Abstract
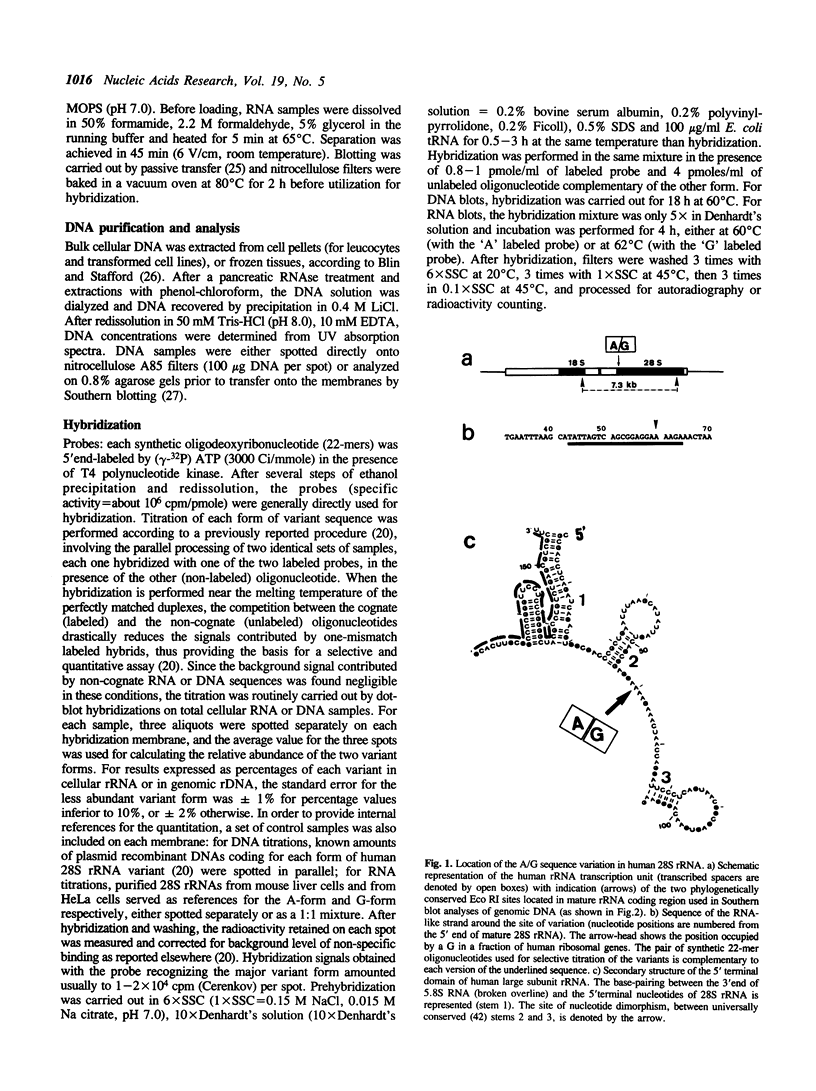
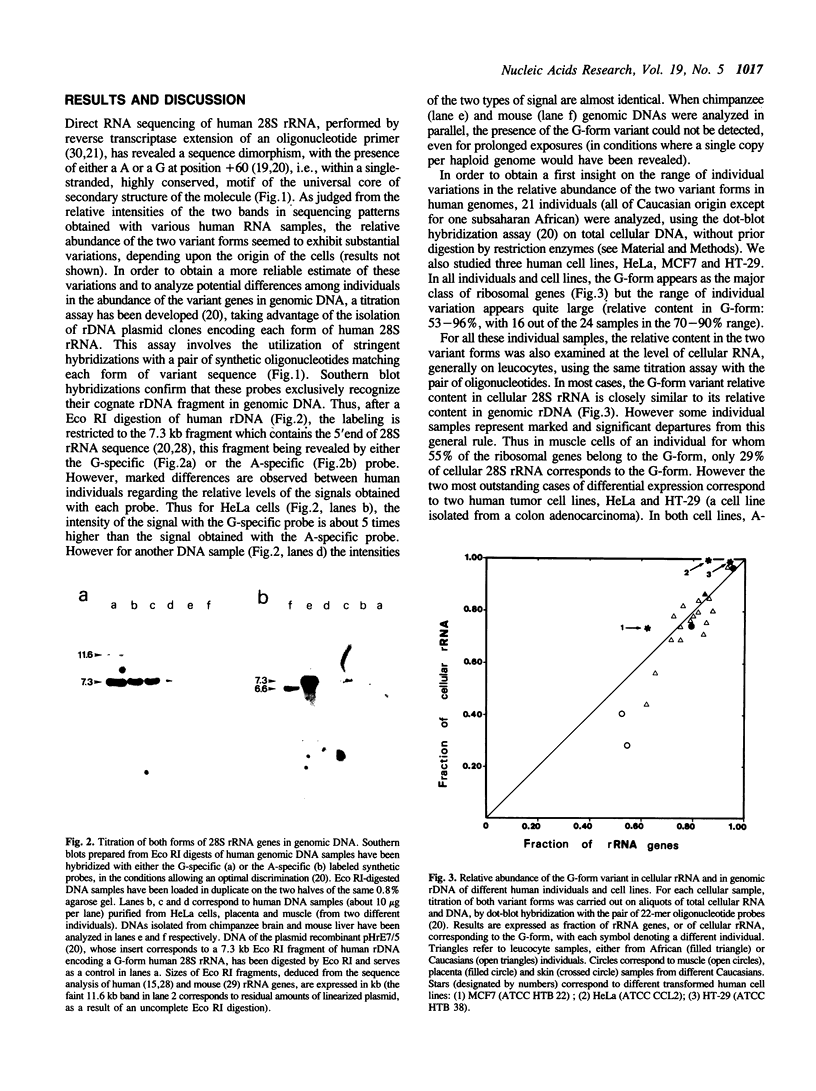
In humans, cellular 28S rRNA displays a sequence dimorphism within an evolutionarily conserved motif, with the presence, at position +60, of either a A (like the metazoan consensus) or a G. The relative abundance of the two forms of variant genes in the genome exhibit large differences among individuals. The two variant forms are generally represented in cellular 28S rRNA in proportion of their relative abundance in the genome, at least for leucocytes. However, in some cases, one form of variant may be markedly underexpressed as compared to the other. Thus, in HeLa cells, A-form genes contribute to only 1% of the cellular content in mature 28S rRNA although amounting to 15% of the ribosomal genes. The differential expression seems to result from different transcriptional activities rather than from differences in pre-rRNA processing efficiency or in stabilities of mature rRNAs. G-form ribosomal genes were not detected in other mammals, including chimpanzee, which suggests that the fixation of this variant type is a rather recent event in primate evolution.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnheim N., Krystal M., Schmickel R., Wilson G., Ryder O., Zimmer E. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7323–7327. doi: 10.1073/pnas.77.12.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Back E., Müller F., Tobler H. Structural organization of the two main rDNA size classes of Ascaris lumbricoides. Nucleic Acids Res. 1984 Feb 10;12(3):1313–1332. doi: 10.1093/nar/12.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerson S. R., Kaufman L. S. Increased rRNA gene activity during a specific window of early pea leaf development. Mol Cell Biol. 1990 Feb;10(2):842–845. doi: 10.1128/mcb.10.2.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. M., Platt T. Pol I transcription: which comes first, the end or the beginning? Cell. 1986 Dec 26;47(6):839–840. doi: 10.1016/0092-8674(86)90795-6. [DOI] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Erickson J. M., Schmickel R. D. A molecular basis for discrete size variation in human ribosomal DNA. Am J Hum Genet. 1985 Mar;37(2):311–325. [PMC free article] [PubMed] [Google Scholar]
- Gonzalez I. L., Chambers C., Gorski J. L., Stambolian D., Schmickel R. D., Sylvester J. E. Sequence and structure correlation of human ribosomal transcribed spacers. J Mol Biol. 1990 Mar 5;212(1):27–35. doi: 10.1016/0022-2836(90)90302-3. [DOI] [PubMed] [Google Scholar]
- Gonzalez I. L., Gorski J. L., Campen T. J., Dorney D. J., Erickson J. M., Sylvester J. E., Schmickel R. D. Variation among human 28S ribosomal RNA genes. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7666–7670. doi: 10.1073/pnas.82.22.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez I. L., Sylvester J. E., Schmickel R. D. Human 28S ribosomal RNA sequence heterogeneity. Nucleic Acids Res. 1988 Nov 11;16(21):10213–10224. doi: 10.1093/nar/16.21.10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J. M., Dover G. A. Molecular coevolution among cryptically simple expansion segments of eukaryotic 26S/28S rRNAs. Mol Biol Evol. 1988 Jul;5(4):377–391. doi: 10.1093/oxfordjournals.molbev.a040505. [DOI] [PubMed] [Google Scholar]
- Hassouna N., Michot B., Bachellerie J. P. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984 Apr 25;12(8):3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. P., Warner J. R. Unusual enhancer function in yeast rRNA transcription. Mol Cell Biol. 1989 Nov;9(11):4986–4993. doi: 10.1128/mcb.9.11.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Volpe A., Simeone A., D'Esposito M., Scotto L., Fidanza V., de Falco A., Boncinelli E. Molecular analysis of the heterogeneity region of the human ribosomal spacer. J Mol Biol. 1985 May 25;183(2):213–223. doi: 10.1016/0022-2836(85)90214-1. [DOI] [PubMed] [Google Scholar]
- Labella T., Schlessinger D. Complete human rDNA repeat units isolated in yeast artificial chromosomes. Genomics. 1989 Nov;5(4):752–760. doi: 10.1016/0888-7543(89)90117-1. [DOI] [PubMed] [Google Scholar]
- LeMeur M., Glanville N., Mandel J. L., Gerlinger P., Palmiter R., Chambon P. The ovalbumin gene family: hormonal control of X and Y gene transcription and mRNA accumulation. Cell. 1981 Feb;23(2):561–571. doi: 10.1016/0092-8674(81)90152-5. [DOI] [PubMed] [Google Scholar]
- Learned R. M., Learned T. K., Haltiner M. M., Tjian R. T. Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell. 1986 Jun 20;45(6):847–857. doi: 10.1016/0092-8674(86)90559-3. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987 May;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Dent C. L., Farrell T. E., Garde J., McCallum F. S., Wakeman J. A. Clones of human ribosomal DNA containing the complete 18 S-rRNA and 28 S-rRNA genes. Characterization, a detailed map of the human ribosomal transcription unit and diversity among clones. Biochem J. 1987 Sep 1;246(2):519–527. doi: 10.1042/bj2460519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F. Structure of mouse rRNA precursors. Complete sequence and potential folding of the spacer regions between 18S and 28S rRNA. Nucleic Acids Res. 1983 May 25;11(10):3375–3391. doi: 10.1093/nar/11.10.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B., Hassouna N., Bachellerie J. P. Secondary structure of mouse 28S rRNA and general model for the folding of the large rRNA in eukaryotes. Nucleic Acids Res. 1984 May 25;12(10):4259–4279. doi: 10.1093/nar/12.10.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoloso M., Liang H. Q., Bachellerie J. P. Titration of variant DNA sequences differing by a single point-mutation by selective dot-blot hybridization with synthetic oligonucleotides. Biochem Biophys Res Commun. 1989 Mar 31;159(3):1233–1241. doi: 10.1016/0006-291x(89)92242-0. [DOI] [PubMed] [Google Scholar]
- Qu H. L., Michot B., Bachellerie J. P. Improved methods for structure probing in large RNAs: a rapid 'heterologous' sequencing approach is coupled to the direct mapping of nuclease accessible sites. Application to the 5' terminal domain of eukaryotic 28S rRNA. Nucleic Acids Res. 1983 Sep 10;11(17):5903–5920. doi: 10.1093/nar/11.17.5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L. H., Nicoloso M., Bachellerie J. P. Phylogenetic calibration of the 5' terminal domain of large rRNA achieved by determining twenty eucaryotic sequences. J Mol Evol. 1988 Dec;28(1-2):113–124. doi: 10.1007/BF02143502. [DOI] [PubMed] [Google Scholar]
- Renalier M. H., Joseph N., Bachellerie J. P. Clones containing variant forms of complete human rRNA genes. Characterization and sequence of their transcription initiation region. FEBS Lett. 1989 Apr 24;247(2):298–302. doi: 10.1016/0014-5793(89)81356-0. [DOI] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence of Xenopus laevis 18S ribosomal RNA inferred from gene sequence. Nature. 1981 May 21;291(5812):205–208. doi: 10.1038/291205a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stewart M. A., Hall L. M., Maden B. E. Multiple heterogeneities in the transcribed spacers of ribosomal DNA from Xenopus laevis. Nucleic Acids Res. 1983 Feb 11;11(3):629–646. doi: 10.1093/nar/11.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware V. C., Tague B. W., Clark C. G., Gourse R. L., Brand R. C., Gerbi S. A. Sequence analysis of 28S ribosomal DNA from the amphibian Xenopus laevis. Nucleic Acids Res. 1983 Nov 25;11(22):7795–7817. doi: 10.1093/nar/11.22.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters A. P., Syin C., McCutchan T. F. Developmental regulation of stage-specific ribosome populations in Plasmodium. Nature. 1989 Nov 23;342(6248):438–440. doi: 10.1038/342438a0. [DOI] [PubMed] [Google Scholar]
- Worton R. G., Sutherland J., Sylvester J. E., Willard H. F., Bodrug S., Dubé I., Duff C., Kean V., Ray P. N., Schmickel R. D. Human ribosomal RNA genes: orientation of the tandem array and conservation of the 5' end. Science. 1988 Jan 1;239(4835):64–68. doi: 10.1126/science.3336775. [DOI] [PubMed] [Google Scholar]