promising targets for pharmaceutical intervention 63, 64. Zhang et al. 61 have constructed a bioluminescent Akt reporter that contains an Akt consensus substrate peptide (Akt-pep) and a domain that binds phosphorylated amino acid residues (FHA2) flanked by the N-terminal (N-Luc) and C-terminal (C-Luc) domains of the firefly luciferase reporter molecule. The reporter has minimal bioluminescence activity when Akt-dependent phosphorylation of the Akt-pep domain results in its interaction with the FHA2 domain. In the absence of Akt activity, the N-Luc and C-Luc domains reassociate, restoring bioluminescence activity (Figure 2). With this reporter construction, Akt activity in cultured cells and tumor xenografts can be monitored quantitatively and dynamically in response to activation or inhibition of receptor tyrosine kinase, inhibition of phosphoinositide 3-kinase, or direct inhibition of Akt.
Figure 2.
Biolumienscent Akt reporter (BAR). A. The domain structure (upper) and the proposed mechanism of action (lower) of the BAR reporter. It involves Akt-dependent phosphorylation of the Akt-pep domain (thick line), which results in its interaction with the FHA2 domain (right). In this form, the reporter has minimal bioluminescence activity. In the absence of Akt activity, the N-Luc and C-Luc domains reassociate, restoring bioluminescence activity (left). B. Bioluminescence imaging of Akt kinase activity. Bioluminescence activity before treatment (time 0) and in response to treatment with API-2 (20 mg/kg or 40 mg/kg) was monitored at various times. Reproduced with permission from ref. 61.
Cyclic adenosine monophosphate (cAMP) is a key second messenger of signal transduction in mammalian cells and cAMP levels have been shown to rise and fall rapdily within the cell following receptor activation and response desensitization 65. A cAMP biosensor has been developed by introducing a cAMP-binding domain B from protein kinase A regulatory subunit type IIβ (RIIβB) into a circularly permuted a circularly permuted form of Photinus pyralis luciferase 66. An evolved version of this biosensor construct was able to accurately track both the magnitude and kinetics of cAMP change in living cell environment 67.
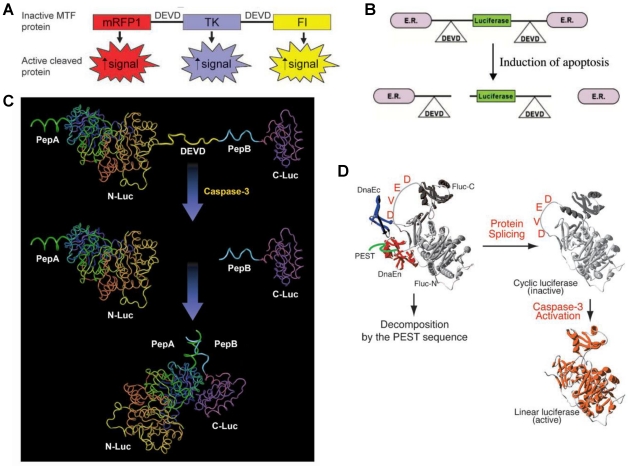
Apoptosis, or programmed cell death, is an active regulatory mechanism functionally opposite but complementary to proliferation. As a tightly regulated multi-step pathway, apoptosis plays a crucial role in many biological processes and various diseases, which are characterized by relative excess of cell death 68. As the cell undergoes apoptosis, there are a number of potential steps in the process that could be imaged using very different imaging modalities. Caspases play a central role in the execution of cell death. Both the intrinsic (mitochondrial) and extrinsic (death receptor) pathways of apoptosis eventually activate several effector caspases 69. Among them, Caspase-3 is one of the key effector caspases which recognizes and cleaves DEVD (aspartic acid-glutamic acid-valine-aspartic acid) peptide sequence that exists in many cellular proteins such as poly(ADPribose) polymerase and lamins 70. Several activatable reporter genes have been reported to image caspase-3 activity in vivo (Figure 3). Laxman et al. 71 constructed a reporter gene vector containing firefly luciferase gene flanked by the ER (residues 281-599 of the modified mouse estrogen receptor sequence) with the DEVD linker. In cells undergoing apoptosis, a caspase-3-specific cleavage of the recombinant product resulted in the restoration of luciferase activity that can be detected in living animals with BLI.
Figure 3.
Activatable reporter gene systems for imaging of caspase 3 activities. A. Activation of caspase-3 cleaves the MTF fusion protein into the three reporter protein components. Upon cleavage, the reporter proteins (mRFP1, FL, andTK) with attenuated activity in fused form (MTF) gain significantly higher activity and can be used for noninvasive imaging of apoptosis. Reproduced with permission from ref. 73. B. Chimeric polypeptides consisting of a reporter molecule fused to the ER resulted in silencing of the reporter activity. Inclusion of a protease cleavage site between these domains provided for protease-mediated activation of the reporter molecule after separation of the silencing domains (i.e., ER). Reproduced with permission from ref. 71. C. The ANLucBCLuc apoptosis imaging reporter constitutes the split luciferase (NLuc and CLuc) domains fused to interacting peptides, peptide A and peptide B, with an intervening caspase-3 cleavage motif. Upon induction of apoptosis, the reporter molecule is proteolytically cleaved by caspase-3 at the DEVD motif. This cleavage enables interaction between pepANLuc and pepBCLuc, thus reconstituting luciferase activity. Reproduced with permission from ref. 61 72. D. For an efficient protein splicing reaction, the C- and N-terminal fragments of DnaE (DnaEc and DnaEn, respectively) are connected with the N- and C-terminal ends of the circularly permuted luciferase. In addition, a PEST sequence was attached to the C-terminal end of the fusion construct to accelerate degradation of a protein. The PEST sequence results in the degradation of only unspliced products because cyclic Fluc does not possess the PEST sequence. Consequently, only cyclic Fluc accumulates inside the cells. If caspase-3 is activated in cells expressing the cyclic Fluc, the Fluc changes into an active form and its luminescence activity is restored. Thus, cells expressing the cyclic Fluc allow monitoring of caspase-3 activity with luminescence signals. Reproduced with permission from ref. 74
Another reporter, ANLucBCLuc , has been designed to image caspase-3 activity in living cells and animals 72. The reporter constituted a fusion of small interacting peptides, peptide A and peptide B, with the NLuc and CLuc fragments of luciferase with a caspase-3 cleavage site (DEVD) between pepANLuc (ANLuc) and pepBCLuc (BCLuc). During apoptosis, caspase-3 cleaves the reporter, enabling the separation of ANLuc from BCLuc. A high-affinity interaction between peptide A and peptide B restores luciferase activity by NLuc and CLuc complementation. Treatment of live cells and mice carrying D54 tumor xenografts with chemotherapeutic agents and radiation resulted in increased bioluminescence activity due to enhanced apoptosis 72. Ray et al. applied a multimodality reporter vector to monitor caspase-3 activation indirectly in live cells and tumors of living animals undergoing apoptosis. In their study, a fusion protein (MTF) was constructed by combining three different reporter proteins-- red fluorescent protein (mRFP1), firefly luciferase (FL), and HSV1-sr39 truncated thymidine kinase (TK)--linked through a caspase-3 recognizable polypeptide linker. Upon apoptosis induction with 8 µM staurosporine, the fusion protein showed significant increases in FL and mRFP1 activity in 293T cells 73. A DEVD containing cyclic luciferase to detect caspases activation has also been reported 74, 75. In these studies, two fragments of DnaE intein were fused to neighboring ends of firefly luciferase connected with a DEVD sequence. After translation into a single polypeptide in living cells, the amino (N) and carboxy (C) terminals of the luciferase are ligated by protein splicing, which resulted in a closed circular polypeptide chain. When the substrate sequence was digested by caspases, the luciferase changed into an active form and restored its activity. Doxil induced apoptosis has been successfully visualized by BLI non-invasively 76.
PET split reporter genes
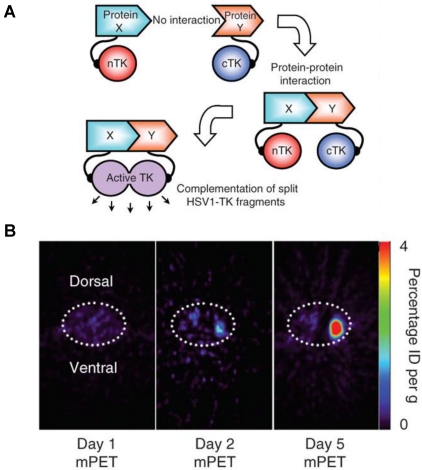
A major disadvantage of optical imaging is the low tissue penetration of light. Owing to absorption and scattering, light in the visible spectrum emitted by fluorescence has only a limited tissue penetration of several hundred micrometers. This problem restricts the application of most optical methods to small animals or to the study of surface structures in humans 77. PET reporter genes have the potential to provide a sensitive and more accurate means of fully quantitative in vivo imaging and precise tomographic localization of protein-protein interactions (PPIs) deeper in the body than optical bioluminescence imaging using split luciferase, since BLI is relatively surface weighted and semiquantitative in nature 78. The HSV-tk enzyme phosphorylates a wide range of nucleoside analogs. Consequently, radiolabeled reporter probes such as 123I-FIAU and 18F-FHBG are transported into cells and are trapped as a result of phosphorylation by HSV-tk to facilitate PET or SPECT imaging 79. Recently, Massoud et al. 78 indentified a split point of the TK enzyme between Thr265 and Ala266 after a partial circular permutation screen. They successfully engineered the first PET based split reporter system to be used for protein fragment complementation assay (PCA) (Figure 4).
Figure 4.
Split thymidine kinase reporter gene for imaging of protein-protein interaction. A. Simplified schematic diagram representing the forward or 'folding' mechanism underlying a proteinfragment complementation assay (PCA). B. Transaxial tomographic microPET images through a representative prone-positioned mouse implanted subcutaneously over the left shoulder with mock-transfected 293T cells, and over the right shoulder with 293T cells stably expressing both nTK(V119C)-FRB and FKBP12-cTK. The mouse was injected with 200 μCi of [18F]-FHBG before imaging on days 1, 2 and 5 into the imaging protocol (that is, after 7 d of initial xenograft growth). Elliptical dotted white line outlines the surface of the mouse's upper thorax. Color intensity is a reflection on probe accumulation after its phosphorylation by the complemented thymidine kinase enzyme. Reproduced with permission from ref. 78.
Design of activatable reporter gene imaging
With regard to protein engineering to design an activatable reporter gene, two main issues need to be considered. One is how to decide the cleave site to make both fragments have no residual activity. Another is how to fully restore the enzyme function after fragment complementation. A detailed knowledge of protein structure, function and mechanism would be of great help in reporter gene engineering 80. For example, the crystal structure of luciferase shows two essentially independent folding domains, the N-terminal domain consisting of residues 1-436 and a C-terminal domain consisting of residues 440-550, connected by a disordered flexible region 81. Constitutive activity of the N terminus fragment has been observed when firefly luciferase was split between amino acid positions 437 and 438 48. Further study revealed that fragmentation of NLuc (2-416) and CLuc (398-550) deleted key structural and active site regions from the N-terminal fragment and, in effect, transferred these regions to the C-terminal fragment. Upon interaction, the CLuc fragment would complement the missing subdomains in the NLuc fragment 53. Compared with the intact enzyme, the interacted fragments or restored protein showed significantly lower activity 48, 53, 74, indicating a “waste” during post-translational activation. Thus, further refinement of the activatable reporter genes is still in need to optimize the function for in vivo applications.
Conclusion
It is foreseeable that activatable reporter gene molecular imaging will play very important roles in studies related to protein-protein interactions and various physiological and pathological pathways. The visual representation, characterization, quantification, and timing of these biological processes in living subjects could create unprecedented opportunities to complement available in vitro or cell culture methodologies 82. Particularly, imaging with activatable reporters may provide a robust platform which would accelerate the development of new drugs that promote or inhibit active protein assembly or target to a particular pathway.
Acknowledgments
The work was supported by the Intramural Research Program of the National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health.
References
- 1.Mankoff DA. A definition of molecular imaging. J Nucl Med. 2007;48:18N. 21N. [PubMed] [Google Scholar]
- 2.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–80. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 3.Massoud TF, Singh A, Gambhir SS. Noninvasive molecular neuroimaging using reporter genes: part I, principles revisited. AJNR Am J Neuroradiol. 2008;29:229–34. doi: 10.3174/ajnr.A0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alam J, Cook JL. Reporter genes: application to the study of mammalian gene transcription. Anal Biochem. 1990;188:245–54. doi: 10.1016/0003-2697(90)90601-5. [DOI] [PubMed] [Google Scholar]
- 5.Gambhir SS, Barrio JR, Herschman HR, Phelps ME. Assays for noninvasive imaging of reporter gene expression. Nucl Med Biol. 1999;26:481–90. doi: 10.1016/s0969-8051(99)00021-9. [DOI] [PubMed] [Google Scholar]
- 6.Cheng SM, Lee SG, Kalyan NK, McCloud S, Levner M, Hung PP. Tissue plasminogen activator (tPA) as a reporter gene in transient gene expression. Gene. 1987;58:299–303. doi: 10.1016/0378-1119(87)90385-4. [DOI] [PubMed] [Google Scholar]
- 7.Ow DW, Jacobs JD, Howell SH. Functional regions of the cauliflower mosaic virus 35S RNA promoter determined by use of the firefly luciferase gene as a reporter of promoter activity. Proc Natl Acad Sci U S A. 1987;84:4870–4. doi: 10.1073/pnas.84.14.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phelps ME. Positron emission tomography provides molecular imaging of biological processes. Proc Natl Acad Sci U S A. 2000;97:9226–33. doi: 10.1073/pnas.97.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross S, Piwnica-Worms D. Spying on cancer: molecular imaging in vivo with genetically encoded reporters. Cancer Cell. 2005;7:5–15. doi: 10.1016/j.ccr.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Niu G, Gaut AW, Ponto LL, Hichwa RD, Madsen MT, Graham MM. et al. Multimodality noninvasive imaging of gene transfer using the human sodium iodide symporter. J Nucl Med. 2004;45:445–9. [PubMed] [Google Scholar]
- 11.Jones T. The imaging science of positron emission tomography. Eur J Nucl Med. 1996;23:807–13. doi: 10.1007/BF00843711. [DOI] [PubMed] [Google Scholar]
- 12.Yaghoubi SS, Barrio JR, Namavari M, Satyamurthy N, Phelps ME, Herschman HR. et al. Imaging progress of herpes simplex virus type 1 thymidine kinase suicide gene therapy in living subjects with positron emission tomography. Cancer Gene Ther. 2005;12:329–39. doi: 10.1038/sj.cgt.7700795. [DOI] [PubMed] [Google Scholar]
- 13.Liang Q, Nguyen K, Satyamurthy N, Barrio JR, Phelps ME, Gambhir SS. et al. Monitoring adenoviral DNA delivery, using a mutant herpes simplex virus type 1 thymidine kinase gene as a PET reporter gene. Gene Ther. 2002;9:1659–66. doi: 10.1038/sj.gt.3301899. [DOI] [PubMed] [Google Scholar]
- 14.Niu G, Krager KJ, Graham MM, Hichwa RD, Domann FE. Noninvasive radiological imaging of pulmonary gene transfer and expression using the human sodium iodide symporter. Eur J Nucl Med Mol Imaging. 2005;32:534–40. doi: 10.1007/s00259-004-1715-6. [DOI] [PubMed] [Google Scholar]
- 15.Ahn BC. Sodium Iodide Symporter for Nuclear Molecular Imaging and Gene Therapy: From Bedside to Bench and Back. Theranostics. 2012;2(4):392–402. doi: 10.7150/thno.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pathak AP, Gimi B, Glunde K, Ackerstaff E, Artemov D, Bhujwalla ZM. Molecular and functional imaging of cancer: Advances in MRI and MRS. Methods Enzymol. 2004;386:3–60. doi: 10.1016/S0076-6879(04)86001-4. [DOI] [PubMed] [Google Scholar]
- 17.Lee S, Biswal S, Lee S. Magnetic Resonance Reporter Gene Imaging. Theranostics. 2012;2:403–12. doi: 10.7150/thno.3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genove G, DeMarco U, Xu H, Goins WF, Ahrens ET. A new transgene reporter for in vivo magnetic resonance imaging. Nat Med. 2005;11:450–4. doi: 10.1038/nm1208. [DOI] [PubMed] [Google Scholar]
- 19.Cohen B, Dafni H, Meir G, Harmelin A, Neeman M. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005;7:109–17. doi: 10.1593/neo.04436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Roessel P, Brand AH. Imaging into the future: visualizing gene expression and protein interactions with fluorescent proteins. Nat Cell Biol. 2002;4:E15–20. doi: 10.1038/ncb0102-e15. [DOI] [PubMed] [Google Scholar]
- 21.Wunderbaldinger P, Bogdanov A, Weissleder R. New approaches for imaging in gene therapy. Eur J Radiol. 2000;34:156–65. doi: 10.1016/s0720-048x(00)00196-0. [DOI] [PubMed] [Google Scholar]
- 22.Beckham JT, Mackanos MA, Crooke C, Takahashi T, O'Connell-Rodwell C, Contag CH. et al. Assessment of cellular response to thermal laser injury through bioluminescence imaging of heat shock protein 70. Photochem Photobiol. 2004;79:76–85. [PubMed] [Google Scholar]
- 23.Lipshutz GS, Flebbe-Rehwaldt L, Gaensler KM. Reexpression following readministration of an adenoviral vector in adult mice after initial in utero adenoviral administration. Mol Ther. 2000;2:374–80. doi: 10.1006/mthe.2000.0136. [DOI] [PubMed] [Google Scholar]
- 24.Szwaya J, Bruseo C, Nakuci E, McSweeney D, Xiang X, Senator D. et al. A novel platform for accelerated pharmacodynamic profiling for lead optimization of anticancer drug candidates. J Biomol Screen. 2007;12:159–66. doi: 10.1177/1087057106297787. [DOI] [PubMed] [Google Scholar]
- 25.Chan CT, Paulmurugan R, Gheysens OS, Kim J, Chiosis G, Gambhir SS. Molecular imaging of the efficacy of heat shock protein 90 inhibitors in living subjects. Cancer Res. 2008;68:216–26. doi: 10.1158/0008-5472.CAN-07-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De A, Gambhir SS. Noninvasive imaging of protein-protein interactions from live cells and living subjects using bioluminescence resonance energy transfer. FASEB J. 2005;19:2017–9. doi: 10.1096/fj.05-4628fje. [DOI] [PubMed] [Google Scholar]
- 27.Cao X, Jia G, Zhang T, Yang M, Wang B, Wassenaar PA. et al. Non-invasive MRI tumor imaging and synergistic anticancer effect of HSP90 inhibitor and glycolysis inhibitor in RIP1-Tag2 transgenic pancreatic tumor model. Cancer Chemother Pharmacol. 2008;62:985–94. doi: 10.1007/s00280-008-0688-8. [DOI] [PubMed] [Google Scholar]
- 28.Hastings JW. Chemistries and colors of bioluminescent reactions: a review. Gene. 1996;173:5–11. doi: 10.1016/0378-1119(95)00676-1. [DOI] [PubMed] [Google Scholar]
- 29.Hsu AR, Cai W, Veeravagu A, Mohamedali KA, Chen K, Kim S. et al. Multimodality molecular imaging of glioblastoma growth inhibition with vasculature-targeting fusion toxin VEGF121/rGel. J Nucl Med. 2007;48:445–54. [PubMed] [Google Scholar]
- 30.Koehne G, Doubrovin M, Doubrovina E, Zanzonico P, Gallardo HF, Ivanova A. et al. Serial in vivo imaging of the targeted migration of human HSV-TK-transduced antigen-specific lymphocytes. Nat Biotechnol. 2003;21:405–13. doi: 10.1038/nbt805. [DOI] [PubMed] [Google Scholar]
- 31.Sun X, Niu G, Chan N, Shen B, Chen X. Tumor hypoxia imaging. Mol Imaging Biol. 2011;13:399–410. doi: 10.1007/s11307-010-0420-z. [DOI] [PubMed] [Google Scholar]
- 32.Shibata T, Giaccia AJ, Brown JM. Development of a hypoxia-responsive vector for tumor-specific gene therapy. Gene Ther. 2000;7:493–8. doi: 10.1038/sj.gt.3301124. [DOI] [PubMed] [Google Scholar]
- 33.Payen E, Bettan M, Henri A, Tomkiewitcz E, Houque A, Kuzniak I. et al. Oxygen tension and a pharmacological switch in the regulation of transgene expression for gene therapy. J Gene Med. 2001;3:498–504. doi: 10.1002/jgm.217. [DOI] [PubMed] [Google Scholar]
- 34.Vordermark D, Shibata T, Brown JM. Green fluorescent protein is a suitable reporter of tumor hypoxia despite an oxygen requirement for chromophore formation. Neoplasia. 2001;3:527–34. doi: 10.1038/sj.neo.7900192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rome C, Couillaud F, Moonen CT. Spatial and temporal control of expression of therapeutic genes using heat shock protein promoters. Methods. 2005;35:188–98. doi: 10.1016/j.ymeth.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 36.Huang Q, Hu JK, Lohr F, Zhang L, Braun R, Lanzen J. et al. Heat-induced gene expression as a novel targeted cancer gene therapy strategy. Cancer Res. 2000;60:3435–9. [PubMed] [Google Scholar]
- 37.Vekris A, Maurange C, Moonen C, Mazurier F, De Verneuil H, Canioni P. et al. Control of transgene expression using local hyperthermia in combination with a heat-sensitive promoter. J Gene Med. 2000;2:89–96. doi: 10.1002/(SICI)1521-2254(200003/04)2:2<89::AID-JGM90>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 38.Borrelli MJ, Schoenherr DM, Wong A, Bernock LJ, Corry PM. Heat-activated transgene expression from adenovirus vectors infected into human prostate cancer cells. Cancer Res. 2001;61:1113–21. [PubMed] [Google Scholar]
- 39.Guilhon E, Quesson B, Moraud-Gaudry F, de Verneuil H, Canioni P, Salomir R. et al. Image-guided control of transgene expression based on local hyperthermia. Mol Imaging. 2003;2:11–7. doi: 10.1162/15353500200302151. [DOI] [PubMed] [Google Scholar]
- 40.Dong D, Dubeau L, Bading J, Nguyen K, Luna M, Yu H. et al. Spontaneous and controllable activation of suicide gene expression driven by the stress-inducible grp78 promoter resulting in eradication of sizable human tumors. Hum Gene Ther. 2004;15:553–61. doi: 10.1089/104303404323142006. [DOI] [PubMed] [Google Scholar]
- 41.Noguchi T, Michihata T, Nakamura W, Takumi T, Shimizu R, Yamamoto M. et al. Dual-color luciferase mouse directly demonstrates coupled expression of two clock genes. Biochemistry. 2010;49:8053–61. doi: 10.1021/bi100545h. [DOI] [PubMed] [Google Scholar]
- 42.Wang F, Niu G, Chen X, Cao F. Molecular imaging of microRNAs. Eur J Nucl Med Mol Imaging. 2011;38:1572–9. doi: 10.1007/s00259-011-1786-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HJ, Chung JK, Hwang do W, Lee DS, Kim S. In vivo imaging of miR-221 biogenesis in papillary thyroid carcinoma. Mol Imaging Biol. 2009;11:71–8. doi: 10.1007/s11307-008-0188-6. [DOI] [PubMed] [Google Scholar]
- 44.Massoud TF, Paulmurugan R, De A, Ray P, Gambhir SS. Reporter gene imaging of protein-protein interactions in living subjects. Curr Opin Biotechnol. 2007;18:31–7. doi: 10.1016/j.copbio.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paulmurugan R, Gambhir SS. An intramolecular folding sensor for imaging estrogen receptor-ligand interactions. Proc Natl Acad Sci U S A. 2006;103:15883–8. doi: 10.1073/pnas.0607385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michnick SW, Remy I, Campbell-Valois FX, Vallee-Belisle A, Pelletier JN. Detection of protein-protein interactions by protein fragment complementation strategies. Methods Enzymol. 2000;328:208–30. doi: 10.1016/s0076-6879(00)28399-7. [DOI] [PubMed] [Google Scholar]
- 47.Wehrman T, Kleaveland B, Her JH, Balint RF, Blau HM. Protein-protein interactions monitored in mammalian cells via complementation of beta -lactamase enzyme fragments. Proc Natl Acad Sci U S A. 2002;99:3469–74. doi: 10.1073/pnas.062043699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci U S A. 2002;99:15608–13. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galarneau A, Primeau M, Trudeau LE, Michnick SW. Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat Biotechnol. 2002;20:619–22. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 50.Kaihara A, Kawai Y, Sato M, Ozawa T, Umezawa Y. Locating a protein-protein interaction in living cells via split Renilla luciferase complementation. Anal Chem. 2003;75:4176–81. doi: 10.1021/ac0300800. [DOI] [PubMed] [Google Scholar]
- 51.Grunberg R, Serrano L. Strategies for protein synthetic biology. Nucleic Acids Res. 2010;38:2663–75. doi: 10.1093/nar/gkq139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–42. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 53.Luker KE, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci U S A. 2004;101:12288–93. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hida N, Awais M, Takeuchi M, Ueno N, Tashiro M, Takagi C. et al. High-sensitivity real-time imaging of dual protein-protein interactions in living subjects using multicolor luciferases. PLoS One. 2009;4:e5868. doi: 10.1371/journal.pone.0005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hardt M, Chantaravisoot N, Tamanoi F. Activating mutations of TOR (target of rapamycin) Genes Cells. 2011;16:141–51. doi: 10.1111/j.1365-2443.2010.01482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pohanka E. New immunosuppressive drugs: an update. Curr Opin Urol. 2001;11:143–51. doi: 10.1097/00042307-200103000-00004. [DOI] [PubMed] [Google Scholar]
- 57.Huang S, Houghton PJ. Resistance to rapamycin: a novel anticancer drug. Cancer Metastasis Rev. 2001;20:69–78. doi: 10.1023/a:1013167315885. [DOI] [PubMed] [Google Scholar]
- 58.Paulmurugan R, Massoud TF, Huang J, Gambhir SS. Molecular imaging of drug-modulated protein-protein interactions in living subjects. Cancer Res. 2004;64:2113–9. doi: 10.1158/0008-5472.can-03-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer. 2004;4:106–17. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- 60.Kim SB, Ozawa T, Watanabe S, Umezawa Y. High-throughput sensing and noninvasive imaging of protein nuclear transport by using reconstitution of split Renilla luciferase. Proc Natl Acad Sci U S A. 2004;101:11542–7. doi: 10.1073/pnas.0401722101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L, Lee KC, Bhojani MS, Khan AP, Shilman A, Holland EC. et al. Molecular imaging of Akt kinase activity. Nat Med. 2007;13:1114–9. doi: 10.1038/nm1608. [DOI] [PubMed] [Google Scholar]
- 62.Lemmon MA, Schlessinger J. Regulation of signal transduction and signal diversity by receptor oligomerization. Trends Biochem Sci. 1994;19:459–63. doi: 10.1016/0968-0004(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 63.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 64.Kondapaka SB, Singh SS, Dasmahapatra GP, Sausville EA, Roy KK. Perifosine, a novel alkylphospholipid, inhibits protein kinase B activation. Mol Cancer Ther. 2003;2:1093–103. [PubMed] [Google Scholar]
- 65.Violin JD, DiPilato LM, Yildirim N, Elston TC, Zhang J, Lefkowitz RJ. beta2-adrenergic receptor signaling and desensitization elucidated by quantitative modeling of real time cAMP dynamics. J Biol Chem. 2008;283:2949–61. doi: 10.1074/jbc.M707009200. [DOI] [PubMed] [Google Scholar]
- 66.Fan F, Binkowski BF, Butler BL, Stecha PF, Lewis MK, Wood KV. Novel genetically encoded biosensors using firefly luciferase. ACS Chem Biol. 2008;3:346–51. doi: 10.1021/cb8000414. [DOI] [PubMed] [Google Scholar]
- 67.Binkowski BF, Butler BL, Stecha PF, Eggers CT, Otto P, Zimmerman K. et al. A luminescent biosensor with increased dynamic range for intracellular cAMP. ACS Chem Biol. 2011;6:1193–7. doi: 10.1021/cb200248h. [DOI] [PubMed] [Google Scholar]
- 68.Schoenberger J, Bauer J, Moosbauer J, Eilles C, Grimm D. Innovative strategies in in vivo apoptosis imaging. Curr Med Chem. 2008;15:187–94. doi: 10.2174/092986708783330647. [DOI] [PubMed] [Google Scholar]
- 69.Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- 70.Grutter MG. Caspases: key players in programmed cell death. Curr Opin Struct Biol. 2000;10:649–55. doi: 10.1016/s0959-440x(00)00146-9. [DOI] [PubMed] [Google Scholar]
- 71.Laxman B, Hall DE, Bhojani MS, Hamstra DA, Chenevert TL, Ross BD. et al. Noninvasive real-time imaging of apoptosis. Proc Natl Acad Sci U S A. 2002;99:16551–5. doi: 10.1073/pnas.252644499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coppola JM, Ross BD, Rehemtulla A. Noninvasive imaging of apoptosis and its application in cancer therapeutics. Clin Cancer Res. 2008;14:2492–501. doi: 10.1158/1078-0432.CCR-07-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ray P, De A, Patel M, Gambhir SS. Monitoring caspase-3 activation with a multimodality imaging sensor in living subjects. Clin Cancer Res. 2008;14:5801–9. doi: 10.1158/1078-0432.CCR-07-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kanno A, Yamanaka Y, Hirano H, Umezawa Y, Ozawa T. Cyclic luciferase for real-time sensing of caspase-3 activities in living mammals. Angew Chem Int Ed Engl. 2007;46:7595–9. doi: 10.1002/anie.200700538. [DOI] [PubMed] [Google Scholar]
- 75.Kanno A, Umezawa Y, Ozawa T. Detection of apoptosis using cyclic luciferase in living mammals. Methods Mol Biol. 2009;574:105–14. doi: 10.1007/978-1-60327-321-3_9. [DOI] [PubMed] [Google Scholar]
- 76.Zhang F, Zhu L, Liu G, Hida N, Lu G, Eden HS. et al. Multimodality imaging of tumor response to doxil. Theranostics. 2011;1:302–9. doi: 10.7150/thno/v01p0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kircher MF, Gambhir SS, Grimm J. Noninvasive cell-tracking methods. Nat Rev Clin Oncol. 2011;8:677–88. doi: 10.1038/nrclinonc.2011.141. [DOI] [PubMed] [Google Scholar]
- 78.Massoud TF, Paulmurugan R, Gambhir SS. A molecularly engineered split reporter for imaging protein-protein interactions with positron emission tomography. Nat Med. 2010;16:921–6. doi: 10.1038/nm.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gambhir SS, Bauer E, Black ME, Liang Q, Kokoris MS, Barrio JR. et al. A mutant herpes simplex virus type 1 thymidine kinase reporter gene shows improved sensitivity for imaging reporter gene expression with positron emission tomography. Proc Natl Acad Sci U S A. 2000;97:2785–90. doi: 10.1073/pnas.97.6.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paulmurugan R, Gambhir SS. Firefly luciferase enzyme fragment complementation for imaging in cells and living animals. Anal Chem. 2005;77:1295–302. doi: 10.1021/ac0484777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996;4:287–98. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- 82.Massoud TF, Paulmurugan R, Gambhir SS. Molecular imaging of homodimeric protein-protein interactions in living subjects. FASEB J. 2004;18:1105–7. doi: 10.1096/fj.03-1128fje. [DOI] [PMC free article] [PubMed] [Google Scholar]