Abstract
Initiation of transcription of most human genes transcribed by RNA polymerase II (RNAP II) requires the formation of a preinitiation complex comprising TFIIA, B, D, E, F, H and RNAP II. The general transcription factor TFIID is composed of the TATA-binding protein and up to 13 TBP-associated factors. During transcription of snRNA genes, RNAP II does not appear to make the transition to long-range productive elongation, as happens during transcription of protein-coding genes. In addition, recognition of the snRNA gene-type specific 3′ box RNA processing element requires initiation from an snRNA gene promoter. These characteristics may, at least in part, be driven by factors recruited to the promoter. For example, differences in the complement of TAFs might result in differential recruitment of elongation and RNA processing factors. As precedent, it already has been shown that the promoters of some protein-coding genes do not recruit all the TAFs found in TFIID. Although TAF5 has been shown to be associated with RNAP II-transcribed snRNA genes, the full complement of TAFs associated with these genes has remained unclear. Here we show, using a ChIP and siRNA-mediated approach, that the TBP/TAF complex on snRNA genes differs from that found on protein-coding genes. Interestingly, the largest TAF, TAF1, and the core TAFs, TAF10 and TAF4, are not detected on snRNA genes. We propose that this snRNA gene-specific TAF subset plays a key role in gene type-specific control of expression.
Keywords: gene regulation, pre-initiation complex, snRNA genes, TAFs, TFIID
Introduction
Transcription is the first step of gene expression and can be strictly regulated to ensure appropriate levels of the gene product. Understanding which factors work together to achieve correct regulation of transcription is critical for a complete understanding of gene regulation. Transcription of protein-coding genes and most small nuclear (sn) RNA genes is performed by RNA polymerase II (RNAP II). Synthesis of pre-mRNA by RNAP II involves assembly of a transcription preinitiation complex (PIC) at the promoter followed by initiation, elongation and termination.1,2 The PIC comprises RNAP II and a set of general transcription factors (GTFs) that include TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH. Formation of the PIC usually begins with TFIID binding to the TATA box, if one is present. TFIID is a multiprotein complex comprised of the TATA-binding protein (TBP) and an associated set of 13 evolutionarily conserved TBP-associated factors (TAFs), and plays a critical role in transcription initiation. TAFs have been subjected to intensive study over the last decade. TAFs from S. cerevisiae, S. pombe, C. elegans, D. melanogaster and H. Sapiens have been identified and their primary sequences show strong conservation.3 Some TAFs are also present in distinct human transcriptional regulatory multiprotein complexes such as the SPT3-TAF9-GCN5 containing complex (STAGA; human SAGA), the TBP-free TAF-containing complex (TFTC) and the p300/CBP associated factor complex (PCAF) (see Table 1).4-8
Table 1. TAF composition in hSTAGA, hTFTC, hPCAF and snTAFc.5,75-77 Comparison between the complement of TAFs in TBP-free complexes.
| COMPLEXES | |||
|---|---|---|---|
|
hSTAGA |
hTFTC |
hPCAF |
snTAFc |
| TAF5L TAF6L TAF9 TAF10 TAF12 |
TAF2 TAF4 TAF5 TAF5L TAF6 TAF6L TAF9 TAF9b TAF10 TAF12 |
TAF5L TAF6L TAF9 TAF10 TAF12 |
TAF5 TAF6 TAF8 TAF9 TAF11 TAF13 |
X-ray crystallography and biochemical experiments have shown that nine TAFs contain a sequence motif homologous to a motif found on histones, the histone fold domain (HFD).9,10 The histone fold-containing TAFs form heterodimers TAF6/9, TAF4/12, TAF 8/10, TAF3/10 and TAF11/13. The histone-like heterodimers associate with TAF5 to form the core subcomplex, which interacts with a subcomplex composed of TAF1, TAF2, TAF7 and TBP to form TFIID.11-13
The structure of TFIID appears to be dynamic, and it has been shown that differences in subunit composition affect TFIID conformation. For example, the incorporation of TAF4b, a TAF4 paralogue, into TFIID induces an open conformation at the lobe involved in interaction with TFIIA and various transcriptional activators.14 TAF2 may not always be present in TFIID and, in its absence, significant domain reorganization within TFIID is observed.15 Therefore, it has been suggested by Cler et al.11 that TFIID has a plastic structure that can be reorganized in response to activator signals, which would help TFIID to stably associate with a wide variety of promoters.
Interestingly, studies using ts13, a hamster cell line with a temperature-sensitive TAF1, demonstrated that mutation of TAF1 has a gene-specific effect. In ts13, an amino acid substitution in TAF1 causes G1-S cell cycle arrest at the non-permissive temperature. It has been demonstrated that transcription directed by the cyclin A promoter, but not the c-Fos promoter, is temperature-sensitive.16 In recent years, biochemical, genetic and molecular strategies have contributed to uncovering many activities of individual TAFs. It has been demonstrated that they can function as determinants of promoter selectivity and co-activators to mediate transcriptional activation.17 Some TAFs can be critical for stability of the complex.18 Yankulov et al.19 propose that different compositions of TFIID respond differently to regulatory signals.
The snRNA genes transcribed by RNAP II, which include the genes for the U1 and U2 spliceosomal RNAs, are structurally different from protein-coding genes. These genes are short, the transcripts are intronless and 3′ end formation is directed by the snRNA gene-specific 3′ box RNA 3′ end processing element rather than by a poly(A) signal. The promoters of these genes minimally comprise an essential Proximal Sequence Element (PSE), with some properties of a TATA box and an enhancer-like Distal Sequence Element (DSE).20
The GTFs TFIIA, TFIIB, TFIIF, TFIIE and TFIIH are implicated in transcription of snRNA genes.21,22 So far, only TAF5 was shown to be present at the promoter of snRNA genes23 and the complement of TAFs associated with these genes is unclear. Interestingly, recombinant TBP is necessary for in vitro transcription from these TATA-less promoters but cannot be substituted by TFIID.24 In addition, the efficiency of in vitro transcription of snRNA genes is very low, particularly in light of their robust transcription in vivo.25 The complement of factors that make up the PIC at snRNA promoters is therefore only partially understood. Interestingly, during expression of snRNA genes, RNAP II does not appear to make the transition to productive elongation during transcription of these genes, as happens during transcription of protein-coding genes.26 In addition, the 3′ box is only recognized if transcription is initiated from a RNAP II-dependent snRNA gene promoter. These gene type-specific properties may, at least in part, be due to differences in factors recruited by the promoter, including TAFs.
In order to better understand the molecular basis for the differential control of transcription and RNA processing in expression of snRNA and protein-coding genes, we have performed a comparative analysis of the TAF association with the well-characterized U2 snRNA and β-actin protein-coding genes in HeLa cells. Our studies provide evidence that the TBP-TAF complex on snRNA genes differs from that found on a typical protein-coding gene. TAF1, TAF2, TAF3, TAF4, TAF7, TAF10 and TAF12, which are present on β−actin gene, are absent from U2 snRNA gene.
These results shed new light on the composition of TAF complexes on two different types of gene transcribed by RNAP II. TBP-TAF complexes might therefore make important contributions to the differences in control of transcription elongation and RNA processing between snRNA and protein-coding genes.
Results
RNAP II and TBP association with the β−actin and U2 snRNA genes
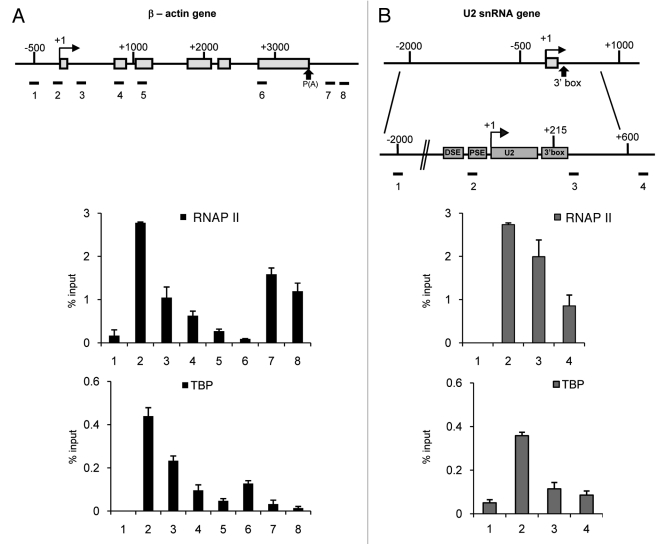
The β-actin protein-coding gene is a single copy gene with a transcription unit of up to 5 kb, encoding transcripts that are spliced and polyadenylated (Fig. 1A).27 Approximately 20 U2 snRNA genes are clustered in 6.2 kb tandem repeats on chromosome 17q21-q22 (Fig. 1B).28,29 The essential PSE is located 50 bp upstream of the transcription start site of the U2 snRNA gene and this element recruits the snRNA-specific PSE-binding transcription factor PTF (also known as SNAPc and PBP).30 The DSE, located 220 bp upstream of the start site of transcription, functions as an enhancer-like element to activate transcription and comprises binding sites for the trans-activating factors Oct-1 and Sp1.31-33 The 3′ box, the snRNA gene-specific RNA 3′ end processing element, is located 9–19 nucleotides downstream of the 3′end of the region encoding the mature snRNA.34
Figure 1. RNAP II and TBP association with the β−actin and U2 snRNA genes. (A) A diagram of the β-actin gene is shown at the top of this and subsequent figures. The arrow represents the transcription start site and exons are denoted by boxes. The results of qPCR analysis of the output of ChIP using anti-RNAP II or anti-TBP antibodies is shown under the diagram. Error bars indicate the standard deviation from at least three independent experiments in this and subsequent figures. Horizontal lines underneath the gene schematics mark the position of the quantitative real-time PCR (qPCR) primer pairs. Primer pairs indicated here were used in all further analyses. (B) A detailed diagram of the U2 snRNA gene is shown at the top of this and subsequent figures. Above this, is shown a schematic of the U2 snRNA gene on the same scale as the β-actin gene schematic in (A). The arrow represents the transcription start site. The distal sequence element (DSE), the proximal sequence element (PSE) in the promoter, the snRNA-encoding region (U2) and the 3′ box processing element are denoted by boxes. The results of qPCR analysis of the output of ChIP using anti-RNAP II or anti-TBP antibodies is shown under the diagram. Error bars indicate the standard deviation from at least three independent experiments in this and subsequent figures. Horizontal lines underneath the gene schematics mark the position of the quantitative real-time PCR (qPCR) primer pairs. Primer pairs indicated here were used in all further analyses.
We have analyzed the association of RNAP II and TBP with the human β-actin and the U2 snRNA genes by high-resolution chromatin immunoprecipitation (ChIP), coupled with quantitative real-time PCR (qPCR) analysis. ChIP localizes RNAP II across the transcribed region of both genes with the highest levels at the promoter region as previously published.26 RNAP II levels remain high over the RNA-encoding region of the U2 snRNA gene (Fig. 1B), whereas the levels across the β-actin gene steadily reduce toward the poly(A) site (Fig. 1A). Interestingly, a second peak of RNAP II is detected on the β-actin gene downstream of the poly(A) site (Fig. 1A, regions 7 and 8), as already described by Gromak et al.27 This may be diagnostic of polymerase pausing,27 looping between this region and the promoter35 or the presence of a downstream promoter. TBP is highest at the start of transcription on both genes, as expected (Fig. 1A and 1B). Interestingly, a second peak of TBP is detected just upstream of the second peak of RNAP II on the β-actin gene (Fig. 1A, region 6).
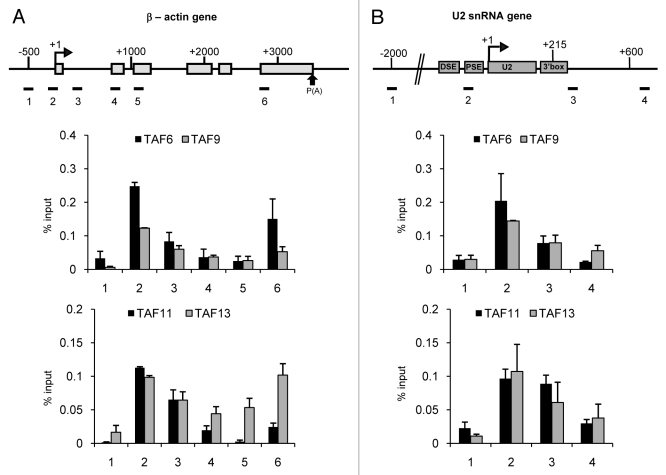
TAF6, TAF9, TAF11 and TAF13 are Associated with the Promoters of the β−actin and U2 snRNA Genes
TAF6, TAF9, TAF11 and TAF13 are detected by ChIP on both the U2 and β−actin genes (Fig. 2A and 2B). TAF6, TAF9, TAF11 and TAF13 on the β-actin gene and TAF6, TAF9, and TAF13 on the U2 gene have a similar profile to TBP on these genes, ie, they peak at the promoter region. Interestingly, all four of these TAFs also show an increase in association with region 6 of the β-actin gene, which is where the second peak of TBP is located (Fig. 2A) and TAF9 and TAF13 decrease again downstream on region 7 (data not shown). TAF11 is also present at a high level close to the U2 snRNA gene 3′ box, suggesting that this TAF interacts differently with the β-actin and U2 snRNA genes. TAF6 forms a heterodimer with TAF9 and TAF11 forms a heterodimer with TAF13.10,13,36 TAF11 and TAF13 do not show the same profile on the U2 snRNA gene (Fig. 2B). Whereas TAF11 is equally high on regions 2 and 3 of the U2 snRNA gene, TAF13 peaks on region 2. This indicates that, although these TAFs normally heterodimerized, they do not interact with U2 snRNA gene promoter in exactly the same way.
Figure 2. TAF6, TAF9, TAF11 and TAF13 are associated with the promoters of the β−actin (A) and U2 snRNA (B) genes. The results of qPCR analysis of the ChIP output using anti-TAF6, anti-TAF9, anti-TAF-11 or anti-TAF13 antibodies are shown.
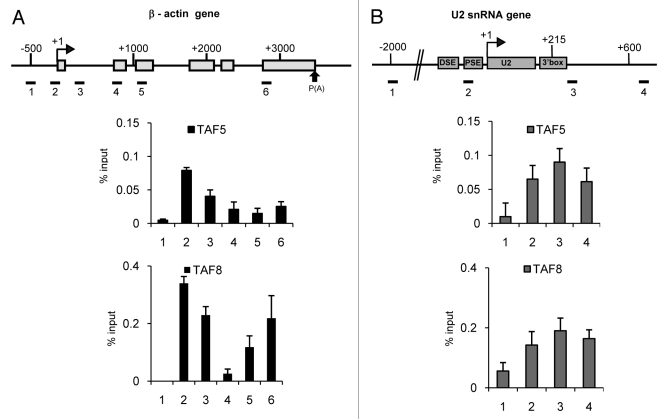
TAF5 and TAF8 Peak at Different Regions of the β−actin and U2 snRNA Genes
TAF5 and TAF8 peak at the promoter of the β-actin gene, as expected (Fig. 3A). They also show an increase in association with region 6 of the β-actin gene. As noted for TAF11, TAF5 and TAF8 show a different distribution on the U2 snRNA gene and instead peak at the 3′ box, suggesting that these TAFs also interact differently with the β-actin and U2 snRNA genes (Fig. 3B), possibly due to differences in the conformation of the complex.
Figure 3. TAF5 and TAF8 show different profiles on the β−actin (A) and U2 snRNA (B) genes. The results of qPCR analysis of the ChIP output using anti-TAF5 or anti-TAF8 antibodies are shown.
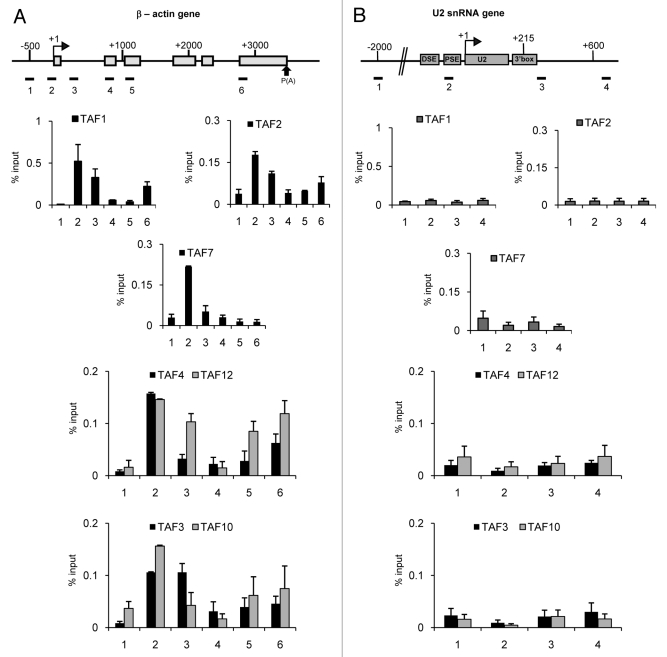
TAF1, TAF2, TAF3, TAF4, TAF7, TAF10 and TAF12 are Absent from the U2 snRNA Gene
Although TAF1, TAF2, TAF3, TAF4, TAF7, TAF10 and TAF12 are readily detected peaking at the β-actin gene promoter or, in the case of TAF3, just downstream, none of these TAFs is detected on either the promoter or the transcribed region of the U2 snRNA gene (Fig. 4A and 4B). This may indicate that these TAFs are absent from the PIC on the U2 snRNA gene or that their conformation precludes crosslinking to the gene. Interestingly, TAF1, TAF2, TAF3, TAF4, TAF10 and TAF12, in common with TBP, show an increase in association with region 6 of the β-actin gene. However, TAF7 does not. In common with TAF9 and TAF13, TAF10 levels decrease again on region 7 (data not shown).
Figure 4. TAF1, TAF2, TAF3, TAF4, TAF7, TAF10 and TAF12 are detected on the β−actin (A) but not on the U2 snRNA (B) gene. The results of qPCR analysis of the ChIP output using anti-TAF1, anti-TAF2, anti-TAF3, anti-TAF4, anti-TAF7, anti-TAF10 or anti-TAF12 antibodies are shown. The results for TAF4/TAF12 and TAF3/TAF10 are merged as those TAFs form heterodimers.
TAF1 and TAF7 are also detected at the promoter of the β-actin gene by ChIP seq (data accessed through ENCODE),37 in accordance with our results. There are no obvious peaks at region 6 in this data set, which may be due to differences in experimental conditions or due to limitations of ChIP Seq in this region.
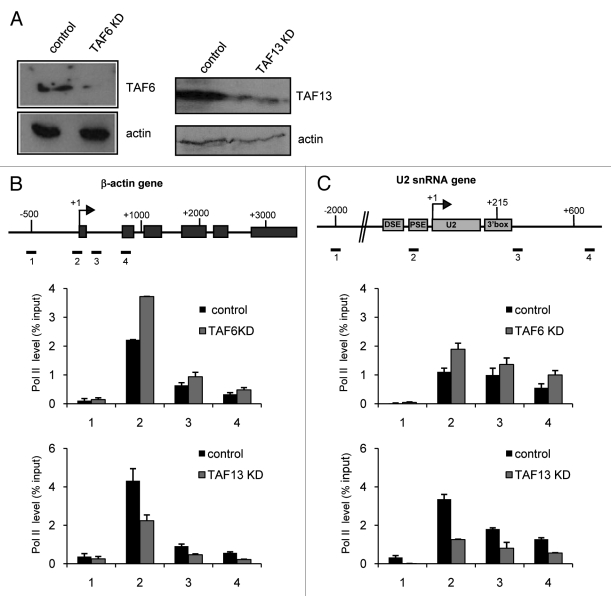
Knockdown of TAF6 and TAF13 Affects the Level of RNAP II on the β−actin and U2 snRNA Genes
In order to determine whether TAF6 and TAF13 association with the β-actin and U2 snRNA genes reflects a functional role in expression of both genes, we have knocked down the level of TAF6 and TAF13 using siRNAs (Fig. 5A) and assayed RNAP II occupancy by ChIP (Fig. 5B and 5C). Knockdown of TAF6 has the surprising effect of increasing the level of RNAP II on both genes. This result indicates that TAF6 plays an important role in transcription of both protein-coding and snRNA genes, but appears to inhibit, rather than favor, recruitment of RNAP II. In contrast, knockdown of TAF13 causes a decrease in the association of RNAP II with the β-actin and U2 snRNA genes, indicating that this TAF is required for transcription of both genes.
Figure 5. Knockdown of TAF6 and TAF13 affects the β−actin and U2 gene RNAP II level. (A) western blot analysis of HeLa whole-cell extracts from control cells or cells transfected with an siRNA specific for TAF6 and TAF13. Antibodies used are noted on the right. Actin is used as a loading control. The results of qPCR analysis of the output of ChIP using anti-RNAP II antibodies before or after TAF6 knockdown (KD) and TAF13 knockdown (KD) for the β-actin (B) and U2 snRNA gene (C) are shown.
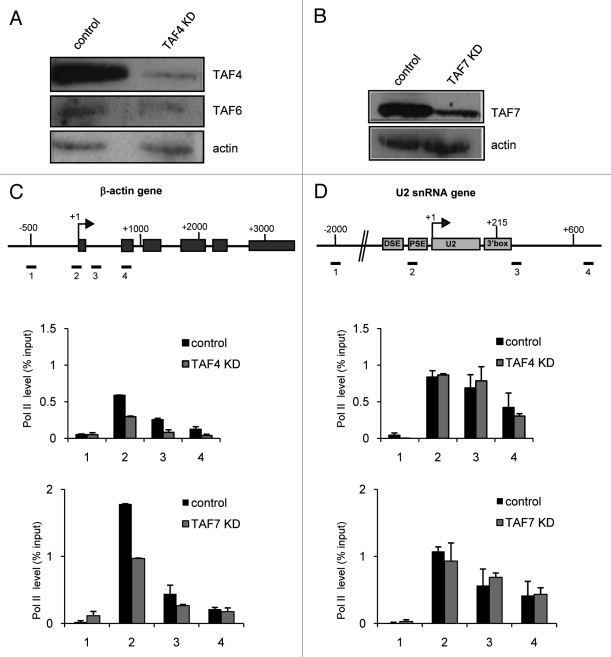
Knockdown of TAF4 or TAF7 Specifically Affects the Level of RNAP II on the β−actin Gene
In order to test the possibility that some TAFs are not recruited to the U2 snRNA gene because they are not required for transcription (Fig. 4), we have performed siRNA-mediated knockdown of TAF4 and TAF7 (Fig. 6). Western blot analysis confirmed that siRNA knockdown of TAF4 and TAF7 was efficient (Fig. 6A and 6B). In TAF4 and TAF7 KD cells the levels of RNAP II on the β-actin gene decrease when compared with the levels observed in control cells (Fig. 6C). In contrast, there is no significant drop of RNAP II on the U2 snRNA genes when either TAF4 or TAF7 are knocked down (Fig. 6D). These results indicate that neither TAF4 nor TAF7 is involved in transcription of the U2 snRNA gene. Interestingly, when TAF4 is lost, there appears to also be some reduction in the level of TAF6 (Fig. 6A). This is consistent with the effect of TAF4 knockdown in Drosophila cells, which leads to degradation of several other TAFs.12 However, the reduction is not sufficient to affect transcription of the U2 snRNA gene (Fig. 6B).
Figure 6. Knockdown of TAF4 or TAF7 specifically affects the level of RNAP II on the β−actin gene. (A) western blot analysis of HeLa whole-cell extracts from control cells or cells transfected with a siRNA specific for TAF4 as indicated. Antibodies used are noted at the right. (B) western blot analysis of HeLa whole-cell extracts from control cells or cells transfected with a siRNA specific for TAF7. Antibodies used are noted at the right. The results of qPCR analysis of the ChIP output using anti-RNAP II antibodies before or after TAF4 or TAF7 knockdown (KD) for the β-actin (C) and U2 snRNA gene (D) are shown.
Taken together, these results support the notion that, in contrast to the β−actin gene, there is only a subset of the full TFIID TAF complement on U2 snRNA genes. The novel snRNA-TAF complex, which we term snTAFc, comprises only TAF5, TAF6, TAF8, TAF9, TAF11 and TAF13.
CPSF is Specifically Associated with the β-actin Gene
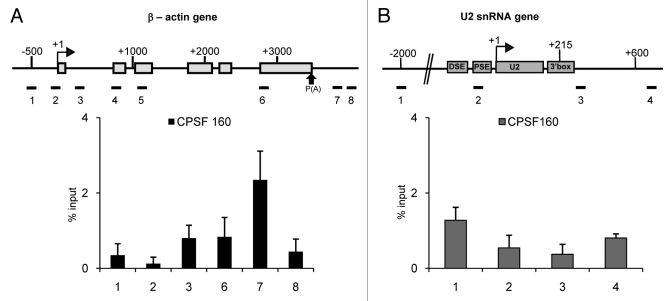
TAFs are implicated in the recruitment of the cleavage-polyadenylation specificity factor, CPSF to protein-coding genes through TFIID. TAF5, TAF7 and TAF12 interact strongly with CPSF160.38 The transcripts from snRNA genes are not polyadenylated and 3′ box-dependent 3′ end formation requires the Integrator complex rather than the factors involved in poly(A) site-dependent cleavage and polyadenylation.39-41 Although TAF5 is detected on the U2 snRNA genes, TAF7 and TAF12 are not (Figs. 3B and 4B). Accordingly, we have analyzed the association of CPSF160 with both the β-actin and snRNA genes (Fig. 7).
Figure 7. CPSF160 is specifically associated with the β-actin gene. The results of the qPCR analysis of the ChIP output using anti-CPSF160 antibody on the β-actin (A) and U2 snRNA gene (B) are shown.
CPSF160 is readily detected by ChIP associated with the β-actin gene, close to the position of the poly(A) signal at +3560 as would be expected since cleavage is generally co-transcriptional (Fig. 7A).42 CPSF160 is, however, is not detected at the promoter region of the β-actin gene, indicating either that interaction between CPSF and TFIID on this gene is not stable or that crosslinking of CPSF160 in this region is not efficient due to lack of contact with or proximity to promoter DNA. In contrast, CPSF73 has been shown to be associated both with the poly(A) site and promoter regions of the c-myc and GAPDH protein-coding genes.16 CPSF160 is not detected at any region of the U2 snRNA gene (Fig. 7B). These results are consistent with a gene-type-specific role of TAF7 and TAF12 in helping to recruit CPSF to protein-coding genes and indicate that TAF5 alone is not sufficient. In line with these results, CPSF73 is not detected on the U2 snRNA gene.16
Discussion
TAFs are critical components of the machinery responsible for the regulated transcription of eukaryotic genes transcribed by RNAP II, and their biological significance has made them the target of extensive study. Understanding the interplay between TAFs and other components of the transcription machinery provides important insights into how transcriptional regulation helps direct the growth and development of eukaryotic organisms. TAFs can function as promoter recognition factors, as co-activators and as enzymatic modifiers of other proteins. Some are important for the stability of complexes they form. Interestingly, the importance of these functions may vary from promoter to promoter.
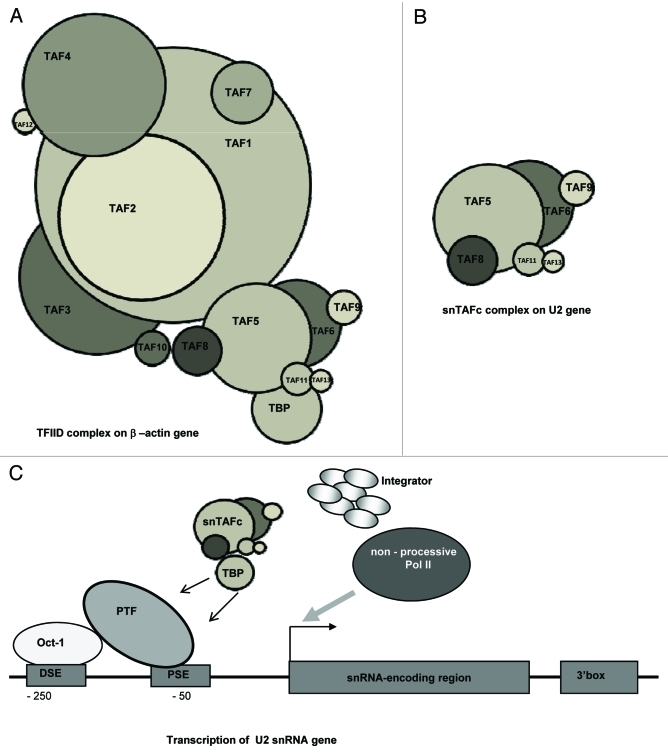
Our data suggest that a subcomplex of TFIID, containing TBP, TAF5, TAF6, TAF8, TAF9, TAF11, and TAF13 is loaded onto snRNA genes (Fig. 8B). Alternatively, TFIID may exist in different conformations on both the U2 and β-actin genes, but with different cross-linking efficiencies for some TAFs depending on promoter structure. However, the existence of a novel subcomplex on the snRNA genes is supported by the finding that knocking down TAF4 and TAF7 only affects β-actin and not U2 snRNA gene transcription. TAFs exist as components of a number of distinct complexes but, interestingly, the proposed snTAFc does not contain the classical core TAFs (TAF4 and TAF10) and lacks the highest-molecular weight TAFs, TAF1 and TAF2 (Fig. 8A and 8B). A complete TFIID complex does not appear to be required for transcription initiation from snRNA gene promoters since knockdown of key TFIID subunits does not arrest transcription of the U2 snRNA gene (Fig. 6). Our ChIP analysis of a complete set of TAFs on the U2 snRNA and β−actin genes therefore provides further evidence for different TFIID compositions at different promoters.
Figure 8. Recruitment of a U2 snRNA gene-specific TBP-TAF complex is essential for proper snRNA gene expression. (A) On the β-actin gene, the histone fold-containing TAFs (TAF3, TAF4, TAF6, TAF8, TAF9, TAF10, TAF11, TAF12 and TAF13) form heterodimers and interact with TAF1, TAF2, TAF5, TAF7 and TBP to form TFIID. Some of the TAFs, e.g., TAF10 may be present in more than one copy11 (B) On the U2 snRNA gene, TAF5, TAF6, TAF8, TAF9, TAF11 and TAF13 associate to form an snRNA gene-specific-TAF complex (snTAFc). (C) On the snRNA gene, TBP associates with PTF and with snTAFc. This may facilitate specific recruitment of the Integrator complex and plays a role in setting the elongation properties of RNAP II, possibly due to the lack of recruitment of key elongation factors.
Interestingly, TAF1 is not present on the U2 snRNA gene (Fig. 4). TAF1 has histone acetyltransferase (HAT) activity that acetylates histone H3 and H443 also acts as a histone-specific ubiquitin-activating/conjugating enzyme to mediate monoubiquitination of histone H1 in vitro.44,45 These activities contribute to the importance of this TAF in gene transcription. For example, transcription of the MHC class I genes is dependent on the HAT activity of TAF1.46-48 TAF7 binds to TAF1 and negatively regulates its HAT activity.48 It has been reported that TAF7 dissociates from the TFIID complex upon recruitment of RNAP II to the assembling PIC. This release is concomitant with phosphorylation of TAF1, which reduces the binding of TAF1 to TAF7. Additional studies have shown that TAF7 functionally interacts with the TFIIH and elongation factor P-TEFb and inhibits their kinase activities. Accordingly, it was proposed that TAF7 acts as a checkpoint regulator and delays transcription until the PIC complex is assembled.49,50 As both TAF1 and TAF7 are absent from the U2 snRNA gene, neither the enzymatic activities of TAF1 nor the kinase interactions of TAF7 are required for expression of this gene. The acetylation of H3 detected at the promoter of the U2 snRNA gene26 must therefore be dependent on other HAT activities and no TAF7-dependent checkpoint can occur on these genes. TAF1 may not be necessary for transcription of the U2 snRNA genes as the transcription unit is nucleosome depleted.26
TAF composition is, at least partially, core promoter dependent.51 The core promoter of genes is defined as the minimal DNA sequence required for accurate transcription initiation in vitro.52 One of the common core promoter elements in human protein-coding genes is a TATA box located approximately 25 bp upstream of the transcription start site. The consensus sequence TATA(A/T)A(A/T)(A/G) is recognized directly by TBP.53 Other core promoter elements, such as the initiator (Inr), the downstream promoter element (DPE), and the downstream core element (DCE) may also be present.54-57 TAFs are thought to significantly widen the range of promoter recognition by TFIID. First, TAF association with TBP helps to overcome chromatin barriers.17 Second, contacts between TAFs and Inrs, DPEs and DCEs can facilitate promoter recognition.58 TAF1/TAF2 recognizes the Inr sequence.59 In addition, UV photo-crosslinking results demonstrate that TAF1 interacts with the DCE sub-element in a sequence-dependent manner.56 Thus, the absence of TAF1 and TAF2 may reflect the absence of an Inr sequence in addition to the lack of functional requirement for these TAFs. In contrast, TAF6 and TAF9, which are found on the U2 snRNA gene, exhibit DPE-binding specificity,60,61 although there is no DPE in U2 snRNA promoter. Thus, TAFs may be differentially required for transcription of the snRNA and β-actin genes or differentially recruited due to differences in the DNA sequence, chromatin structure at the promoters and the complement of other factors at the promoter.
A number of reports demonstrate that some TAFs directly interact with activators. For example, human TAF4 interacts with Sp1 and CREB,62 TAF7 interacts with Sp1, YY1, USF, CTF, adenovirus E1A protein and HIV-1 Tat,2,17 NTF-1 interacts with TAF2,63 TAF9 interacts with the activation domain of Herpes viral protein 16 (VP16)64 and TAF1, TAF3, TAF6 and TAF9 interact with the tumor suppressor protein p53.65,66 Those interactions suggest that TAFs can function as co-activators to bridge DNA-bound activators and the PIC. In agreement with this model, it has been demonstrated that mutations of Sp1 that disrupt interaction with TAFs affect activation of transcription.67 Thus, activators may target different TAFs to recruit TFIID to promoters. In addition, holo-TFIID may integrate different signals from different activators. Interestingly, VP16 and p53 activate transcription of protein-coding genes but not snRNA genes,68 although TAF6 and TAF9 are present on U2 snRNA genes (Fig. 2). In addition, Sp1 can activate transcription of snRNA genes68 although TAF4 and TAF7 are absent. This indicates that not all activators are compatible with snRNA gene promoters, that TAF6 and TAF9 are not sufficient to mediate activation by VP16 and p53 and that activation by Sp1 occurs in the absence of interaction with TAF4 and TAF7.
It is particularly striking that TAF4 and TAF10 are missing from the U2 snRNA gene promoter, as these play primary roles in TFIID assembly. Knockdown of Drosophila TAF4 leads to degradation of most of the TAFs12 and TAF10 gene disruption in mice leads to disassembly of TFIID.69 TAF5, which is associated with the U2 snRNA genes, is also implicated in TFIID structure. Thus, snTAFs must not require TAF4 and TAF10 for assembly and stability. TBP is recruited to snRNA genes by PTF—as an integral part of SNAPc.70 The snTAFc may be recruited as a new TBP-free complex, with a different composition from TFTC, STAGA, PCAF/GCN5 (see Table 1). Alternatively, snTAFc may not be stable off DNA and may assemble on the snRNA gene promoter through interactions with PTF and TBP. Recognition of the snRNA gene-specific 3′ box RNA 3′ end processing element only occurs if transcription is initiated from a RNAP II-dependent snRNA gene promoter,26,71 implicating specialized transcription complexes in the processing reaction. In addition, RNAP II initiated at the promoter of snRNA genes does not appear to make the transition to productive elongation and lack the H3K36me3 mark of productive elongation.26 The selective recognition of the 3′ box and the lack of processivity of RNAP II transcribing snRNA genes might therefore be, in some way, connected to the specific TAF composition of the PIC.
Transcripts from snRNA genes, in contrast to mRNA genes, are not polyadenylated. Dantonel et al.38 showed that cleavage-polyadenylation specificity factor CPSF associates with the PIC through TFIID. Neither TAF7 nor TAF12, which interact strongly with CPSF-160, is recruited to the U2 snRNA gene promoter (Fig. 7). The snRNA gene-specific TAF complex may therefore play a central role in facilitating gene-type-specific RNA processing.
Based on our results, we propose a model in which recruitment of a subset of TAFs to snRNA genes underpins a cascade of events critical for proper gene expression (Fig. 8C). PTF binding to the PSE facilitates stable recruitment of TBP, which in turn helps to recruit an snRNA gene-specific subset of TAFs (snTAFc) involved in specific recruitment of, for example, the Integrator complex. In addition, snTAFc may lack TAFs involved in recruiting elongation factors, thereby helping to determine the elongation potential of RNAP II.
The significance of the presence of the second peak of TBP and several TAFs within the gene but upstream of the poly(A) site of the β-actin gene is not clear. This may be diagnostic for a gene loop. However, loops are generally found between the promoter and the pol(A) site/region of transcription termination.35 In addition, the second peak of RNAP II does not coincide with the second peak of TBP and TAFs. Thus, the second peak of RNAP II may reflect pausing at the end of the gene and the TBP/TAF peak may mark the position of a second promoter where an inactive PIC lacking TAF7 has assembled.
Materials and Methods
Chromatin Immunoprecipitation
ChIP analysis was performed as described previously.72 DNA was sonicated to give fragments of less than 500 bp. PTFγ ChIP on the U2 snRNA gene was used to determine that the resolution was within 200 bp as described.26 ChIP samples were analyzed by qPCR using QuantiTect SYBR Green PCR (Qiagen) using the primers indicated in Table 2. Antibodies against RNAP II (N-20), TBP (sc-421), TAF1 (sc-735), TAF4 (sc-736), TAF7 (sc-101167), TAF10 (sc-102125), TAF13 (sc-15264) and CPSF160 (sc-28872) were obtained from Santa Cruz Biotechnology. Antibodies against TAF2 (NBP1–21371) and TAF3 (A302–360A) were obtained from Novus Biologicals and Bethyl laboratories, respectively. Anti-TAF5, anti- TAF6, anti-TAF8, anti-TAF9, anti-TAF11 were produced in the laboratory of RGR and anti- TAF6, anti-TAF8, anti-TAF9, anti-TAF11 have been previously described.73
Table 2. Sequence or primers used for qPCR analysis.
| Name | β-actin gene | |
|---|---|---|
| |
Forward |
Reverse |
| 1 |
GCTGCGGCTGGGTAGGTTTG |
CACTTAGAAGTCGCAGGACC |
| 2 |
CCAATCAGCGTGCGCCGTTCCGA |
GGTGTGGACGGGCGGCGGATC |
| 3 |
GGGCAACCGGCGGGGTCTTT |
ACGCAGTTAGCGCCCAAAGG |
| 4 |
CACAGCGCGCCCGGCTATTC |
AGCCAGCTCCCCTACCTGGT |
| 5 |
CCCCATCGAGCACGGCATCGTC |
CACCTGGGTCATCTTCTCGCGGT |
| 6 |
CATTGCTCCTCCTGAGCGCAAGTA |
TTGCGGTGGACGATGGAGGGGCC |
| 7 |
GGGACTATTTGGGGGTGTCT |
TCCCATAGGTGAAGGCAAAG |
| 8 |
TGGGCCACTTAATCATTCAAC |
CCTCACTTCCAGACTGACAGC |
|
Name |
U2 gene |
|
| |
Forward |
Reverse |
| 1 |
GGAGCGGAGCGTTCTCTGTCTCCCC |
AGAGTGTGAGCCCTCATTCACGCCC |
| 2 |
ATGAGAGTGGGACGGTGA |
CACTTGATCTTAGCCAAAAGG |
| 3 |
ACGAGTCCTGTGACGCGCCGGCTTG |
CTCCGGGTGGGTCCCATTCCTTTAA |
| 4 | CCTCCCCGCCTCTCCCTCGCTC | GGACAAATAGCCAACGCATGCGG |
siRNA-mediated Knockdown
siRNAs targeting TAF4, TAF6, TAF7 and TAF13 were purchased from Dharmacon and transfected into HeLa cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Western blot analysis
Western blotting was performed as described previously74 using antibodies against TAF4 (sc-736, Santa Cruz), TAF6 (Roeder), TAF7 (sc-101167, Santa Cruz), TAF13 (sc-15264) and actin (sc-1615, Santa Cruz).
Acknowledgments
We would like to thank the members of Murphy lab for discussions and critical comments. We would also like to thank Laszlo Tora for his generous support throughout this project. The work was supported by grants from the Wellcome Trust to SM, from the EPA Trust to JZ and by NIH CA129325 grant to RGR.
Glossary
Abbreviations:
- ChIP
chromatin immunoprecipitation
- CPSF
the cleavage-polyadenylation specificity factor
- DCE
downstream core element
- DPE
downstream promoter element
- DSE
distal sequence element
- GTFs
general transcription factors
- HAT
histone acetyltransferase
- mRNA
messenger RNA
- PA
polyadenylation
- PIC
preinitiation complex
- RNAP II
RNA polymerase II
- PSE
Proximal Sequence Element
- qPCR
quantitative real-time PCR
- snRNA
small nuclear RNA genes
- SYBR
SybrGreen
- TAFs
TBP-associated factors
- TBP
TATA-binding protein
- TC
transcription complex
- TFTC
the TBP-free TAF-containing complex
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/19783
References
- 1.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang CM, Roeder RG. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–6. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 3.Tora L. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 2002;16:673–5. doi: 10.1101/gad.976402. [DOI] [PubMed] [Google Scholar]
- 4.Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, 3rd, et al. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/S0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 5.Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, et al. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/S0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 6.Brand M, Yamamoto K, Staub A, Tora L. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J Biol Chem. 1999;274:18285–9. doi: 10.1074/jbc.274.26.18285. [DOI] [PubMed] [Google Scholar]
- 7.Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, et al. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–95. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demény MA, Soutoglou E, Nagy Z, Scheer E, Jànoshàzi A, Richardot M, et al. Identification of a small TAF complex and its role in the assembly of TAF-containing complexes. PLoS One. 2007;2:e316. doi: 10.1371/journal.pone.0000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birck C, Poch O, Romier C, Ruff M, Mengus G, Lavigne AC, et al. Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell. 1998;94:239–49. doi: 10.1016/S0092-8674(00)81423-3. [DOI] [PubMed] [Google Scholar]
- 10.Gangloff YG, Romier C, Thuault S, Werten S, Davidson I. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem Sci. 2001;26:250–7. doi: 10.1016/S0968-0004(00)01741-2. [DOI] [PubMed] [Google Scholar]
- 11.Cler E, Papai G, Schultz P, Davidson I. Recent advances in understanding the structure and function of general transcription factor TFIID. Cell Mol Life Sci. 2009;66:2123–34. doi: 10.1007/s00018-009-0009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright KJ, Marr MT, 2nd, Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc Natl Acad Sci U S A. 2006;103:12347–52. doi: 10.1073/pnas.0605499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gangloff YG, Sanders SL, Romier C, Kirschner D, Weil PA, Tora L, et al. Histone folds mediate selective heterodimerization of yeast TAF(II)25 with TFIID components yTAF(II)47 and yTAF(II)65 and with SAGA component ySPT7. Mol Cell Biol. 2001;21:1841–53. doi: 10.1128/MCB.21.5.1841-1853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu WL, Coleman RA, Grob P, King DS, Florens L, Washburn MP, et al. Structural changes in TAF4b-TFIID correlate with promoter selectivity. Mol Cell. 2008;29:81–91. doi: 10.1016/j.molcel.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papai G, Tripathi MK, Ruhlmann C, Werten S, Crucifix C, Weil PA, et al. Mapping the initiator binding Taf2 subunit in the structure of hydrated yeast TFIID. Structure. 2009;17:363–73. doi: 10.1016/j.str.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–8. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–78. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 18.Näär AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- 19.Yankulov K, Blau J, Purton T, Roberts S, Bentley DL. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–59. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 20.Egloff S, O’Reilly D, Murphy S. Expression of human snRNA genes from beginning to end. Biochem Soc Trans. 2008;36:590–4. doi: 10.1042/BST0360590. [DOI] [PubMed] [Google Scholar]
- 21.Kuhlman TC, Cho H, Reinberg D, Hernandez N. The general transcription factors IIA, IIB, IIF, and IIE are required for RNA polymerase II transcription from the human U1 small nuclear RNA promoter. Mol Cell Biol. 1999;19:2130–41. doi: 10.1128/mcb.19.3.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, et al. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol. 2009;29:5455–64. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oelgeschläger T. Regulation of RNA polymerase II activity by CTD phosphorylation and cell cycle control. J Cell Physiol. 2002;190:160–9. doi: 10.1002/jcp.10058. [DOI] [PubMed] [Google Scholar]
- 24.Bernués J, Simmen KA, Lewis JD, Gunderson SI, Polycarpou-Schwarz M, Moncollin V, et al. Common and unique transcription factor requirements of human U1 and U6 snRNA genes. EMBO J. 1993;12:3573–85. doi: 10.1002/j.1460-2075.1993.tb06031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunderson SI, Knuth MW, Burgess RR. The human U1 snRNA promoter correctly initiates transcription in vitro and is activated by PSE1. Genes Dev. 1990;4(12A):2048–60. doi: 10.1101/gad.4.12a.2048. [DOI] [PubMed] [Google Scholar]
- 26.Egloff S, Al-Rawaf H, O’Reilly D, Murphy S. Chromatin structure is implicated in “late” elongation checkpoints on the U2 snRNA and beta-actin genes. Mol Cell Biol. 2009;29:4002–13. doi: 10.1128/MCB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol. 2006;26:3986–96. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Arsdell SW, Weiner AM. Human genes for U2 small nuclear RNA are tandemly repeated. Mol Cell Biol. 1984;4:492–9. doi: 10.1128/mcb.4.3.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westin G, Lund E, Murphy JT, Pettersson U, Dahlberg JE. Human U2 and U1 RNA genes use similar transcription signals. EMBO J. 1984;3:3295–301. doi: 10.1002/j.1460-2075.1984.tb02293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadowski CL, Henry RW, Lobo SM, Hernandez N. Targeting TBP to a non-TATA box cis-regulatory element: a TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev. 1993;7:1535–48. doi: 10.1101/gad.7.8.1535. [DOI] [PubMed] [Google Scholar]
- 31.Murphy S, Yoon JB, Gerster T, Roeder RG. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol Cell Biol. 1992;12:3247–61. doi: 10.1128/mcb.12.7.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattaj IW, Lienhard S, Jiricny J, De Robertis EM. An enhancer-like sequence within the Xenopus U2 gene promoter facilitates the formation of stable transcription complexes. Nature. 1985;316:163–7. doi: 10.1038/316163a0. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka M, Grossniklaus U, Herr W, Hernandez N. Activation of the U2 snRNA promoter by the octamer motif defines a new class of RNA polymerase II enhancer elements. Genes Dev. 1988;2(12B):1764–78. doi: 10.1101/gad.2.12b.1764. [DOI] [PubMed] [Google Scholar]
- 34.Uguen P, Murphy S. The 3′ ends of human pre-snRNAs are produced by RNA polymerase II CTD-dependent RNA processing. EMBO J. 2003;22:4544–54. doi: 10.1093/emboj/cdg430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hampsey M, Singh BN, Ansari A, Lainé JP, Krishnamurthy S. Control of eukaryotic gene expression: gene loops and transcriptional memory. Adv Enzyme Regul. 2011;51:118–25. doi: 10.1016/j.advenzreg.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leurent C, Sanders S, Ruhlmann C, Mallouh V, Weil PA, Kirschner DB, et al. Mapping histone fold TAFs within yeast TFIID. EMBO J. 2002;21:3424–33. doi: 10.1093/emboj/cdf342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 38.Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 39.Hernandez N. Formation of the 3′ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 1985;4:1827–37. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baillat D, Hakimi MA, Näär AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–76. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 41.Egloff S, Zaborowska J, Laitem C, Kiss T, Murphy S. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell. 2012;45:111–22. doi: 10.1016/j.molcel.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–8. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 43.Dunphy EL, Johnson T, Auerbach SS, Wang EH. Requirement for TAF(II)250 acetyltransferase activity in cell cycle progression. Mol Cell Biol. 2000;20:1134–9. doi: 10.1128/MCB.20.4.1134-1139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pham AD, Sauer F. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science. 2000;289:2357–60. doi: 10.1126/science.289.5488.2357. [DOI] [PubMed] [Google Scholar]
- 45.Maile T, Kwoczynski S, Katzenberger RJ, Wassarman DA, Sauer F. TAF1 activates transcription by phosphorylation of serine 33 in histone H2B. Science. 2004;304:1010–4. doi: 10.1126/science.1095001. [DOI] [PubMed] [Google Scholar]
- 46.Howcroft TK, Raval A, Weissman JD, Gegonne A, Singer DS. Distinct transcriptional pathways regulate basal and activated major histocompatibility complex class I expression. Mol Cell Biol. 2003;23:3377–91. doi: 10.1128/MCB.23.10.3377-3391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weissman JD, Brown JA, Howcroft TK, Hwang J, Chawla A, Roche PA, et al. HIV-1 tat binds TAFII250 and represses TAFII250-dependent transcription of major histocompatibility class I genes. Proc Natl Acad Sci U S A. 1998;95:11601–6. doi: 10.1073/pnas.95.20.11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gegonne A, Weissman JD, Singer DS. TAFII55 binding to TAFII250 inhibits its acetyltransferase activity. Proc Natl Acad Sci U S A. 2001;98:12432–7. doi: 10.1073/pnas.211444798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gegonne A, Weissman JD, Lu H, Zhou M, Dasgupta A, Ribble R, et al. TFIID component TAF7 functionally interacts with both TFIIH and P-TEFb. Proc Natl Acad Sci U S A. 2008;105:5367–72. doi: 10.1073/pnas.0801637105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gegonne A, Weissman JD, Zhou M, Brady JN, Singer DS. TAF7: a possible transcription initiation check-point regulator. Proc Natl Acad Sci U S A. 2006;103:602–7. doi: 10.1073/pnas.0510031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Green MR. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem Sci. 2000;25:59–63. doi: 10.1016/S0968-0004(99)01527-3. [DOI] [PubMed] [Google Scholar]
- 52.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–35. [PubMed] [Google Scholar]
- 53.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7(7B):1291–308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 54.Kaufmann J, Smale ST. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 1994;8:821–9. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- 55.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–79. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 56.Lee DH, Gershenzon N, Gupta M, Ioshikhes IP, Reinberg D, Lewis BA. Functional characterization of core promoter elements: the downstream core element is recognized by TAF1. Mol Cell Biol. 2005;25:9674–86. doi: 10.1128/MCB.25.21.9674-9686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Purnell BA, Emanuel PA, Gilmour DS. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes Dev. 1994;8:830–42. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- 58.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/S0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 59.Chalkley GE, Verrijzer CP. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J. 1999;18:4835–45. doi: 10.1093/emboj/18.17.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shao H, Revach M, Moshonov S, Tzuman Y, Gazit K, Albeck S, et al. Core promoter binding by histone-like TAF complexes. Mol Cell Biol. 2005;25:206–19. doi: 10.1128/MCB.25.1.206-219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sengupta T, Cohet N, Morlé F, Bieker JJ. Distinct modes of gene regulation by a cell-specific transcriptional activator. Proc Natl Acad Sci U S A. 2009;106:4213–8. doi: 10.1073/pnas.0808347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X, Truckses DM, Takada S, Matsumura T, Tanese N, Jacobson RH. Conserved region I of human coactivator TAF4 binds to a short hydrophobic motif present in transcriptional regulators. Proc Natl Acad Sci U S A. 2007;104:7839–44. doi: 10.1073/pnas.0608570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen JL, Attardi LD, Verrijzer CP, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 64.Kim DH, Lee SH, Nam KH, Chi SW, Chang I, Han KH. Multiple hTAF(II)31-binding motifs in the intrinsically unfolded transcriptional activation domain of VP16. BMB Rep. 2009;42:411–7. doi: 10.5483/BMBRep.2009.42.7.411. [DOI] [PubMed] [Google Scholar]
- 65.Allende-Vega N, Saville MK, Meek DW. Transcription factor TAFII250 promotes Mdm2-dependent turnover of p53. Oncogene. 2007;26:4234–42. doi: 10.1038/sj.onc.1210209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bereczki O, Ujfaludi Z, Pardi N, Nagy Z, Tora L, Boros IM, et al. TATA binding protein associated factor 3 (TAF3) interacts with p53 and inhibits its function. BMC Mol Biol. 2008;9:57. doi: 10.1186/1471-2199-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gill G, Pascal E, Tseng ZH, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci U S A. 1994;91:192–6. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Das G, Hinkley CS, Herr W. Basal promoter elements as a selective determinant of transcriptional activator function. Nature. 1995;374:657–60. doi: 10.1038/374657a0. [DOI] [PubMed] [Google Scholar]
- 69.Mohan WS, Jr., Scheer E, Wendling O, Metzger D, Tora L. TAF10 (TAF(II)30) is necessary for TFIID stability and early embryogenesis in mice. Mol Cell Biol. 2003;23:4307–18. doi: 10.1128/MCB.23.12.4307-4318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henry RW, Sadowski CL, Kobayashi R, Hernandez N. A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerase II and III. Nature. 1995;374:653–6. doi: 10.1038/374653a0. [DOI] [PubMed] [Google Scholar]
- 71.Hernandez N, Weiner AM. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986;47:249–58. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- 72.Medlin J, Scurry A, Taylor A, Zhang F, Peterlin BM, Murphy S. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J. 2005;24:4154–65. doi: 10.1038/sj.emboj.7600876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Raha T, Cheng SW, Green MR. HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol. 2005;3:e44. doi: 10.1371/journal.pbio.0030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Medlin JE, Uguen P, Taylor A, Bentley DL, Murphy S. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J. 2003;22:925–34. doi: 10.1093/emboj/cdg077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–57. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- 76.Wieczorek E, Brand M, Jacq X, Tora L. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature. 1998;393:187–91. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]
- 77.Martinez E, Kundu TK, Fu J, Roeder RG. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998;273:23781–5. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]