Cell polarity is a fundamental feature of all cell types and is essential for cell division, cell migration, and vectorial transport of cell fate determinants within a cell (1). In recent years, application of cytological methods to study protein, chromosome, and episome localization in chemically fixed and live bacterial cells has signaled the advent of prokaryotic cell biology and has provided us with new, sometimes startling, imagery of the spatial organization within a bacterial cell (2). Proteins are found to be positioned at specific sites in bacteria such as the cell pole, the cell equator, and the division septum. Such site-specific protein targeting appears to be central to the regulation of DNA replication, chromosome segregation, cell division, cell differentiation, and chemotaxis. Recent studies on a group of proteins in Escherichia coli, termed Min, have revealed an extraordinary oscillatory behavior that governs their cellular location. The dynamic behavior of the Min proteins creates transient domains of subcellular asymmetry in the E. coli cell and such cell polarization is critical to the ability of the Min system to ensure that a cell divides unerringly in the middle. In a recent issue of PNAS, Fu et al. (3) describe a remarkable cell polarization feature of the MinE protein that localizes as an off-center ring (E-ring) and as a polar zone (PZ) that extends from the ring to the proximal cell pole. Time-lapse microscopy of live E. coli cells expressing a MinE-green fluorescent protein fusion reveals that the membrane-associated E-ring and the PZ form a mobile unit, with the PZ shrinking as the E-ring moves toward the proximal pole. Upon reaching the polar extremity, the PZ and the ring disappear, only to reappear at the opposite pole. The sequence of MinE assembly, poleward movement, dispersion, and reassembly is repeated many times in each cell division cycle, with a pole-to-pole oscillation frequency of ≈2–3 min. To put such protein acrobatics in perspective, we summarize current knowledge of the Min system and discuss the major unanswered questions concerning the polarization process and division site selection in bacterial cells.
Unraveling the mechanisms involved in the acrobatic behavior and localization of the Min proteins promises to illuminate our understanding of a remarkable mechanism for generating and maintaining cell polarity.
The site specificity of cell division occurring at the equator of E. coli is regulated by the products of the min locus: MinC, MinD, and MinE (4). Abrogation of the Min system causes a large fraction of cells to divide near the poles instead of the middle, creating one viable bacterium with two copies of the genome and one “minicell” that is devoid of DNA. MinC and MinD, together, form a binary inhibitor of cell division that can act nonspecifically at all sites throughout the cell. Recently, a biologically active MinD- green fluorescent protein fusion protein was found to localize as a membrane-associated PZ in half of the cell in a MinE-dependent manner and to undergo an extremely rapid (periodicity being only tens of seconds) pole-to-pole oscillation in live E. coli cells (5). MinC also was shown to co-oscillate with MinD in the presence of MinE (6, 7). MinE provides topological specificity to cell division by antagonizing the inhibitory effect of MinCD at midcell. Thus, it seemed logical when Raskin and de Boer (8) found that a functional MinE-green fluorescent protein chimera localized to a ring-like structure (E-ring) at a site adjacent to the cell center in E. coli. Formation of the E-ring was shown to be independent of the essential division protein FtsZ that assembles into a distinct cytokinetic ring (Z-ring) at midcell. This finding implied that the E-ring was stationary near midcell and could somehow shield Z-ring assembly at the medial division site from being disrupted by the MinCD complex. This view now changes with the demonstration by Fu et al. (3) that the off-center E-ring is a mobile structure tracking toward the proximal cell pole that also harbors the MinCD PZ. It is evident that the poleward migration of the E-ring stimulates the retraction of the MinCD PZ, which then rapidly relocates to the opposite cell pole.
The 88-aa MinE protein contains two distinct functional domains: the N terminus (residues 1–32) is responsible for the anti-MinCD function, whereas the C terminus (residues 32–88) is the topological specificity domain (TSD) that presumably tethers the protein near the cell center (9). Recently, the solution structure of the MinE TSD was solved and was shown to be an antiparallel homodimer, forming a novel α-helix–β-sheet sandwich (9). The antiparallel arrangement of the TSD monomers suggested that the N-terminal anti-MinCD domains may project on either side of a MinE dimer. On this basis, King, Rothfield, and colleagues (9) proposed a model wherein the bipolar orientation of the dimers in the E-ring, which was at the time presumed to be a static structure near midcell, could disrupt MinCD complexes as they approach the cell center in their oscillatory path. Based on their new findings that the E-ring is mobile, King, Rothfield, and colleagues (3) conclude that the postulated bipolar orientation of MinE dimers is unlikely to be the mechanistic basis for antagonizing MinCD action. Further work is required to determine the physiological importance of the antiparallel monomer arrangement in the MinE dimer.
The results of Fu et al. (3) raise many questions. How is the site adjacent to midcell chosen for E-ring assembly and what is the mechanism of assembly? How does the E-ring move vectorially and what signals its dissolution at a site close to the polar extremity? How do the PZs comprising MinE as well as MinCD shrink? What are the determinants for dissociation of MinCD or MinE from a pole and relocation to the opposite pole?
It was suggested previously that MinE could localize near the cell septum by interacting with a putative topological marker that functions as a midcell signpost (9). The new results of Fu et al. (3) argue against a stationary signpost. The authors have proposed two alternative models for E-ring assembly. In the first model, after MinD delivers MinE to the membrane (8), MinE is postulated to associate with a topological target or a specific receptor initiating assembly of the E-ring near mid-cell (3, 10). It remains unknown whether the topological marker itself or some other component confers mobility to the E-ring. In the second model, MinE molecules are postulated to concentrate at the medial edge of the MinE PZ, and thereby appear as a ring, without requiring association with a specific receptor. In this case, as the authors suggest, retraction of the PZ could stimulate E-ring movement. The first model seems more plausible because the ca. 200 molecules of the 88-aa MinE polypeptide present in an E. coli cell (11) are insufficient to generate a continuous ring around the cell circumference without being interspersed with one or more accessory components (10).
What might define the site for E-ring assembly? Regamey et al. (12) recently have shown that the midcell nucleation site for Z-ring assembly in Bacillus subtilis is occluded by the replisome until ca. 80% of chromosome replication is complete. Furthermore Yu and Margolin (13) previously had shown that a nucleoid-free zone at midcell is an important prerequisite for Z-ring assembly in E. coli. Similar spatial constraints also may regulate E-ring assembly, because E-rings form adjacent to Z-rings. A close look at the time lapse micrographs of Fu et al. (3) suggests the tantalizing possibility that the sites of assembly and dissolution of the E-ring may correspond to the medial and polar edges of a segregated nucleoid in half of a predivisional cell. Could the nucleoid edge provide a spatial cue for the off-center E-ring assembly? The E-ring could assemble at the medial edge of a nucleoid, perhaps using the stationary replisome as a reference point, and move toward the proximal pole to collapse at the polar edge of the nucleoid (Fig. 1).
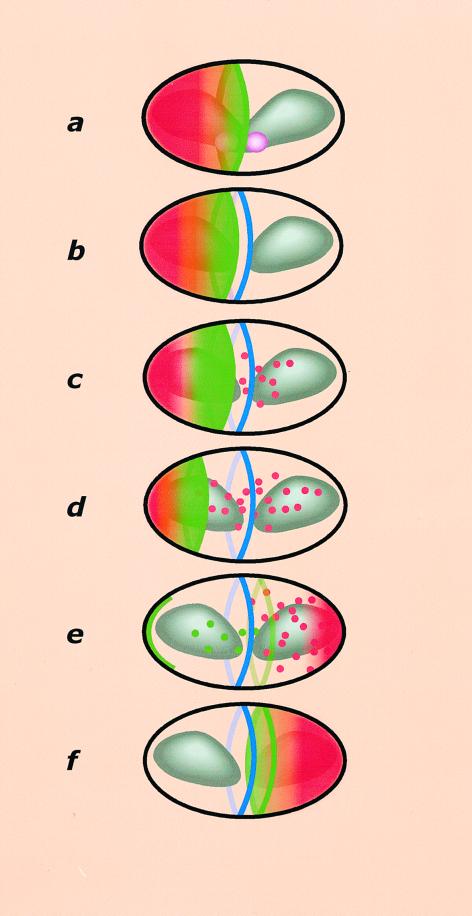
Figure 1.
A model for MinCD and MinE dynamics in E. coli. Representation of a single oscillation of MinCD and MinE. (a) Membrane-associated MinCD complexes (red) are present as a PZ at one end of the cell. MinE is present both as a ring structure (E-ring) near the cell center (green ring) and as a PZ (green shade) extending from the E-ring into the MinCD PZ. The nucleoid is shown as a bi-lobed structure (gray) in the final stages of replication. The associated replisomes are illustrated as two spheres (purple) flanking the cell center. The space between the replicated nucleoids, specifically the medial edge of a nucleoid, is depicted to be the site for E-ring assembly (see text). (b) The FtsZ protein assembles as a ring (Z-ring in blue) at the center of the cell distinct from the E-ring. (c and d) The E-ring and its PZ are shown migrating toward the pole containing MinCD. The poleward front of the MinE PZ dislodges MinCD from the cell membrane, resulting in the shrinkage of the MinCD PZ and relocation of MinCD complexes (red spheres) to the opposite end of the cell. (e) The E-ring and the MinE PZ are shown collapsed into a cortical band at the cell pole (green arc). Free MinE (green spheres) is shown forming a new ring in the opposite half of the cell, stimulated by a newly established MinCD PZ (see text). (f) End of a single MinE oscillation, which is repeated every 2–3 min.
Fu et al. (3) suggest that vectorial movement of the E-ring, culminating in the appearance of MinE as a cortical band at the pole, serves to dislodge MinCD complexes and force their relocation to the opposite pole (Fig. 1). Surprisingly, a truncated MinE variant, containing only the anti-MinCD domain, which is unable to assemble into an E-ring, is capable of supporting MinD oscillation (14). However, the frequency of MinD oscillation in this case is significantly slower, resulting in inappropriate polar septation. Thus, the assembly and movement of the E-ring appear to be important determinants for the fidelity of midcell septation.
How does MinD promote E-ring assembly and MinE oscillation? MinD is believed to bring MinE to the membrane (8). It is possible that MinD then hands off MinE to receptors that are accessible in the internucleoid zone at midcell, thus helping to initiate E-ring assembly. How does the E-ring move and does MinD play a role in the movement? Although MinD is likely to provide the driving force for its own oscillation along with that of MinC, via its ATPase activity (10, 15), it is unlikely to catalyze E-ring movement because there is no prolonged contact between the E-ring and the MinD PZ. The E-ring may not be a closed structure but could have ends for capture and release of subunits by a treadmilling mechanism. Treadmilling coupled with adherence of the E-ring to the membrane surface could provide the directionality for its poleward migration. Treadmilling of actin filaments plays an active role in cell motility (16). Similarly, treadmilling is believed to power the movement of cortical actin patches in nonmotile budding yeast (17, 18).
A key observation in the Fu et al. (3) paper is that the MinE and MinD PZs colocalize to the same end of the cell because only half of the cell was fluorescent when both proteins fused to green fluorescent protein, were coexpressed. This implied that oscillation of MinE and MinD is synchronous. There is, however, a striking disparity in the reported oscillation frequency of MinD (average 20 sec; ref. 5) relative to that of MinE (128 ± 39 sec). Moreover, Fu et al. (3) observed that a nascent E-ring forms in the distal half of a significant fraction (25%) of cells before disappearance of the original E-ring at the other cell pole. This finding strongly suggests that MinD must be present at the distal pole to promote E-ring assembly. The faster oscillation exhibited by MinD may allow it to precede MinE to the opposite pole. Labeling of MinD and MinE with different fluorophores, which should allow their oscillations in the cell to be followed in real time, can resolve this issue.
Unraveling the mechanisms involved in the acrobatic behavior and localization of the Min proteins promises to illuminate our understanding of a remarkable mechanism for generating and maintaining cell polarity.
Acknowledgments
We thank the National Institutes of Health (A.W.) and the U.S. Defense Advanced Research Projects Agency (D.R.C.) for supporting our work.
Footnotes
See companion article on page 980 in issue 3 of volume 98.
References
- 1.Drubin D, editor. Cell Polarity. Oxford: Oxford Univ. Press; 2000. [Google Scholar]
- 2.Shapiro L, Losick R. Cell. 2000;100:89–98. doi: 10.1016/s0092-8674(00)81686-4. [DOI] [PubMed] [Google Scholar]
- 3.Fu X, Shih Y-L, Yan Z, Rothfield L. Proc Natl Acad Sci USA. 2001;98:980–985. doi: 10.1073/pnas.031549298. . (First Published January 23, 2001; 10.1073/pnas.031549298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Boer P A, Crossley R E, Rothfield L I. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 5.Raskin D M, de Boer P A. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raskin D M, de Boer P A. J Bacteriol. 1999;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z, Lutkenhaus J. Mol Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- 8.Raskin D M, de Boer P A. Cell. 1997;91:685–694. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 9.King G, Shih Y-L, Maciejewski M, Bains N, Pan B, Rowland S L, Mullen G, Rothfield L I. Nat Struct Biol. 2000;7:1013–1017. doi: 10.1038/80917. [DOI] [PubMed] [Google Scholar]
- 10.RayChaudhuri D, Gordon G, Wright A. Nat Struct Biol. 2000;7:997–999. doi: 10.1038/81014. [DOI] [PubMed] [Google Scholar]
- 11.Zhao C R, de Boer P A, Rothfield L I. Proc Natl Acad Sci USA. 1995;92:4313–4317. doi: 10.1073/pnas.92.10.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Regamey A, Harry E J, Wake R G. Mol Microbiol. 2000;38:423–434. doi: 10.1046/j.1365-2958.2000.02130.x. [DOI] [PubMed] [Google Scholar]
- 13.Yu X C, Margolin W. Mol Microbiol. 1999;32:315–326. doi: 10.1046/j.1365-2958.1999.01351.x. [DOI] [PubMed] [Google Scholar]
- 14.Rowland S L, Fu X, Sayed M A, Zhang Y, Cook W R, Rothfield L I. J Bacteriol. 2000;182:613–619. doi: 10.1128/jb.182.3.613-619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Boer P A, Crossley R E, Hand A R, Rothfield L I. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlier M F. Curr Opin Cell Biol. 1998;10:45–51. doi: 10.1016/s0955-0674(98)80085-9. [DOI] [PubMed] [Google Scholar]
- 17.Doyle T, Botstein D. Proc Natl Acad Sci USA. 1996;93:3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waddle J A, Karpova T S, Waterston R H, Cooper J A. J Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]