SUMMARY
Fibrinogen, upon enzymatic conversion to monomeric fibrin, provides the building blocks for fibrin polymer, the scaffold of blood clots and thrombi. Little has been known about the force-induced unfolding of fibrin(ogen), even though it is the foundation for the mechanical and rheological properties of fibrin, which are essential for hemostasis. We determined mechanisms and mapped the free energy landscape of the elongation of fibrin(ogen) monomers and oligomers through combined experimental and theoretical studies of the nanomechanical properties of fibrin(ogen), using atomic force microscopy-based single-molecule unfolding and simulations in the experimentally relevant timescale. We have found that mechanical unraveling of fibrin(ogen) is determined by the combined molecular transitions that couple stepwise unfolding of the γ chain nodules and reversible extension-contraction of the α-helical coiled-coil connectors. These findings provide important characteristics of the fibrin(ogen) nanomechanics necessary to understand the molecular origins of fibrin viscoelasticity at the fiber and whole clot levels.
INTRODUCTION
Fibrin is a polymer that provides the scaffold for protective hemostatic plugs and obstructive thrombi. Human fibrin is formed from fibrinogen (Fg), a blood plasma protein, consisting of pairs of Aα chains, Bβ chains, and γ chains, linked by S-S bonds (Figure 1) (Weisel, 2005). Two distal globular regions and one central globular region of fibrin(ogen) are connected by two α-helical coiled-coils; each end region contains the β- and γ-nodules associated via noncovalent interactions (Figure 1). The β- and γ-nodules are homologous and each of them contains three crystallographically distinct globular domains (Yee et al., 1997; Spraggon et al., 1997; Brown et al., 2000; Madrazo et al., 2001). Fg is partially cleaved by thrombin, which triggers formation of a three-dimensional fibrous network called a fibrin clot (Weisel, 2005).
Figure 1. Human Fibrinogen Structure (PDB Entry: 3GHG).
(A) A schematic representation of the Fg molecule in the naturally folded and fully unfolded states. The molecule is constrained at the C-terminal part of one γ chain (γIle394), and the mechanical force (f) is applied to the C-terminal part of the other γ chain (gGly395).
(B) Structural details showing the central nodule, γ-nodules, β-nodules, disulfide rings, and the γ-γ-crosslinking sites. Dimensions are shown as in the compact crystal structure, and the contour lengths of various structural elements are shown in the fully unfolded state assuming a contour length per residue of 0.38 nm.
The mechanical properties of fibrin are essential for the ability of fibrin clots to stop bleeding and affect the danger of obstructive thrombi that cause heart attack and stroke (Weisel, 2004). Despite such critical importance, the structural basis of clot mechanics is not well understood (Weisel, 2008). The rising attention to this problem has resulted in a number of recent papers describing unique viscoelastic properties of the whole fibrin clot and single fibers in correlation with structural rearrangements (Liu et al., 2006, 2010; Guthold et al., 2007; Carlisle et al., 2010; Piechocka et al., 2010; Hudson et al., 2010; Falvo et al., 2010). Elongation of fibrin molecules is generally considered an important mechanism helping to accommodate strain, but there is no consensus regarding the particular structures of fibrin(ogen) involved. Depending on methodological approaches, different groups have suggested the following molecular origins of the remarkable extensibility of fibrin(ogen): (1) unfolding of the coiled-coil connectors (Brown et al., 2007; Lim et al., 2008); (2) unfolding of the globular γ-nodule (Averett et al., 2008, 2009); (3) straightening and unfolding of the αC region (Collet et al., 2005; Houser et al., 2010; Tsurupa et al., 2009; Doolittle and Kollman, 2006; Falvo et al., 2008), and (4) some combination (Guthold et al., 2007; Liu et al., 2010; Falvo et al., 2010; Brown et al., 2009; Purohit et al., 2011).
Because of the complexity of fibrin(ogen) molecules (Figure 1), the force-driven elongation involves unraveling of various compact structures, but it is virtually impossible to experimentally resolve the structural underpinnings on scales <1 nm (Hyeon et al., 2006a; Zhmurov et al., 2011). In contrast, biomolecular simulations help provide a detailed nanoscale picture of structural rearrangements and offer meaningful interpretations for the experimental data of mechanical protein unfolding (Barsegov et al., 2006; Raman et al., 2007). However, the main challenge is to computationally generate the unfolding dynamics within the experimental timescale using physiologically relevant forces (Zhmurov et al., 2010a).
Here we report our results of combined experimental and theoretical studies of fibrin(ogen) nanomechanics, obtained using atomic force microscopy (AFM)-based single-molecule unfolding and simulations of forced unfolding of fibrin(ogen) and its derivatives. This paper is a significant advancement over previously published preliminary work on the unfolding of fibrinogen oligomers (Brown et al., 2007) with additional AFM experiments, including new molecular variants, and novel approaches to data analysis and modeling. Importantly, the experimental measurements have been analyzed in comparison with the computational protein unfolding performed under identical conditions of the mechanical force-ramp. The main finding is that molecular elongation of fibrin(ogen) is largely determined by the combined sequential unfolding transitions in the C-terminal γ chain nodules and limited reversible extension-contraction of the α-helical coiled-coil connectors.
RESULTS
AFM Unfolding Experiments
Single-molecule AFM unfolding was performed with either fibrinogen monomers (Fg) or linear single-chain fibrinogen oligomers (Fgn) cross-linked via the C-termini of the γ chains using factor XIIIa (see Section I.1 in Supplemental Experimental Procedures available online). Oligomers were formed to generate reproducible data, as demonstrated in other unfolding studies on artificial protein tandems (Dietz and Rief, 2004). The proteins were allowed to adsorb on mica followed by their upward extension from the surface by an AFM tip, with registration of the force-extension profile. We have gathered many data traces, from which meaningful force-extension curves have been selected based on criteria: (1) the well-resolved first and desirable last desorption peaks with at least one unfolding peak between them (the last peak might be absent if the desorption force is comparable to the unfolding force); (2) repeated and reproducible signal pattern. The approach used to analyze experimental force-distance curves is presented in Section I.2 in Supplemental Experimental Procedures.
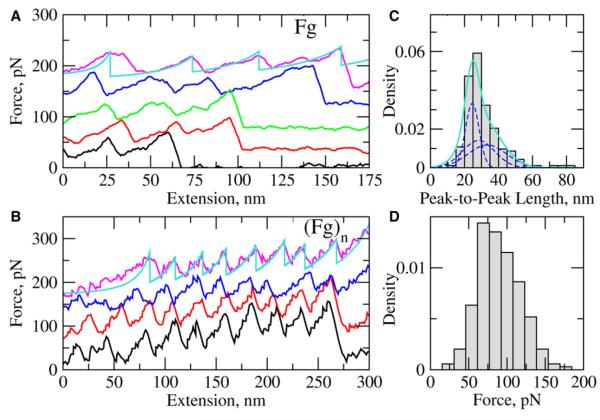
All of the experimental force-distance curves both for Fg and Fgn were sawtooth-like, reflecting sharp increases in the mechanical tension experienced by the polypeptide chain as a result of force application followed by steep decreases in tension due to the onset of unfolding transitions. No distinct plateaus were revealed in the force-extension curves despite purposeful scrutiny. A number of representative force-distance curves are shown in Figure 2 and additional typical raw data traces are presented in Figure S1, which is available with this article online. To quantify the force-extension curves, they were fit with a wormlike chain model (Figure 2), which enabled us to extract force peak values (f) and peak-to-peak distances (x). Statistical analysis of the unfolding data showed that the average forces and peak-to-peak distances for Fg (f ≈ 89 pN and x ≈ 33 nm) and Fgn (f ≈ 90 pN and x ≈ 30 nm) were very similar (Table 1). The molecular spring constants for Fg and Fgn were calculated using the wormlike chain fits to the experimental force spectra (Figure 2) by evaluating the derivative, K = df/fx, with similar values of K ≈ 3.7 pN/nm and 4.9 pN/nm for Fg and Fgn, respectively. On the basis of the similarity of the parameters, we have combined the data sets for unfolding of Fg and Fgn and plotted them as the histograms of peak-to-peak distances (Figure 2C) and peak forces (Figure 2D). For the combined data set, the average peak force, peak-to-peak distance, and the spring constant were f ≈ 90 pN, x ≈ 31 nm, and K ≈ 4.5 pN/nm, respectively. Hence, Fg and Fgn respond similarly to mechanical stress and the unfolding transitions in Fg and in Fgn are likely to occur within the same structural domains.
Figure 2. Unfolding of Fibrinogen In Vitro with AFM.
(A and B) Experimental force-distance curves obtained at a pulling velocity vf = 1.0 mm/s for fibrinogen monomer Fg (A) and oligomers Fgn (B). See Figure S1 for more examples. All the curves that appear above the lowest curve are offset for clarity. In this figure and in Figures 3A and 3B, Figures 4A–4C, and Figure 5B the smooth overlaying curves represent numerical fits obtained using the wormlike chain model.
(C and D) The histograms of the peak-to-peak distances (number of data points n = 186) and peak forces (n = 226) are shown in (C) and (D), respectively. The bin size (Δx ≈ 6 nm and Δf ≈ 15.5 pN) has been estimated using the Freedman-Diaconis rule for optimal bandwidth selection (Bura et al., 2009). In (C), the solid curve represents the superposition of three Gaussian probability densities of unfolding forces (dashed lines) for the transitions of type 1, 2, and 3, displayed in Figure 3.
Table 1.
Unfolding Data Obtained from Dynamic Force Measurements In Vitro (AFM) and In Silico (SOP-GPU) for Fibrin(ogen) Monomer and Single-Chain Oligomers
| System | Fibrinogen Monomer (Fg) f, pN; x, nm; K, pN/nm |
Fibrinogen Oligomers (Fgn) f, pN; x, nm; K, pN/nm |
Combined Set (Fg + Fgn) f, pN; x, nm; K, pN/nm |
|---|---|---|---|
| Experiment | 89.1 ± 22.5; 32.8 ± 8.2; 3.7 ± 1.2 | 90.2 ± 29.1; 30.1 ± 11.1; 4.9 ± 0.8 | 90.1 ± 27.5; 31.1 ± 11.1; 4.5 ± 0.7 |
| Simulations | 134.2 ± 23.7; 28.6 ± 9.4; 8.7 ± 2.2 | 112.4 ± 34.1; 27.3 ± 10.2; 6.0 ± 1.7 | 127.4 ± 28.9; 28.2 ± 9.5; 7.0 ± 1.6 |
Presented are the average values and standard deviations for the peak forces (f), peak-to-peak distances (x), and molecular spring constants (K), extracted from the experimental and simulated force-extension curves. Theoretical estimates for single-chain oligomers are given for the fibrinogen dimer (Fg2).
Because the unfolding geometry of Fgn is specified by γ-γ-crosslinking, the observed sawtooth patterns are likely due to unfolding of either the coiled-coils or the globular C-terminal portions of the γ chains (Figure 1). (Potentially, the central nodule could also unfold but it is highly constrained by disulphide bonds.). The peak-to-peak distance is roughly equal to the contour length of the unfolded structural element minus the length of the element when it was folded. Each coiled-coil consists of 111 or 112 amino acid residues of the Aα, Bβ, and γ chains, which, when fully unfolded, form a ~42 nm long thread (assuming a contour length per residue of 0.38 nm) corresponding to an expected peak-to-peak length of 25 nm (extension due to unfolding minus 17 nm initial folded length). This was in moderately good agreement with the experimental value of ~31 nm (Table 1). In contrast, the C-terminal γ-nodules each consist of 204 potentially “unfoldable” amino acid residues (not including the disulfide loop or the chain beyond the first crosslinking site) with an expected peak-to-peak length of ~82 nm, significantly longer than the average unfolding length observed. However, each γ-nodule has three compact domains, which may unfold separately. Then the estimated unfolding lengths are ~7 nm for the N-terminal domain (19 residues, γ143–γ153 and γ182–γ191), ~40 nm for the central domain (106 residues, γ192–γ286 and γ380–γ392 of the β strand insert), and ~30 nm for the C-terminal domain (79 residues, γ287–γ326 and γ339–γ379) (Yee et al., 1997). Given the very small size of the domains in the folded state, their expected fully unfolded (contour) lengths are almost equal to the elongations resulting from the unfolding transitions. Of the three globular domains composing the γ-nodule, the C-terminal domain seems to be the most likely candidate for unfolding because its calculated contour length (30 nm) is quite close to the average experimental elongation of 31 nm. Unfortunately, the spatial resolution of AFM does not allow discrimination between elongation because of unfolding of the coiled-coils and the domains of the γ-nodule.
Indirect evidence for preferable unfolding of the γ-nodule came from the AFM data for unfolding of monomeric Fg. The force-distance curves for monomeric Fg displayed up to 4 forcesignals, each seemingly corresponding to a separate unfolding event (Figure 2A; Figure S1A). Because one fibrinogen molecule has two coiled-coil connectors and two γ-nodules, each with multidomain structure (Figure 1), those several peaks imply that fibrinogen unfolding involves consecutive unraveling of multiple structural elements, either in the γ-nodules alone or in the γ-nodules and coiled-coils, but not solely in the coiled-coils, which are expected to unravel in a single step each. Of course, there were more force signals observed in the sawtooth patterns for oligomeric Fgn compared to monomeric Fg because there are more structural repeats in Fgn. Additional force peaks in the force-distance curves for Fgn might emerge because of disruption of the D-D junctions connecting Fg in the single-chain oligomers through the γ-γ crosslinking.
To test a potential role of the unstructured αC region in the mechanical stability of fibrin(ogen), we performed AFM unfolding on fibrinogen and its derivatives without the αC region. One of the samples was a monomer of recombinant fibrinogen variant truncated at position 251 of the Aα chain (Fg α251) (Collet et al., 2005). The other one represented end-to-end oligomers made of fibrinogen I-9, a proteolytically cleaved normal fraction of fibrinogen lacking the αC region (Mosesson and Sherry, 1966). Notably, the absence of the αC region affected neither qualitatively nor quantitatively the sawtooth pattern of protein unfolding. The average unfolding force (f = 89 ± 23 pN) and peak-to-peak distance (x = 29 ± 8 nm) were statistically indistinguishable from the full-length fibrinogen monomer and/or oligomers, suggesting that the αC region was not involved in the mechanical response of fibrin(ogen).
Pulling Simulations
To provide a basis for interpretation of the experimental data, we turned to computer-based modeling. The force measurements in silico were performed on the isolated γ-nodule, the γ-nodule with 25 adjoining residues from the β-nodule, the complex of the γ- and β-modules, the γ-γ-crosslinked double-D fragment (D-dimer), fibrinogen monomer (Fg), and dimer (Fg2). The crystal structures for human fibrinogen were taken from the PDB entry 3GHG. The structural model and pulling setup for each system are presented in Supplemental Experimental Procedures (Section II.1). The crystallographic data on fibrinogen structure are incomplete because there are missing unstructured portions. These are residues 1–26, 1–57, and 1–13 at the N termini of the Aα,Bβ, and γ chains, respectively, and residues 201–610, 459–461, and 395– 411 at the C termini of the Aα,Bβ, and γ chains, respectively (Yee et al., 1997; Spraggon et al., 1997; Brown et al., 2000; Madrazo et al., 2001). Accordingly, these portions have not been included in the virtual structures. We employed the self-organized polymer (SOP) model (Hyeon et al., 2006a) and computations on Graphics Processing Units (GPUs) (Experimental Procedures). This enabled us to span the experimental 0.1–0.4 s timescale (Table S2). The SOP model has been used to describe allosteric transitions in GroEL (Hyeon et al., 2006b) and dynamics of myosin V (Tehver and Thirumalai, 2010). We parameterized the SOP model using all-atom Molecular Dynamics (MD) simulations (Section II.2 in Supplemental Experimental Procedures).
The experimental force-distance curves for Fg and Fgn (Figure 2) and theoretical force-extension profiles for Fg and Fg2 (Figure 3) were very similar. Statistical analysis of the simulated data, averaged over 5 long 0.2 s-simulation runs (Table S2), revealed average peak-to-peak distances of x ≈ 29 nm for Fg and x ≈ 27 nm for Fg2, which were in good agreement with the experimental estimates of this parameter (Table 1). The average peak forces (i.e., f ≈ 134 pN for Fg and f ≈ 112 pN for Fg2) obtained from simulations were somewhat higher compared to the experimental values. This could be due to either the over-stabilization of the tertiary structure described by the SOP energy function, which lifts up the baseline in the force-extension curves, or insufficient sampling. Consequently, the molecular spring constants for Fg and Fg2, K ≈ 8.7 pN/nm and 6.0 pN/nm, and for the combined set of data, K ≈ 7.0 pN/nm, were larger than their experimental counterparts (Table 1). However, because the agreement observed for peak-to-peak distances matters most, the agreement between the experimental and simulated force spectra is quite good.
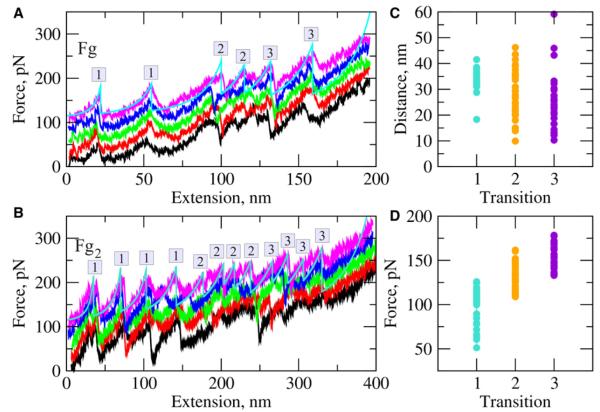
Figure 3. Unfolding of Fibrinogen In Silico.
(A and B) Representative force-extension profiles for fibrinogen monomer Fg (A) and dimer Fg2 (B), respectively, obtained at a pulling velocity vf = 1.0 μm/s. All the curves that appear above the lowest curve are offset for clarity.
(C and D) Plots show the peak-to-peak distances and peak forces for the transitions of type 1–3 (see Figure S2 and Table S3). The peaks corresponding to these transitions are numbered as 1, 2, and 3 in the force spectra (A and B). A movie detailing the unfolding transitions in the dimer Fg2 is in Movie S1.
Structural analysis showed that the sequences of molecular transitions for Fg and Fg2 were identical. We describe the unfolding mechanism for one half of the Fg molecule (illustrated explicitly in Figure 4D and Figure S2A) because the unfolding transitions in the other half of the molecule were the same. First, partial ~10–15 nm extension of the coiled-coils occurs. Then, the C-terminal β strand (residues γ380–392) in the central domain of each γ-nodule is pulled out, followed by unraveling of residues γ234–311. This event marks the break-up of each γ-nodule into two globular parts, one of which (residues γ311–381) roughly corresponds to the C-terminal P-domain and the other (residues γ139–234) includes the N-terminal domain, and leads to an average elongation of 33.4 nm. This is a combined result of the unfolding of 89 residues of the γ chain and partial unraveling of the coiled-coils. We refer to this event as the type 1 transition. Second, the C-terminal residues γ311–326 and γ339–380 of the γ-nodule unravel, which leads to an average elongation of 28.3 nm (type 2 transition), a combination of the unfolding of 56 residues and partial extension of the coiled-coils. Finally, the N-terminal residues γ139–153 and γ182–234 of the γ-nodule unravel, resulting in an average elongation of 24.5 nm, a sum of the unfolding of 66 residues, and partial extension of the coiled-coil connectors (type 3 transition). Each transition of type 1–3 is accompanied by 5–7-nm-long spring-like contraction of the coiled-coils (Figure S2B). The transition of type 2 and 3 occur second or third with an equal probability and the β-nodule is not perturbed in simulations or experiments, because the tension does not propagate along the backbone of the β-nodule. We performed a theoretical fit of the experimental histogram of peak-to-peak distances (Section I.2 in Supplemental Experimental Procedures), using the sum of equally weighted Gaussian density functions Gi(x) (i = 1, 2, and 3), each centered at the average distance of 33.4, 28.3, and 24.5 nm and having standard deviations of 4.0, 9.5, and 11.3 nm, which correspond to the computationally observed transitions of types 1, 2, and 3, respectively (Figure 3C). The theoretical density curve describes well the experimental histogram of molecular extensions (Figure 2C).
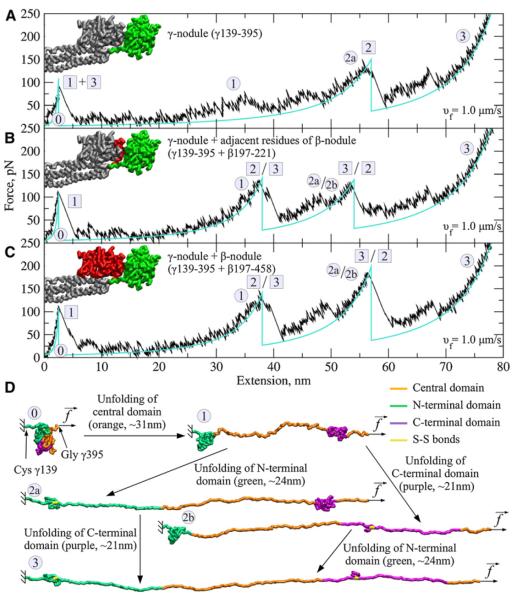
Figure 4. Forced Unfolding of the Fibrin(ogen) γ-Nodule In Silico.
(A–C) Panels show force-extension profiles for the γ-nodule alone (A), for the γ-nodule and adjacent portion of the β-chain (B), and for the complex of the γ- and β-nodules (C). In each structure, the simulated portion is colorized, and the “truncated” portion, not used in the simulations, is shown in gray. The force peaks correspond to the transitions of type 1–3 (see Table S3).
(D) The unfolding steps and intermediate conformations observed for the γ-nodule. Structures 0, 1, 2a, 2b, and 3 correspond to the accordingly numbered force peaks in the force-distance curves (A–C). Also shown are the maximal extensions, corresponding to the peak-to-peak distances in the force-distance curves (see Figure S2).
The distal D region of fibrin(ogen) contains contacting γ- and β-nodules formed by the C-terminal parts of the γ and Bβ chains, respectively. To probe the role of interactions between the γ- and β-nodules, we carried out pulling simulations of an isolated compact portion of the γ-nodule (residues γ139–395), the same portion of the γ-nodule and adjacent residues from the β-nodule (β197–221), and the major part of the D region, which includes the compact globular portions of the γ-nodule (residues γ139–395) and the β-nodule (residues β197–458). Unlike in the full-length fibrinogen, the force spectra for the γ-nodule alone show only two force peaks (Figure 4A). The first peak force of 90 pN at an extension of ~3 nm corresponds to the onset of unfolding of the central and N-terminal parts of the γ-nodule (residues γ139–153, γ182–311, and γ380–392), which occur simultaneously. This results in an average elongation of ~55 nm. The second peak force of ~125 pN corresponds to unfolding of the C-terminal portion of the γ-nodule (residues γ311–326 and γ339–380). Adding to the system 25 residues β197–221 from the β-nodule results in an additional third force peak (Figure 4B), observed at an average extension of ~38 nm and characterized by an average peak force of ~125 pN. Including the entire β-nodule (β- and γ-nodules held together) does not further change the unfolding pattern, but leads to additional stabilization of the γ-nodule (Figure 4C), which is reflected in the stronger 165 pN force signal at ~55 nm average extension.
Additional mechanical stabilization in Fgn might come from the interactions at the D-D interface. Pulling simulations on the D-dimer showed that disruption of the D-D interface induced a conformational transition from the “closed” state, in which the D-D junction is hidden, to the “open” state, in which the D-D interface opens up (Figure 5A). The force of this transition was only ~40 pN with a ~5 nm separation distance (Figure 5B; Figure S3). These results imply that in Fgn the D-D interface breaks up first, which does not alter the overall unfolding mechanism. We also found that the central nodule of the E region is only slightly deformed at a force >300 pN after the γ-nodules had unraveled and the coiled-coils were fully extended. The resulting elongation, measured by the γ23–γ23 distance between residues γ23 in the symmetric γ-chains, was only ~7 nm (Figure 6). A movie detailing all these unfolding steps in the fibrinogen dimer is provided in the Supplemental Information (Movie S1).
Figure 5. Forced Unfolding of the Fibrin(ogen) D-Dimer In Silico (See Also Figure S3).
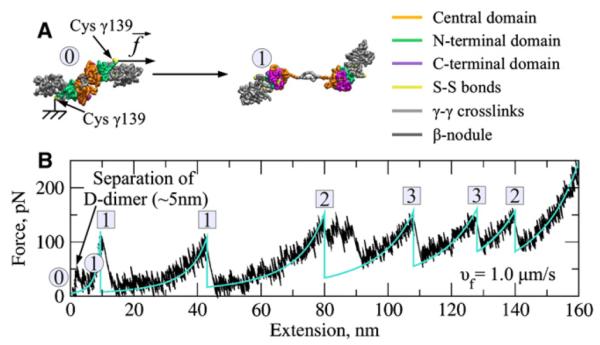
(A) D-dimer undergoing the conformational transition from the “closed” state (structure 0) to the “open” state (structure 1).
(B) A force-extension profile for the D-dimer obtained at a pulling velocity vf = 1.0 mm/s. Disruption of the D-D junction is characterized by the weak first peak ( ~50 pN) and short chain extension ( ~3–5 nm). Also shown are the peak-to-peak distances and peak forces for transitions of type 1–3 (see Figure S2 and Table S3).
Figure 6. Forced Unfolding of the Central Nodule of Fibrin(ogen)’s E Region.
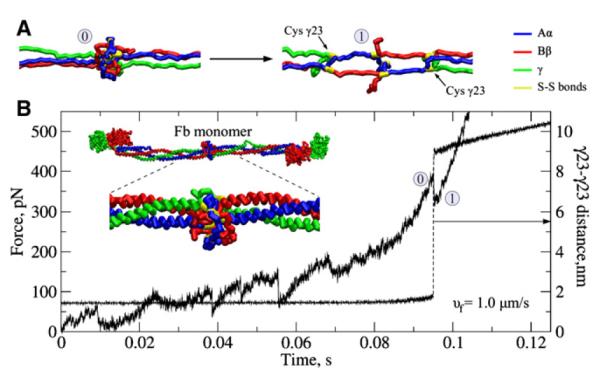
(A) Representative structures of the central nodule prior (structure 0) and after (structure 1) the unfolding transition.
(B) The time-dependence of the mechanical tension in the central nodule (solid curve; left y axis), and its extension along the direction of pulling force (dashed curve; right y axis), quantified by the distance between the two γ23Cys residues of the opposite γ chains. A sudden ~7 nm jump in the extension, observed at ~0.095 s, is due to the central domain unfolding. The inset shows the fibrin(ogen) structure with the central nodule and the adjacent coiled-coil regions.
We estimated the total energy change and work performed. For the D region, D-dimer, and Fg, we compared the energy for the fully extended state (at f= 150 pN) with the energy for the folded state (f= 0). Neglecting energy dissipation, we evaluated the total work as the area under the force-distance curves (Figures 4C, 5B, and 3A). Because in simulations we maintained the conditions of constant pressure (water density) and temper ature, the total work performed on the system is expended to change the Gibbs free energy ΔG= ΔH − TΔS. The enthalpy change ΔH (disruption of the native contacts), work (w), and entropy change TΔS (chain elongation) are shown in Table 2.
Table 2.
Numerical Estimates of the Thermodynamic Parameters (with Standard Deviations) for the Fibrinogen Forced Unfolding at Room Temperature (T = 300 K)
| System | D Region | Coiled-Coils | Fg Monomer | D-Dimer |
|---|---|---|---|---|
| ΔH, kcal/mol | 692 ± 10 | 136 ± 20 | 1520 ± 15 | 17.5 ± 4 |
| w, kcal/mol | 790 ± 17 | 184 ± 34 | 1764 ± 29 | 18.7 ± 3.7 |
| TΔS, kcal/mol | −98 ± 17 | −48 ± 34 | −244 ± 29 | — |
Shown are the average enthalpy change (ΔH), work performed on the system (w), and entropy change (TΔS) for unfolding of the D-region, coiled-coil connectors, and entire Fg monomer, and for the disruption of D-D self-association interface.
DISCUSSION
We have provided a structural foundation for fibrin mechanics at the molecular scale (~10–100 nm) and at different levels of complexity varying from the isolated structural elements (γ-nodule, D region, and D-dimer) to the full-length fibrin(ogen) monomer and oligomers. This information is important to understand fibrin(ogen) nanomechanics and has far reaching implications for the (patho)physiology of fibrin clots and thrombi, the key contributors to life-threatening hemostatic and thrombotic disorders. Using single-molecule AFM unfolding along with a Cα-based SOP model and Langevin simulations accelerated ~200-fold on graphics processors, we were able to directly compare the theoretical and experimental data, generated under the same force-load, to unambiguously interpret the experimental protein unfolding patterns.
The central result of our studies is that fibrin(ogen) forced unfolding is a collective process, which involves a number of mechanically coupled structural elements of the fibrin(ogen) molecule (Figure 1). Mainly, the process involves reversible extension-contraction of the α-helical coiled-coils, connecting the distal γ-nodules and the central nodule, and sequential dissociation and unraveling of the compact globular structural elements in the γ-nodules. In each γ-nodule, first, the C-terminal β strand (residues γ380–392) peels off from the five-stranded antiparallel β sheet in the central domain. This is followed by unraveling of residues γ234–311, leading to the separation of the γ-nodule into two globular parts, the C-terminal portion (residues γ311–380) and the N-terminal part (residues γ139–234). Each of these domains might unfold second or third with an equal probability (Figure 4). These consecutive transitions of type 1, 2, and 3 summarized in Table S3 correspond to the force signals observed in the simulated force spectra at an average fibrin(ogen) elongation of ~33, ~28, and ~25 nm (Figure 3). It is especially important that the experimental unfolding force spectra (Figure 2; Figure S1) and simulated force-extension profiles (Figure 3), indeed, look very similar and that the experimental and theoretical estimates of the average peak-to-peak distance of ~31 and ~28 nm agree quite well (Table 1). It is noteworthy that fibrin(ogen) and its smaller and larger derivatives invariably followed the same reproducible unfolding patterns, indicating that the overall unfolding mechanism is robust (deterministic) rather than variable (stochastic).
Detachment of the C-terminal β strand and its structural consequences observed in simulations confirm the “pull-out hypothesis” (Yakovlev et al., 2000a; Yakovlev et al., 2001), stating that the β strand (residues γ380–392) inserted into the central domain of the γ-nodule, can be removed (“pulled-out”) without destroying its compact structure; yet, such structure without the β strand insert is significantly destabilized. Our results show that the applied force first pulls out the β strand, after which the γ-nodule becomes unstable and falls apart (type 1 transition).
There is a good agreement between the results of thermal and force-driven unfolding of the γ-nodule. The enthalpy of melting of fibrinogen molecules ΔHm = 1365 kcal/mol measured by (Privalov and Medved, 1982) at pH 8.5 is relatively close to the combined enthalpy of forced unfolding of two D regions, 1384 kcal/mol, but is smaller than the enthalpy of mechanical unfolding of the full-length fibrin(ogen) monomer, ΔHFg = 1520 kcal/mol, determined in this study (Table 1). Our estimates are relatively close to the value of ΔHm = 1452 kcal/mol observed at pH = 6.0 by (Mihalyi and Donovan, 1985). The small disagreement is likely due to the difference between the mechanisms of thermal denaturation and of mechanical elongation and unfolding.
The α-helical coiled-coils in fibrin(ogen) act as highly elastic molecular capacitors. Reversible unfolding-refolding transitions enable the coiled-coils to store the mechanical energy and smooth out the effect of external perturbation. The synergy between the coiled-coil connectors and γ-nodules makes coiled-coils able to gradually accumulate mechanical tension before and reduce tension after an unfolding transition (of types 1–3) has occurred in the γ-nodules. The dynamic signature of this coupling is large, 5–7 nm stochastic fluctuations in the length of coiled-coils, which accompany unfolding transitions in the γ-nodules (Figure S2B), and large standard deviations of the peak-to-peak distances especially for the transitions of type 2 and 3 (Figure 3C; Table S3). In contrast with this new notion on the mechanical role of the coiled-coil-connectors, it has been suggested previously (Brown et al., 2007; Lim et al., 2008) that this region is fully unfolded and is primarily responsible for force-induced elongation of fibrin(ogen). However, this hypothesis seems to be unjustified for the following reasons. First, as discussed above, the complexity of fibrinogen requires that unfolding of domains of the γ-nodule also be considered. Second, the experimental AFM traces reported in one study (Lim et al., 2008) displayed unrealistic nanoNewton pulling forces. Third, in MD simulations (Lim et al., 2008), the pulling force was applied to the end of the coiled-coil region, and hence, no information about possible unfolding in the γ-nodule could be gathered. Fourth, the force-loading rate used in these simulations (Lim et al., 2008) was ~8 orders of magnitude higher than in their AFM experiments, making experimental and simulation results difficult to compare.
In addition to the major roles played by the γ-nodules and the coiled-coil connectors, the β-nodule, which remains unaffected by mechanical tension, was found to provide intramolecular stabilization to the γ-nodule through noncovalent interdomain interactions, which agrees with earlier findings by Yakovlev et al. (2000b) and Medved et al. (2001). This stabilizing effect is reflected in the emergence of an additional force signal in the force-extension curves for the (γ-nodule):(β-nodule) complex (Figure 4). The central nodule in the E region is the most mechanically stable portion of the fibrin(ogen) molecule as it yields last at >300 pN pulling force, long after the γ-nodules are fully unfolded and the coiled-coil connectors are maximally stretched. Because of the presence of disulfide rings (Figure 1), forced elongation of the central nodule leads to a small ~7nm extension of the fibrin(ogen) molecule (Figure 6). There were missing parts of the human fibrinogen molecule (i.e., at the N- and C-terminal portions of the Aα,Bβ, and γ chains), which were not resolved in the crystal structure (3GHG) and hence not included in the simulations. However, these parts do not seem to directly affect the fibrin(ogen) unfolding mechanism because they do not possess compact structures detectable by X-ray crystallography. Hence, these parts are not expected to generate force signals, although, potentially, they might affect unfolding scenarios indirectly.
The αC region has been suggested to be involved in fibrin unfolding and there is considerable experimental evidence that the αC regions play an important role in determining the mechanical properties of fibrin clots (Collet et al., 2005; Houser et al., 2010; Tsurupa et al., 2009; Doolittle and Kollman, 2006; Falvo et al., 2008). Clots made from α251 fibrinogen are considerably less stiff than those made from normal fibrinogen and show more plastic deformation (Collet et al., 2005). Furthermore, the mechanical properties of individual fibers made from fibrinogen of different animal species are strongly dependent on the lengthof the αC regions of their fibrin (Falvo et al., 2008, 2010). Finally, several models for the mechanical properties of fibrin suggest a role for the αC regions in the observed macroscopic stiffness (Falvo et al., 2010; Purohit et al., 2011). We tested the potential importance of this portion of the molecule by probing monomeric Fg α251 and oligomeric Fg I-9, fibrinogen derivatives without the αC region (Collet et al., 2005; Mosesson and Sherry, 1966). The AFM unfolding patterns of both truncated fibrinogen variants were indistinguishable from the full-length Fg, indicating that the αC regions are not involved in the unfolding of fibrinogen monomers or single-chain oligomers. However, there is no conflict between the literature and experiments reported here, because the effects of this part of the molecule are most likely to be mediated through αC-αC interactions, which are present in the fibers/clots but not in Fg or Fgn. In addition to the αC-αC interaction, there are at least two more important features of a fibrin oligomer not considered in this study, which are the A:a and B:b knob-hole interactions. Further work is in progress with more complex molecular models, such as double-stranded fibrin oligomers containing the knob-hole bonds and the αC-polymers.
The distance x‡ between the minimum (folded state) and the transition state barrier (maximum) in the free energy landscape is an important molecular characteristic of mechanical unfolding of proteins. We estimated x‡ for fibrinogen unfolding by analyzing transient structures of the D region (γ-nodule-β-nodule complex) for the folded state and the transition state for each transition of type 1–3. We found that these transitions were characterized by a long x‡ ≈ 1 nm. We tested whether intermolecular interactions in oligomeric and polymeric fibrin could serve as an additional source of mechanical stability. Because the D-D interface was found to open first at an average force of ~40 pN, which resulted in an average extension of ~5nm (Figure 5), this transition does not seem to alter the mechanics and kinetic pathways of unfolding of fibrin(ogen) monomers and oligomers. Hence, most likely, individual fibrin(ogen) molecules forming fibrin(ogen) polymers unfold independently.
Our results are directly comparable to the parameters of fibrin stretching observed in the experiments on single fibrin fibers, in which the estimated mechanical force exerted on fibrin(ogen) monomer was found to be about 140 pN (Liu et al., 2010). This is a force large enough to unfold the γ-nodules and partially stretch the coiled-coils, as inferred from our AFM experiments and pulling simulations (Figures 2A and 3A). According to our data, at tensile forces of ~125-165 pN the fibrin(ogen) monomer is extended by ~160-170 nm (Figure 1), which is 360%–380% extension of the initial 45 nm length of the molecule. Hence, taking into account the half-staggered geometry of fibrin protofibrils forming a single fibrin fiber, we find that in a fibrin fiber at 100% strain, either a fraction (~26%–28%) of fibrin monomers forming fibrin protofibrils are fully unfolded, or all fibrin units are partially stretched on average by ~45 nm (100%-elongation), or elongation of fibrin fibers is a combination of these states.
Distinguishing between these possibilities precisely is challenging, but previous small angle X-ray scattering measurements on stretched fibrin clots (Brown et al., 2009) show that even at strains up to 1.0, there remains a population of molecules with the characteristic 22.5 nm spacing of folded half-staggered fibrin. This observation is inconsistent with significant gradual extension of the coiled-coils because this would lead to an increase in this spacing and therefore a shift in the X-ray diffraction peak. Although there was no shift in the diffraction pattern, there was an increase in the peak width indicative of an increase in sample disorder. This was attributed to a two-state like unfolding of the coiled-coils, but our new results suggest the alternative interpretation that the stretching and increased disorder are primarily due to unfolding of domains within the γ-nodules. Because coiled-coils in other systems have previously been shown to refold rapidly (Schwaiger et al., 2002), the unfolding of globular domains in the γ-nodule might also explain the slow refolding observed after fibrin clot relaxation (Brown et al., 2009).
These results are essential for understanding the mechanical properties of blood clots, where it has been demonstrated that unfolding is necessary to account for fibrin’s unusual properties. According to a recent constitutive model for the mechanical properties of fibrin, initially fibrin stiffness is small because early stages of tensile deformation are accommodated primarily by alignment of fibers along the direction of strain (Brown et al., 2009; Purohit et al., 2011). However, significant amounts of unfolding start at strains of about 0.15–0.2, and strain stiffening occurs at strains around 1.0 because of chain stretching. Direct experimental evidence for unfolding of fibrin in clots comes from both observation of changes in X-ray fiber diffraction from a pattern consistent with an α-helical coiled-coil to β sheet transition (Bailey et al., 1943) and measurements of changes in staining by Congo Red, specific for β sheet structure (Purohit et al., 2011). All of these results are entirely consistent with the new results on unfolding of fibrin(ogen) presented here. The general features of the simulated atomic force microscopy unfolding experiment derived from a discrete version of the model (Purohit et al., 2011) are qualitatively in agreement with these results, but the peak forces are smaller, because the calculations assume equilibrium with no rate dependence, and the distances are slightly different because of the model parameters.
The mechanical unfolding of the D region and full-length fibrinogen monomer is mostly an enthalpy driven process (Table 2). The enthalpy required to disrupt the network of native contacts stabilizing the D region is ΔHD ≈ 692 kcal/mol, which is the sum of ΔH1 ≈ 257 kcal/mol (type 1 transition), ΔH2 ≈ 218 kcal/mol kcal/mol (type 2 transition), and ΔH3 ≈ 217 kcal/mol (type 3 transition). The enthalpy change associated with unraveling of the D regions is roughly half the value for the full-length fibrinogen monomer (ΔHFg = 1520 kcal/mol). We found that ΔH accounts for ~90% of the change in Gibbs free energy (ΔG) for the D-dimer and Fg, whereas the entropic contribution (TΔS) is only ~10% (Table 2). The entropy change for forced elongation of the coiled-coils accounts for ~25% of ΔG. Hence, most of the energy change is due to unraveling of the D region, and only ~10% is due to elongation of the coiled-coils.
To conclude, we have provided important information and structural characteristics of the dynamic mechanical behavior of fibrin(ogen) at different spatial scales. The molecular mechanism of fibrin(ogen) elongation is shown to be based on the stepwise unraveling of the γ-nodules concomitant with partial stretching and contraction of the coiled-coil connectors. The adjacent β-nodule provides stabilization for the γ-nodule, and, because interdomain interactions at the D-D junction are weak, individualfibrin(ogen) monomers undergo independent unfolding transitions in fibrin(ogen) oligomers and polymers. The GPU-based computational acceleration makes direct comparison of the experimental and simulation results of dynamic force measurements possible. Hence, dynamic signatures for unfolding transitions observed in silico can be used to provide meaningful interpretation and modeling of the force peaks obtained in vitro and to unmask unfolding mechanisms.
EXPERIMENTAL PROCEDURES
Atomic Force Microscopy
Details of sample preparation are given in Section I.1 in Supplemental Experimental Procedures. For the AFM experiments, 50 μl of a 50 μg/ml fibrinogen monomer or oligomerized fibrinogen solution in 20 mM HEPES buffer (pH 7.4) containing 100 mM NaCl and 3 mM CaCl2 were pipetted onto freshly cleaved mica and allowed to adsorb for 10 minutes before being rinsed three times with 50 μl of buffer. Force-extension curves were collected using a Multimode AFM with a Nanoscope E controller (Digital Instruments, Santa Barbara, CA) and MLCT silicon nitride cantilevers (Veeco, Santa Barbara, CA). The cantilever spring constant was calibrated using the thermal fluctuation method (Hutter and Bechhoefer, 1993).
Self-Organized Polymer (SOP) Model
The SOP model (Hyeon et al., 2006a) has proved to describe well the mechanical properties of proteins, including green fluorescent protein (Mickler et al., 2007), and kinesin (Hyeon and Onuchic, 2007). Each residue is described by a single interaction center (Cα-atom). The potential energy function of the protein conformation USOP specified in terms of the coordinates {r} = r1, r2, …, rN (N is the total number of residues) is given by . The finite extensible nonlinear elastic (FENE) potential with the spring constant k = 14 N/m and the tolerance in the change of a covalent bond distance R0 = 2 Å describes the backbone chain connectivity. The distance between residues i and i+1, is ri,i+1, and r0i,i+1 is its value in the native (PDB) structure. We used the Lennard-Jones potential to account for the noncovalent interactions that stabilize the native folded state. We assumed that if the noncovalently linked residues i and j (∣i-j∣ > 2) are within the cut-off distance RC =8 Å in the native state, then Δij = 1, and zero otherwise. The value of εn quantifies the strength of the nonbonded interactions. All the nonnative interactions ( are treated as repulsive. A constraint is imposed on the bond angle formed by residues i, i+1, and i+2 by including the repulsive potential with parameters εr = 1 kcal/mol and σ = 3.8 Å, which determine the strength and the range of the repulsion.
Pulling Simulations on a GPU
Graphics processing units (GPUs) offer a significant gain in the computational speed compared to more traditional simulations on central processors (Zhmurov et al., 2011; Stone et al., 2007; Anderson et al., 2008; Friedrichs et al., 2009). We utilized the SOP model and Langevin simulations (Dima and Joshi, 2008; Veitshans et al., 1997; Zhmurov et al., 2010b) on a GPU (SOP-GPU package; Section II.3 in Supplemental Experimental Procedures) (Zhmurov et al., 2010a). It took ~10 days of “GPU-time” to generate five 0.2 s trajectories (5 3 109 simulation steps) for the fibrinogen monomer (N = 1913 amino acids) on a single GPU GeForce GTX 480 (NVidia) using the many-runs-per-GPU approach (Zhmurov et al., 2010a). For comparison, it would take ~5 years to obtain 5 trajectories on a single CPU core Intel Xeon E5440. The unfolding dynamics at T = 300K were obtained by integrating the Langevin equations for each Cα-particle position ri, i = 1,2,…,N, in the overdamped limit, ηdri/dt = - ∂U/∂ri + gi(t). Here, U=USOP − fX is the total potential energy, which includes a contribution from an applied pulling force (fX), gi is the Gaussian random force, and η is the friction coefficient. To mimic the force-ramp measurement for each system, the N-terminus of the molecule was constrained, and a time-dependent force f(t) = rf t (rf is the force-loading rate) was applied to the C-terminal along the end-to-end vector X (Section II.1 in Supplemental Experimental Procedures). Equations of motion were propagated with the time step Δt =40 ps (Veitshans et al., 1997). Numerical values of εn were determined using all-atom MD simulations (Section II.2 in Supplemental Experimental Procedures). Pulling simulations were performed using bulk water viscosity (η = 7.0 3 105 pN ps/nm) and the experimental value of the cantilever spring constant κ = 70 pN/nm and pulling speed νf = 1.0 μm/s (rf = κνf = 70 nN/s). The SOP model parametrization is presented in Table S1; the computational performance is summarized in Table S2. Simulated force-extension curves are analyzed in Section II.4 in Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
Fibrinogen α251 was a generous gift of Susan T. Lord, and fibrinogen fraction I-9 was kindly provided by Michael W. Mosesson. The authors thank Leonid Medved for his insight and valuable comments. This work was supported by the American Heart Association (grant 09SDG2460023 to V.B.), the Russian Ministry of Education and Science (grant 02-740-11-5126 to V.B.), the National Science Foundation (grant MCB-0845002 to R.I.D.), and the National Institutes of Health (grants HL030954 and HL090774 to J.W.W.).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes three tables, three figures, one movie, and Supplemental Experimental Procedures and can be found with this article online at doi:10.1016/j.str.2011.08.013.
REFERENCES
- Anderson JA, Lorenz CD, Travesset A. General purpose molecular dynamics simulations fully implemented on graphics processing units. J. Comp. Phys. 2008;227:5342–5359. 10.1016/j.jcp.2008.01.047. [Google Scholar]
- Averett LE, Schoenfisch MH, Akhremitchev BB, Gorkun OV. Kinetics of the multistep rupture of fibrin ‘A-a’ polymerization interactions measured using atomic force microscopy. Biophys. J. 2009;97:2820–2828. doi: 10.1016/j.bpj.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averett LE, Geer CB, Fuierer RR, Akhremitchev BB, Gorkun OV, Schoenfisch MH. Complexity of “A-a” knob-hole fibrin interaction revealed by atomic force spectroscopy. Langmuir. 2008;24:4979–4988. doi: 10.1021/la703264x. [DOI] [PubMed] [Google Scholar]
- Bailey K, Astbury WT, Rudall KM. Fibrinogen and fibrin as members of the keratin-myosin group. Nature. 1943;151:716–717. [Google Scholar]
- Barsegov V, Klimov DK, Thirumalai D. Mapping the energy landscape of biomolecules using single molecule force correlation spectroscopy: theory and applications. Biophys. J. 2006;90:3827–3841. doi: 10.1529/biophysj.105.075937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AEX, Litvinov RI, Discher DE, Weisel JW. Forced unfolding of coiled-coils in fibrinogen by single-molecule AFM. Biophys. J. 2007;92:L39–L41. doi: 10.1529/biophysj.106.101261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AEX, Litvinov RI, Discher DE, Purohit PK, Weisel JW. Multiscale mechanics of fibrin polymer: gel stretching with protein unfolding and loss of water. Science. 2009;325:741–744. doi: 10.1126/science.1172484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Volkmann N, Jun G, Henschen-Edman AH, Cohen C. The crystal structure of modified bovine fibrinogen. Proc. Natl. Acad. Sci. USA. 2000;97:85–90. doi: 10.1073/pnas.97.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bura E, Zhmurov A, Barsegov V. Nonparametric density estimation and optimal bandwidth selection for protein unfolding and unbinding data. J. Chem. Phys. 2009;130:015102–015117. doi: 10.1063/1.3050095. [DOI] [PubMed] [Google Scholar]
- Carlisle CR, Sparks EA, Der Loughian C, Guthold M. Strength and failure of fibrin fiber branchpoints. J. Thromb. Haemost. 2010;8:1135–1138. doi: 10.1111/j.1538-7836.2010.03824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet J-P, Moen JL, Veklich YI, Gorkun OV, Lord ST, Montalescot G, Weisel JW. The alphaC domains of fibrinogen affect the structure of the fibrin clot, its physical properties, and its susceptibility to fibrinolysis. Blood. 2005;106:3824–3830. doi: 10.1182/blood-2005-05-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz H, Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. USA. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dima RI, Joshi H. Probing the origin of tubulin rigidity with molecular simulations. Proc. Natl. Acad. Sci. USA. 2008;105:15743–15748. doi: 10.1073/pnas.0806113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle RF, Kollman JM. Natively unfolded regions of the vertebrate fibrinogen molecule. Proteins. 2006;63:391–397. doi: 10.1002/prot.20758. [DOI] [PubMed] [Google Scholar]
- Falvo MR, Gorkun OV, Lord ST. The molecular origins of the mechanical properties of fibrin. Biophys. Chem. 2010;152:15–20. doi: 10.1016/j.bpc.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falvo MR, Millard D, O’Brien ET, 3rd, Superfine R, Lord ST. Length of tandem repeats in fibrin’s alphaC region correlates with fiber extensibility. J. Thromb. Haemost. 2008;6:1991–1993. doi: 10.1111/j.1538-7836.2008.03147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichs MS, Eastman P, Vaidyanathan V, Houston M, Legrand S, Beberg AL, Ensign DL, Bruns CM, Pande VS. Accelerating molecular dynamic simulation on graphics processing units. J. Comput. Chem. 2009;30:864–872. doi: 10.1002/jcc.21209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthold M, Liu W, Sparks EA, Jawerth LM, Peng L, Falvo M, Superfine R, Hantgan RR, Lord ST. A comparison of the mechanical and structural properties of fibrin fibers with other protein fibers. Cell Biochem. Biophys. 2007;49:165–181. doi: 10.1007/s12013-007-9001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser JR, Hudson NE, Ping L, Brien ET, 3rd, Superfine R, Lord ST, Falvo MR. Evidence that αC region is origin of low modulus, high extensibility, and strain stiffening in fibrin fibers. Biophys. J. 2010;99:3038–3047. doi: 10.1016/j.bpj.2010.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson NE, Houser JR, O’Brien ET, 3rd, Taylor RM, 2nd, Superfine R, Lord ST, Falvo MR. Stiffening of individual fibrin fibers equitably distributes strain and strengthens networks. Biophys. J. 2010;98:1632–1640. doi: 10.1016/j.bpj.2009.12.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter JL, Bechhoefer J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993;64:1868–1873. 10.1063/1.1143970. [Google Scholar]
- Hyeon C, Onuchic JN. Internal strain regulates the nucleotide binding site of the kinesin leading head. Proc. Natl. Acad. Sci. USA. 2007;104:2175–2180. doi: 10.1073/pnas.0610939104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyeon C, Dima RI, Thirumalai D. Pathways and kinetic barriers in mechanical unfolding and refolding of RNA and proteins. Structure. 2006a;14:1633–1645. doi: 10.1016/j.str.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Hyeon C, Lorimer GH, Thirumalai D. Dynamics of allosteric transitions in GroEL. Proc. Natl. Acad. Sci. USA. 2006b;103:18939–18944. doi: 10.1073/pnas.0608759103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim BBC, Lee EH, Sotomayor M, Schulten K. Molecular basis of fibrin clot elasticity. Structure. 2008;16:449–459. doi: 10.1016/j.str.2007.12.019. [DOI] [PubMed] [Google Scholar]
- Liu W, Carlisle CR, Sparks EA, Guthold M. The mechanical properties of single fibrin fibers. J. Thromb. Haemost. 2010;8:1030–1036. doi: 10.1111/j.1538-7836.2010.03745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Jawerth LM, Sparks EA, Falvo MR, Hantgan RR, Superfine R, Lord ST, Guthold M. Fibrin fibers have extraordinary extensibility and elasticity. Science. 2006;313:634. doi: 10.1126/science.1127317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrazo J, Brown JH, Litvinovich S, Dominguez R, Yakovlev S, Medved L, Cohen C. Crystal structure of the central region of bovine fibrinogen (E5 fragment) at 1.4-A resolution. Proc. Natl. Acad. Sci. USA. 2001;98:11967–11972. doi: 10.1073/pnas.211439798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medved L, Tsurupa G, Yakovlev S. Conformational changes upon conversion of fibrinogen into fibrin: the mechanisms of exposure of cryptic sites. Ann. NY Acad. Sci. 2001;936:185–204. doi: 10.1111/j.1749-6632.2001.tb03505.x. [DOI] [PubMed] [Google Scholar]
- Mickler M, Dima RI, Dietz H, Hyeon C, Thirumalai D, Rief M. Revealing the bifurcation in the unfolding pathways of GFP by using single-molecule experiments and simulations. Proc. Natl. Acad. Sci. USA. 2007;104:20268–20273. doi: 10.1073/pnas.0705458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalyi E, Donovan JW. Clotting of fibrinogen. 2. Calorimetry of the reversal of the effect of calcium on clotting with thrombin and with ancrod. Biochemistry. 1985;24:3443–3448. doi: 10.1021/bi00335a008. [DOI] [PubMed] [Google Scholar]
- Mosesson MW, Sherry S. The preparation and properties of human fibrinogen of relatively high solubility. Biochemistry. 1966;5:2829–2835. doi: 10.1021/bi00873a008. [DOI] [PubMed] [Google Scholar]
- Piechocka IK, Bacabac RG, Potters M, Mackintosh FC, Koenderink GH. Structural hierarchy governs fibrin gel mechanics. Biophys. J. 2010;98:2281–2289. doi: 10.1016/j.bpj.2010.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalov PL, Medved LV. Domains in the fibrinogen molecule. J. Mol. Biol. 1982;159:665–683. doi: 10.1016/0022-2836(82)90107-3. [DOI] [PubMed] [Google Scholar]
- Purohit PK, Litvinov RI, Brown AEX, Discher DE, Weisel JW. Protein unfolding accounts for the unusual mechanical behavior of fibrin networks. Acta Biomater. 2011;7:2374–2383. doi: 10.1016/j.actbio.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman EP, Barsegov V, Klimov DK. Folding of tandem-linked domains. Proteins. 2007;67:795–810. doi: 10.1002/prot.21339. [DOI] [PubMed] [Google Scholar]
- Schwaiger I, Sattler C, Hostetter DR, Rief M. The myosin coiled-coil is a truly elastic protein structure. Nat. Mater. 2002;1:232–235. doi: 10.1038/nmat776. [DOI] [PubMed] [Google Scholar]
- Spraggon G, Everse SJ, Doolittle RF. Crystal structures of fragment D from human fibrinogen and its crosslinked counterpart from fibrin. Nature. 1997;389:455–462. doi: 10.1038/38947. [DOI] [PubMed] [Google Scholar]
- Stone JE, Phillips JC, Freddolino PL, Hardy DJ, Trabuco LG, Schulten K. Accelerating molecular modeling applications with graphics processors. J. Comput. Chem. 2007;28:2618–2640. doi: 10.1002/jcc.20829. [DOI] [PubMed] [Google Scholar]
- Tehver R, Thirumalai D. Rigor to post-rigor transition in myosin V: link between the dynamics and the supporting architecture. Structure. 2010;18:471–481. doi: 10.1016/j.str.2010.01.019. [DOI] [PubMed] [Google Scholar]
- Tsurupa G, Hantgan RR, Burton RA, Pechik I, Tjandra N, Medved L. Structure, stability, and interaction of the fibrin(ogen) alphaC-domains. Biochemistry. 2009;48:12191–12201. doi: 10.1021/bi901640e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitshans T, Klimov DK, Thirumalai D. Protein folding kinetics: timescales, pathways and energy landscapes in terms of sequence-dependent properties. Fold. Des. 1997;2:1–22. doi: 10.1016/S1359-0278(97)00002-3. [DOI] [PubMed] [Google Scholar]
- Weisel JW. The mechanical properties of fibrin for basic scientists and clinicians. Biophys. Chem. 2004;112:267–276. doi: 10.1016/j.bpc.2004.07.029. [DOI] [PubMed] [Google Scholar]
- Weisel JW. Fibrinogen and fibrin. Adv. Protein Chem. 2005;70:247–299. doi: 10.1016/S0065-3233(05)70008-5. [DOI] [PubMed] [Google Scholar]
- Weisel JW. Biophysics: enigmas of blood clot elasticity. Science. 2008;320:456–457. doi: 10.1126/science.1154210. [DOI] [PubMed] [Google Scholar]
- Yakovlev S, Loukinov D, Medved L. Structural and functional role of the β-strand insert (g 381-390) in the fibrinogen γ-module: a “pull out” hypothesis. Ann. N Y Acad. Sci. 2001;936:122–124. [PubMed] [Google Scholar]
- Yakovlev S, Litvinovich S, Loukinov D, Medved L. Role of the β-strand insert in the central domain of the fibrinogen γ-module. Biochemistry. 2000a;39:15721–15729. doi: 10.1021/bi001836h. [DOI] [PubMed] [Google Scholar]
- Yakovlev S, Makogonenko E, Kurochkina N, Nieuwenhuizen W, Ingham K, Medved L. Conversion of fibrinogen to fibrin: mechanism of exposure of tPA- and plasminogen-binding sites. Biochemistry. 2000b;39:15730–15741. doi: 10.1021/bi001847a. [DOI] [PubMed] [Google Scholar]
- Yee VC, Pratt KP, Côté HCF, Trong IL, Chung DW, Davie EW, Stenkamp RE, Teller DC. Crystal structure of a 30 kDa C-terminal fragment from the γ chain of human fibrinogen. Structure. 1997;5:125–138. doi: 10.1016/s0969-2126(97)00171-8. [DOI] [PubMed] [Google Scholar]
- Zhmurov A, Dima RI, Barsegov V. Order statistics theory of unfolding of multimeric proteins. Biophys. J. 2010b;99:1959–1968. doi: 10.1016/j.bpj.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhmurov A, Dima RI, Kholodov Y, Barsegov V. Sop-GPU: accelerating biomolecular simulations in the centisecond timescale using graphics processors. Proteins. 2010a;78:2984–2999. doi: 10.1002/prot.22824. [DOI] [PubMed] [Google Scholar]
- Zhmurov A, Rybnikov K, Kholodov Y, Barsegov V. Generation of random numbers on graphics processors: forced indentation in silico of the bacteriophage HK97. J. Phys. Chem. B. 2011;115:5278–5288. doi: 10.1021/jp109079t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.