Abstract
VEGF is a pivotal pro-angiogenic growth factor and its dosage decisively impacts vascularization. We recently identified a CTCF-dependent chromatin insulator that critically restrains the transcriptional induction of VEGF and angiogenesis. We postulate that CTCF may exert enhancer blocking by mediating chromatin looping and/or RNA polymerase pausing at the VEGF locus.
Keywords: angiogenesis, boundary, chromatin looping, CTCF, enhancer blocking, insulator, polymerase pausing, VEGF
A chromatin insulator that restrains VEGF transcription and angiogenesis
Angiogenesis refers to new capillary growth from pre-existing blood vessels. Vascular formation is a fundamental physiological process that is tightly controlled by a fine balance between angiogenic inducers and inhibitors. Deregulation of angiogenesis is implicated in a number of pathologic states.1 In cancer, the balance between pro- and anti-angiogenic activities shifts toward a more angiogenic state, resulting in excessive and sustained synthesis of new vasculature, which facilitates the growth and spread of cancer cells. Vascular endothelial growth factor A (VEGF-A or simply VEGF) is a potent pro-angiogenic factor and a key molecule that orchestrates the formation and function of vascular networks.2 VEGF induces endothelial cell proliferation, migration, and vessel sprouting. The action of VEGF is dose-dependent and excess VEGF is sufficient to cause abnormal neovascularization. The importance of VEGF in angiogenesis is highlighted by the fact that targeting VEGF has been the major approach of anti-angiogenesis therapy.3 Advancing our understanding of the regulatory mechanisms central to angiogenesis, including control of VEGF levels, will provide useful insights into potential manipulation of the process for therapeutic gains. VEGF transcription is dynamically induced in response to a variety of stimuli, such as hypoxia and estrogens. A number of transcription factors, including the hypoxia-inducible factor (HIF), are activated under these conditions and bind to enhancer elements of the VEGF gene to stimulate its transcription.4-6 However, very little is known about potential mechanisms that may confine the induction of VEGF within an appropriate magnitude for proper angiogenesis. Because a precise dose of VEGF is critical for vascularization, disruption of such mechanisms may contribute to pathological angiogenesis.
Chromatin insulators are regulatory DNA elements that partition the genome into independent chromatin domains and prevent inappropriate interactions between adjacent domains. When placed between enhancers and a promoter, insulators function as enhancer blockers to interfere with gene activation.7 The vertebrate zinc finger transcription factor CTCF is the most characterized insulator-binding protein that demonstrates enhancer blocking activity and is a key genome organizer.7,8 CTCF binding sites in the genome extensively overlap with boundaries between active and repressive chromatin domains.9 A global CTCF-mediated chromatin interactome study further validates that CTCF organizes the genome into epigenetically distinct domains by forming chromatin loops.10
We recently identified a CTCF-dependent insulator in the proximal promoter of VEGF.11 Both CTCF and its partner protein Cohesin bind to a -0.6kb site upstream of the transcription start site (TSS) of VEGF. According to the ENCODE Consortium chromatin immunoprecipitation (ChIP) database, binding of CTCF to this site is invariant in many human cell types and mouse cells (not shown). This insulator is positioned between the TSS and many upstream enhancers, including the hypoxia responsive element (HRE) that is located at -1kb and several distal estrogen responsive elements (EREs). The insulator restricts the action of HIF and estrogen receptor (ER) in a CTCF-dependent manner.11 In cells depleted of CTCF, induction of VEGF by hypoxia or estrogen becomes much more robust. During development of the retina, VEGF expression and angiogenesis is primarily activated by cellular oxygen tension (physiological hypoxia).12 In retinal progenitor-derived cells, VEGF is transcribed in the ganglion cell layer (GCL) and the inner nuclear layer (INL), but not in the outer nuclear layer (ONL) that is avascular.13 Interestingly, CTCF exhibits a similar expression pattern (not shown). Depletion of CTCF in retinal progenitors results in excess angiogenesis in vivo, most notably, ectopic vascular penetration into the ONL.11 This observation suggests that CTCF is present in VEGF-producing retinal cells where it limits the extent of VEGF induction by physiological cues, ensuring that VEGF is not overproduced. Taken together, CTCF acts as an enhancer blocker to restrain the induction of VEGF and govern normal angiogenesis. Consistently, several missense mutants of CTCF identified in cancer lose the enhancer blocking activity at the VEGF locus,11 which conceivably confer increased angiogenic potential on cancer cells.
Potential molecular bases underlying CTCF-mediated enhancer-blocking chromatin insulation
Chromatin insulator interferes with the communication between a promoter and enhancers. Several models have been proposed to explain the action of enhancer-blocking insulators, including the chromatin loop domain model and the promoter decoy model, which are not necessarily mutually exclusive.14 CTCF may use such mechanisms to dampen transcription of VEGF activated by enhancers.
CTCF-mediated chromatin loops interfere with enhancer-promoter communication
Gene activation can be stimulated by enhancer elements located far from promoters. Distal enhancers can physically interact with their cognate promoters, although the molecular mechanism responsible for the enhancer-promoter juxtapositions remains elusive.15 Three-dimensional genome topology has been increasingly recognized to play a key role in gene transcription.16 A popular model for how insulators may block enhancers is that insulator sites interact with each other and/or with nuclear structural elements to form chromatin loops, which may separate enhancers and promoters into topologically distinct domains.17 This may have a steric effect that blocks enhancers from contacting their designated promoters.
CTCF can interact with each other to form clusters and therefore create closed loop domains. Genome-wide analysis of CTCF-associated chromatin interactome demonstrates that a small fraction of the CTCF binding sites in the genome (less than 10%) mediate looping interactions, although it is unknown what governs the selection of such CTCF sites for pairing.10 In addition to the proximal promoter, the ENCODE ChIP studies have uncovered multiple CTCF binding sites at the VEGF locus, including those in the introns and far upstream regions (Fig. 1), which may lead to a variety of possible intrachromosomal loop conformations. Based on the CTCF-mediated interactome map in mouse embryonic stem (ES) cells,10 the -9kb region upstream of VEGF (corresponding to the -11kb CTCF site of the human VEGF gene) connects to the -100kb upstream site, which is in the RSPH9 gene. This intrachromosomal loop configuration probably secludes a couple EREs and other potential distal enhancers from the VEGF promoter, but may not affect proximal enhancers such as HRE.11 Moreover, CTCF also establishes interchromosomal contacts involving the VEGF locus in mouse ES cells.10 The -58kb region upstream of VEGF, which is located on mouse chromosome 17, interacts with the PPP2R5A gene (encoding for protein phosphatase 2 regulatory subunit B) located on chromosome 1; and the -45kb region of VEGF, where a non-coding RNA (AK142185) is transcribed, interacts with the myosin light chain kinase (MYLK) gene on chromosome 16. These interchromosomal interactions may render the participating genes co-localized in the nucleus to form certain “chromatin hubs.” It remains to be investigated whether these genes share common regulatory mechanisms and how they may modulate transcription of VEGF and the activity of its enhancers.
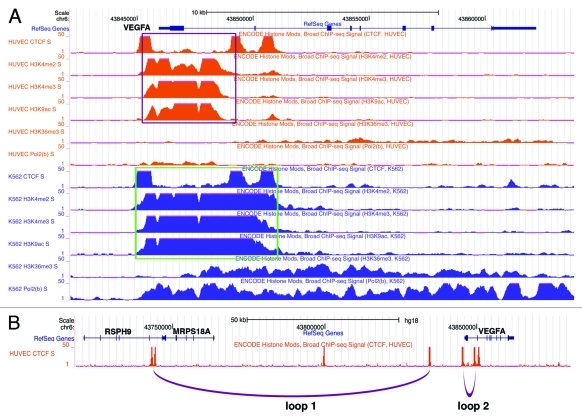
Figure 1. Binding of CTCF and RNA polymerase II, histone modifications, and chromatin looping at the VEGF locus. (A) CTCF and RNAP II binding and histone marks at the VEGF gene (based on the ENCODE ChIP assays). Potential chromatin domains are boxed. (B) Putative CTCF-mediated intrachromosomal looping interactions at the VEGF locus. Loop 1 is based on CTCF interactome in mouse ES cells. Loop 2 is inferred from histone modification patterns. Interchromosomal interactions are not shown.
CTCF-connected loops divide chromatin into distinct domains that often exhibit unique histone modification patterns.10 Conversely, a chromatin domain showing a uniform epigenetic signature may exist as a loop. We notice that there is a homogeneous domain marked by di- and tri-methylation of histone H3 lysine 4 (H3K4) and H3 acetylation between CTCF binding sites at -0.6 kb and intron 1 of VEGF in HUVEC cells (Fig. 1A), implying possible loop formation between these two CTCF sites. This loop should effectively exclude the VEGF promoter from most enhancers (including HRE), therefore fulfilling the task of enhancer blocking. Intriguingly, this domain is enriched for active histone marks. It is curious whether it may also poise the gene for activation. Low abundance binding of RNA polymerase II (RNAP II) seems to be limited to this domain as well (Fig. 1A). In K562 cells, a similar but extended domain emerges between the CTCF sites at -0.6 kb and intron 2 (Fig. 1A). Based on the abundance of RNAP II binding and trimethylated H3K36, a mark for transcription elongation, VEGF is more actively transcribed in K562 than in HUVEC, although it is unclear whether this is related to the CTCF-mediated differential domain formation. In these two cells and many other cell types, the CTCF sites at -0.6 kb and intron 1 or 2 of VEGF represent sharp boundaries with regard to active histone marks. Therefore, we postulate that CTCF may establish two intrachromosomal loops around the VEGF locus in human cells (Fig. 1B): loop 1 forms between the -11kb and -100kb sites upstream of VEGF, and loop 2 between -0.6kb and intron 1/2. Loop 2 spans less than 5 kb and is not included in the global CTCF interactome study which only considers loops larger than 10 kb.10 These potential CTCF-mediated loops, in particular loop 2, separate the VEGF promoter from enhancers and may interfere with distal enhancers' interaction with the promoter.
CTCF binding sites act as decoy promoters to pause RNA polymerase
Recent studies have revealed striking similarities between insulators and promoters.18 Global epigenetic mapping shows that CTCF binding sites are deprived of nucleosomes, and that nucleosomes flanking CTCF sites are enriched in the histone variant H2A.Z and several specific histone modifications, in particular methylation of H3K4.19,20 These histone signatures around CTCF sites are also well-established epigenetic feature of TSS, suggesting that CTCF sites may closely resemble promoters at the molecular level. According to the promoter decoy model,21 CTCF sites may function as decoy promoters to compete with bona fide promoters for interaction with enhancers, thereby trapping enhancers and reducing their efficiency in transcriptional activation.
The unusual likeness between CTCF sites and promoters may also allow them to vie for recruitment of the transcription machinery. It has been reported that CTCF interacts with RNAP II and, indeed, many intergenic CTCF sites are bound by both CTCF and RNAP II.22 However, recruitment of RNAP II to CTCF sites does not appear to lead to productive transcription. CTCF prefers to associate with the hypophosphorylated RNAP II, which is typically present in the initiation complex, rather than the hyperphosphorylated form, which is in the transcription elongation state.22 In fact, CTCF sites can cause pausing of the elongating RNAP II for alternative splicing.23 Furthermore, CTCF sites commonly generate so-called transcription initiation RNAs (tiRNAs), which are nuclear localized 18 nucleotide RNAs produced by RNAP II.24 The tiRNAs enriched at CTCF sites are reminiscent of short transcripts derived from stalled RNAP II,25 hence possibly reflecting the pausing of RNAP II. Collectively, it is tempting to suggest that CTCF sites recruit but retain RNAP II, thereby preventing its productive transcription elongation. At the VEGF 5′ region, the -0.6 kb CTCF site may mimic TSS and trap RNAP II. In HUVEC cells, RNAP II seems to be contained in a chromatin domain between the CTCF sites at -0.6 kb and intron 1 of VEGF (Fig. 1A). It remains to be determined whether any tiRNAs or short RNAs are generated from the -0.6 kb CTCF site upstream of VEGF.
Regulation/dysregulation of CTCF-mediated insulator activity
The insulator activity may be dynamic. In principle, insulator-mediated chromatin loops may be regulated at the level of DNA binding by insulator-associated proteins or activities of such proteins required for looping. Dysregulated CTCF may contribute to pathologic conditions.
CTCF expression is significantly reduced in some breast cancer cells,26 making it possible that there is not sufficient amount of CTCF to carry out its normal function. CTCF is also mutated somatically in several types of cancers (these cells also lose the wild type CTCF allele),27 and we have shown some of these missense mutants fail to bind to the VEGF promoter for enhancer blocking.11 CTCF has a paralogue known as BORIS or CTCFL.28 BORIS has a central multi-zinc finger domain homologous to CTCF's, and is capable of binding to CTCF sites. However, BORIS does not have chromatin insulation activity. BORIS is transiently expressed during spermatogenesis, but is silenced in normal human tissues. It has been reported that expression of BORIS is re-activated in many cancer types,28 raising the possibility that it may compete with CTCF for DNA binding and hinder CTCF's enhancer-blocking activity in cancer cells. All these mechanisms may impair DNA binding of CTCF in cancer cells. Therefore, CTCF-dependent enhancer blocking (including at the VEGF locus) may be compromised in cancer, conferring increased angiogenic potential and growth advantage on cancer cells.
Binding of CTCF to certain sites is sensitive to DNA methylation. At the imprinting control region (ICR) of the IGF2-H19 locus, the binding site for CTCF on the paternal allele is methylated. This prevents binding of CTCF to DNA, leading to loss of enhancer blocking.29,30 The CTCF site at the -0.6 kb region of VEGF contains a single CG dinucleotide. It may worth examining whether the site may be methylated under any circumstances, and whether such methylation may disrupt CTCF binding.
Nucleosome positioning also affects the binding of CTCF to DNA. CTCF always binds to extended linker DNA regions between nucleosomes.20 It has been proposed that at the IGF2-H19 ICR, intrinsic nucleosome positioning sequences (NPSs) determine local nucleosome positions, ensuring the CTCF sites to be located in linker regions and constitutively accessible for CTCF.31 It is shown that RNA transcription through a CTCF binding site results in the repositioning of a nucleosome over the site and consequently, CTCF eviction.32 The nucleosome positioning pattern at the VEGF promoter has not been characterized, and it is unknown whether it may influence CTCF binding.
Regulation of CTCF's insulation activity may be beyond its DNA binding. Insulator sites interact with one another to form chromatin loops. Only a small subset of CTCF-bound sites connect with each other.10 The pairing of CTCF sites is possibly determined by the composition of the CTCF-containing protein complex, covalent modifications of CTCF, local chromatin features, and factors bound adjacently. Each of these may promote or hinder chromatin looping and thus modulate the function of CTCF, even if the binding of CTCF to DNA is not affected. Posttranslational modifications of CTCF, such as poly(ADP-ribosyl)ation or PARlation, are essential for CTCF's insulation activity.33 PARlation of CTCF is decreased in cancer.34 Partner proteins of CTCF may influence loop configuration. In fly, upon induction of heat shock genes, occupancy of insulator-binding proteins such as CTCF is not notably affected during heat shock, however, localization of the partner CP190 is altered.35 Indeed, CP190 recruitment to insulator sites crucially stabilizes the chromatin loops and is a key regulatory step in controlling insulator function.35 Binding of CTCF to the VEGF locus apparently persists across many human cell lines. It is unclear whether co-occupancy of its partner factors and loop formation may differ in different cells. When cells are under hypoxia and VEGF is highly transcribed, CTCF remains bound to the VEGF promoter region, and there is no significant change in the molecular weight of CTCF (not shown). It remains to be determined whether transcription of VEGF accompanies changes in local chromatin loop formation. On the other hand, CTCF-mediated enhancer blocking might be constitutive. For instance, CTCF's insulation activity is not undermined by hypoxia, as the hypoxic induction of VEGF is augmented by depletion of CTCF.11
Conclusion and Outlook
CTCF possesses enhancer blocking activity. Binding of CTCF to the proximal promoter of VEGF, which encodes a prominent angiogenic regulator, diminishes VEGF enhancer activities and limits its transcriptional induction by pro-angiogenic stimuli. CTCF deficiency leads to over-production of VEGF and hyperactivation of angiogenesis. This phenomenon has been observed in some cancer cells. Based on the ENCODE ChIP data, CTCF also binds to the promoters of a few additional notable vascular genes, in particular signaling molecules (ligands) such as angiopoietin (Angpt) 1 and 2, and SDF1 (or CXCL12). It is interesting to investigate whether CTCF acts as an enhancer-blocking factor at those loci as well. Given CTCF's essential role in global genome organization, and its nearly ubiquitous expression, induction of angiogenesis seems to be always restrained in most normal cells. Therefore, CTCF-mediated chromatin insulation functions as a built-in attenuating mechanism to prevent excessive angiogenesis.
CTCF may achieve enhancer blocking by regulating local chromatin state as well as long distance, higher order chromatin structures, including intra- and inter-chromosomal interactions and 3-dimensional nuclear localization. The exact biochemical nature of CTCF-mediated enhancer blocking remains largely elusive. Ultimately, a better understanding of how enhancers activate a promoter will help elucidate the basis of enhancer blocking.
Acknowledgments
J.L. is supported by grants from the National Cancer Institute (R01CA137021) and Florida Bankhead-Coley Cancer Research Program (09BN-12–23092 and 2BT01).
Glossary
Abbreviations:
- ChIP
chromatin immunoprecipitation
- ERE
estrogen responsive element
- GCL
ganglion cell layer
- H3K4
histone H3 lysine 4
- HRE
hypoxia responsive element
- ICR
imprinting control region
- INL
inner nuclear layer
- NPS
nucleosome positioning sequence
- ONL
outer nuclear layer
- RNAP II
RNA polymerase II
- tiRNA
transcription initiation RNA
- TSS
transcription start site
- VEGF
vascular endothelial growth factor
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/19634
References
- 1.Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3:401–10. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 2.Ferrara N. Vascular endothelial growth factor and age-related macular degeneration: from basic science to therapy. Nat Med. 2010;16:1107–11. doi: 10.1038/nm1010-1107. [DOI] [PubMed] [Google Scholar]
- 3.Chung AS, Lee J, Ferrara N. Targeting the tumour vasculature: insights from physiological angiogenesis. Nat Rev Cancer. 2010;10:505–14. doi: 10.1038/nrc2868. [DOI] [PubMed] [Google Scholar]
- 4.Loureiro RM, D’Amore PA. Transcriptional regulation of vascular endothelial growth factor in cancer. Cytokine Growth Factor Rev. 2005;16:77–89. doi: 10.1016/j.cytogfr.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Hirota K, Semenza GL. Regulation of angiogenesis by hypoxia-inducible factor 1. Crit Rev Oncol Hematol. 2006;59:15–26. doi: 10.1016/j.critrevonc.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Hyder SM. Sex-steroid regulation of vascular endothelial growth factor in breast cancer. Endocr Relat Cancer. 2006;13:667–87. doi: 10.1677/erc.1.00931. [DOI] [PubMed] [Google Scholar]
- 7.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–7. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–8. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang M, Chen B, Lin T, Li Z, Pardo C, Pampo C, et al. Restraint of angiogenesis by zinc finger transcription factor CTCF-dependent chromatin insulation. Proc Natl Acad Sci U S A. 2011;108:15231–6. doi: 10.1073/pnas.1104662108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gariano RF, Gardner TW. Retinal angiogenesis in development and disease. Nature. 2005;438:960–6. doi: 10.1038/nature04482. [DOI] [PubMed] [Google Scholar]
- 13.Scott A, Powner MB, Gandhi P, Clarkin C, Gutmann DH, Johnson RS, et al. Astrocyte-derived vascular endothelial growth factor stabilizes vessels in the developing retinal vasculature. PLoS One. 2010;5:e11863. doi: 10.1371/journal.pone.0011863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bulger M, Groudine M. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev Biol. 2010;339:250–7. doi: 10.1016/j.ydbio.2009.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Splinter E, de Laat W. The complex transcription regulatory landscape of our genome: control in three dimensions. EMBO J. 2011;30:4345–55. doi: 10.1038/emboj.2011.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–13. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 18.Raab JR, Kamakaka RT. Insulators and promoters: closer than we think. Nat Rev Genet. 2010;11:439–46. doi: 10.1038/nrg2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geyer PK. The role of insulator elements in defining domains of gene expression. Curr Opin Genet Dev. 1997;7:242–8. doi: 10.1016/S0959-437X(97)80134-7. [DOI] [PubMed] [Google Scholar]
- 22.Chernukhin I, Shamsuddin S, Kang SY, Bergström R, Kwon YW, Yu W, et al. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol. 2007;27:1631–48. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–9. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taft RJ, Hawkins PG, Mattick JS, Morris KV. The relationship between transcription initiation RNAs and CCCTC-binding factor (CTCF) localization. Epigenetics Chromatin. 2011;4:13. doi: 10.1186/1756-8935-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–8. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green AR, Krivinskas S, Young P, Rakha EA, Paish EC, Powe DG, et al. Loss of expression of chromosome 16q genes DPEP1 and CTCF in lobular carcinoma in situ of the breast. Breast Cancer Res Treat. 2009;113:59–66. doi: 10.1007/s10549-008-9905-8. [DOI] [PubMed] [Google Scholar]
- 27.Filippova GN, Qi CF, Ulmer JE, Moore JM, Ward MD, Hu YJ, et al. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter tts DNA-binding specificity. Cancer Res. 2002;62:48–52. [PubMed] [Google Scholar]
- 28.Martin-Kleiner I. BORIS in human cancers - A review. Eur J Cancer. 2011 Oct 21. [Epub ahead of print] PubMed PMID: 22019212. [DOI] [PubMed]
- 29.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–5. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 30.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–9. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 31.Kanduri M, Kanduri C, Mariano P, Vostrov AA, Quitschke W, Lobanenkov V, et al. Multiple nucleosome positioning sites regulate the CTCF-mediated insulator function of the H19 imprinting control region. Mol Cell Biol. 2002;22:3339–44. doi: 10.1128/MCB.22.10.3339-3344.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lefevre P, Witham J, Lacroix CE, Cockerill PN, Bonifer C. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol Cell. 2008;32:129–39. doi: 10.1016/j.molcel.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet. 2004;36:1105–10. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- 34.Docquier F, Kita GX, Farrar D, Jat P, O’Hare M, Chernukhin I, et al. Decreased poly(ADP-ribosyl)ation of CTCF, a transcription factor, is associated with breast cancer phenotype and cell proliferation. Clin Cancer Res. 2009;15:5762–71. doi: 10.1158/1078-0432.CCR-09-0329. [DOI] [PubMed] [Google Scholar]
- 35.Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, et al. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol Cell. 2011;44:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]