Abstract
Atg4 is required for cleaving Atg8, allowing it to be conjugated to phosphatidylethanolamine on phagophore membranes, a key step in autophagosome biogenesis. Deconjugation of Atg8 from autophagosomal membranes could be also a regulatory step in controlling autophagy. Therefore, the activity of Atg4 is important for autophagy and could be a target for therapeutic intervention. In this study, a sensitive and specific method to measure the activity of two Atg4 homologs in mammalian cells, Atg4A and Atg4B, was developed using a fluorescence resonance energy transfer (FRET)-based approach. Thus LC3B and GATE-16, two substrates that could be differentially cleaved by Atg4A and Atg4B, were fused with CFP and YFP at the N- and C-terminus, respectively, allowing FRET to occur. The FRET signals decreased in proportion to the Atg4-mediated cleavage, which separated the two fluorescent proteins. This method is highly efficient for measuring the enzymatic activity and kinetics of Atg4A and Atg4B under in vitro conditions. Applications of the assay indicated that the activity of Atg4B was dependent on its catalytic cysteine and expression level, but showed little changes under several common autophagy conditions. In addition, the assays displayed excellent performance in high throughput format and are suitable for screening and analysis of potential modulators. In summary, the FRET-based assay is simple and easy to use, is sensitive and specific, and is suitable for both routine measurement of Atg4 activity and high-throughput screening.
Keywords: Atg4, Atg8, FRET assay, GATE-16, LC3B
Introduction
Macroautophagy (hereafter referred to as autophagy) is a major intracellular degradation process conserved in all eukaryotic cells. Autophagy removes superfluous components, damaged organelles, misfolded proteins and certain intracellular pathogens via double-membraned autophagosomes, which degrade their content after fusing with lysosomes.1,2 Autophagy can also contribute to cellular dysfunction and cell death under specific conditions.3 The roles of autophagy in both cell survival and cell death indicate that the autophagy process can be an important therapeutic target for diseases such as neurodegeneration and cancer.2,4,5
Autophagosome biogenesis requires two ubiquitin-like conjugation systems: the Atg12–Atg5 and the Atg8–phosphotidylethanolamine (PE) systems.6-8 The cysteine protease Atg4 cleaves newly synthesized Atg8 to reveal a glycine residue required for covalent attachment to PE (lipidation), and removes conjugated Atg8 from the autophagosome membranes (delipidation) to facilitate autophagosome biogenesis and Atg8 recycling.9-11
While yeast express single genes encoding Atg4 and Atg8, there are six human Atg8 homologs. The LC3 subfamily of Atg8 includes MAP1A/B light chain 3 (LC3A, LC3B and LC3C), and the GABARAP subfamily consists of γ-aminobutyric acid receptor-associated protein (GABARAP), Golgi-associated ATPase enhancer of 16 kDa (GATE-16/GABARAPL2) and Atg8L (GABARAPL1).8,12,13 It seems that molecules of the two subfamilies can differ in their functions in autophagosome biogenesis.14 In mammals there are four Atg4 homologs.15 Atg4B is likely the principal human Atg4 homolog. It is able to cleave each of the human Atg8-related proteins tested so far in vitro and in living cells.8,11,16,17 Studies have also shown that Atg4A is a potent protease for the GABARAP subfamily, but not the LC3 subfamily, of Atg8 substrates, whereas Atg4C and Atg4D seem to possess marginal activities toward the Atg8 homologs of both subfamilies.17,18
Blockage of Atg4-mediated Atg8 processing could be an effective way to interfere with autophagy. Toward that end it would be critical to develop an efficient, sensitive and specific method to measure Atg4 activity. Several methods have been used in previous studies. Assessment of Atg4 activity has been mainly based on an SDS-PAGE-based assay,9,17,19,20 which can be cumbersome and highly variable with a relatively low detection sensitivity. These methods would be only suitable for in vitro analysis and cannot be formatted for high-throughput analysis. Several fluorescence or luminescence-based assays, which utilize substrates that emit signals after cleavage by Atg4, have been developed for in vivo use or for high throughput screening.21,22 While versatile and sensitive, these methods are prone to nonspecific interference due to their design.
Here, we report the development of a novel Atg4 assay based on fluorescence resonance energy transfer (FRET). We used cyan fluorescence protein (CFP) as the donor and yellow fluorescence protein (YFP) as the receptor, which were fused to the N-and C-termini of the Atg8 substrates, respectively. Cleavage of the fusion protein by Atg4A or Atg4B led to a proportional loss of FRET signals. We found this assay sensitive, specific and reproducible for routine analysis of the Atg4 activity and kinetics, and for high throughput screening.
Results
FRET-Atg8 fusion substrates can be cleaved by Atg4
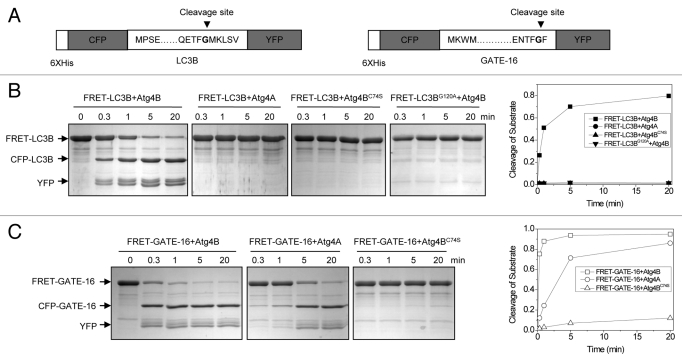
When a suitable pair of fluorophores is brought within close proximity (10–100 Å) of one another, excitation of the donor molecule can result in energy transfer to the acceptor fluorophore.23 CFP and YFP are one of the most common pairs used for FRET constructs. The structures of LC3B and GATE-16 suggest that the N terminus and the C terminus of these proteins are spatially close to each other.24,25 We hypothesized that C- and N-terminal fusions of YFP and CFP to the full-length LC3B or GATE-16 would allow FRET to occur, and that cleavage of the products would lead to the separation of the two fluorescent moieties, thus causing the loss of the FRET signals (Fig. 1A). We verified that these fusion proteins, collectively termed FRET-Atg8, could be efficiently processed by Atg4A and Atg4B using the well-established SDS-PAGE method (Fig. 1B and C). FRET-LC3B was rapidly cleaved by Atg4B, generating the CFP-LC3B and the YFP fragments. This reaction was specific since the substrate could not be processed by Atg4A or the Atg4B catalytic mutant, Atg4BC74S. Consistently, the cleavage site mutant, FRET-LC3BG120A, was not digested by Atg4B (Fig. 1B). In contrast, both Atg4A and Atg4B, but not Atg4BC74S, effectively cleaved FRET-GATE-16 (Fig. 1C). Under the experimental conditions tested, nearly all FRET-GATE-16 and up to 80% of FRET-LC3B could be processed within 20 min (Fig. 1B and C), and nearly all FRET-LC3B could be processed within 60 min (data not shown). These results were consistent with previous studies,17 indicating that the fusion molecules remained effective substrates for Atg4A and/or Atg4B.
Figure 1. Verification of the cleavage of FRET substrates by Atg4 using SDS-PAGE. (A) Schematic representation of the FRET-LC3B and FRET-GATE-16 constructs. The cleavage site of LC3B and GATE-16 at the glycine (G) residue is indicated. (B and C) FRET-LC3B, FRET-LC3BG120A (B) or FRET-GATE-16 (C) (5 µg) were incubated with Atg4B, Atg4BC74S or Atg4A (0.25 µg), respectively. The reaction was stopped at different times using SDS-PAGE sample buffer. The substrates and the cleaved products were separated by SDS-PAGE and examined by CBB staining. Representative gel images from three independent experiments are shown in the left panels. Percentage of cleaved substrate was calculated based on the densitometry of protein bands of the representative experiments (right panels).
Cleavage of FRET-Atg8 substrates by Atg4 can be specifically measured with changes in the FRET signal
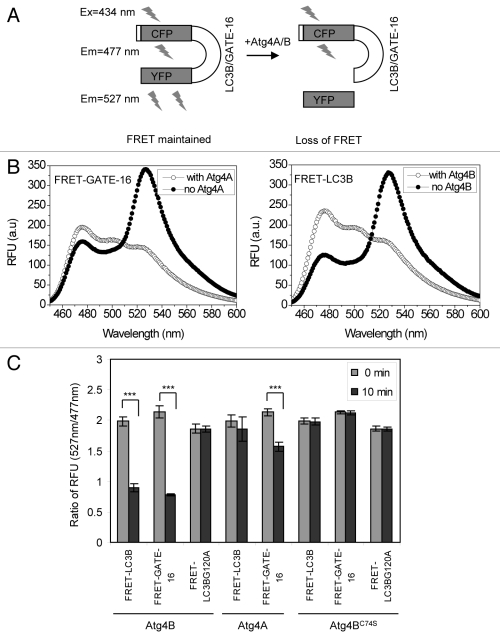
We then determined whether the fusion molecules could generate FRET signals that were sensitive to cleavage (Fig. 2A). FRET-GATE-16 and FRET-LC3B were examined by fluorescence spectrometry in the absence or presence of Atg4B. Both the donor (CFP) and the acceptor (YFP) emission wavelengths can be measured when excited at the wavelength of 434 nm. Thus, the full spectrum scan indicated that both fusion proteins had a major emission peak at 527 nm, and a minor emission peak at 477 nm in the absence of Atg4A or Atg4B as expected, indicating that the FRET signals were maintained (Fig. 2B). However, when an Atg4 enzyme was present, the 527 nm/477 nm ratio decreased from 1.65 to 0.69 for FRET-LC3B, and from 1.8 to 0.6 for FRET-GATE-16 (Fig. 2B).
Figure 2. Verification of the cleavage of FRET substrate by Atg4 using the FRET-based assay. (A) Schematic representation of the assay principle. The FRET signal (λem = 527 nm) is reduced as the result of cleavage, which separates the CFP (donor) moiety from the YFP (acceptor) moiety. (B) The fluorescence emission spectra of FRET-GATE-16 or FRET-LC3B before and after cleavage. Substrate (500 μg/ml) were mixed with buffer, Atg4A (100 μg/ml) or Atg4B (2 μg/ml) in a cuvette in the volume of 0.5 ml for 30 min. Data from representative experiments were collected on a Cary Eclipse spectrophotometer. The excitation wavelength was 434 nm. Emission peaked at 477 nm with Atg4 present, but at 527 nm with no Atg4 present in the reactions. In the presence of the corresponding Atg4 enzyme, the ratio of 527 nm/477 nm for FRET-GATE-16 decreased from 1.8 to 0.6 and that for FRET-LC3B was reduced from 1.65 to 0.69. (C) FRET-LC3B, FRET-GATE-16 and FRET-LC3BG120A (100 μg/ml) were incubated with Atg4A, Atg4B, or Atg4BC74S (2 μg/ml) in a volume of 200 μl for 10 min. The fluorescence ratios of 527 nm/477 nm at the beginning and at the 10 min point were determined. Data represent the mean ± SD from three independent experiments. ***p < 0.001 (paired t-test, panel C).
The relative fluorescence unit (RFU) ratio of 527 nm to 477 nm could thus be a sensitive measurement of the Atg4 cleavage activity. To examine this hypothesis, we compared the FRET signal (ratio of 527 nm/477 nm) during the cleavage of the fusion proteins by Atg4A, Atg4B or Atg4BC74S (Fig. 2C). Atg4B significantly reduced the FRET signal of FRET-LC3B and FRET-GATE-16 in 10 min, whereas Atg4BC74S did not. On the other hand, Atg4A significantly reduced the FRET signal of FRET-GATE-16 but not that of FRET-LC3B, consistent with the fact that FRET-LC3B cannot be cleaved by Atg4A. The reduction of FRET signal was not due to nonspecific degradation of the FRET substrates, as no FRET reduction was observed in the absence of Atg4 enzymes or if the substrate contained the cleavage site mutant (LC3BG120A) (Fig. 2C). Thus, these results indicated that the FRET-based assay was specific and sensitive to Atg4-mediated cleavage.
Measurement of the kinetics of Atg4A and Atg4B using the FRET-based assay
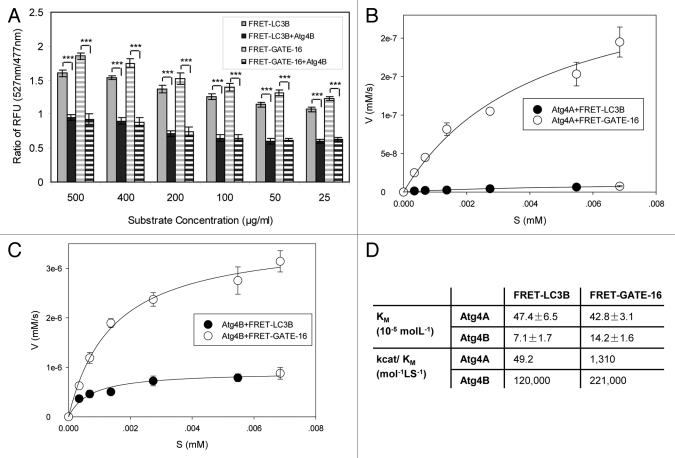
We had previously determined the kinetic parameters of all four Atg4 homologs toward LC3B and GATE-16 using the SDS-PAGE-based method.17 To determine whether the FRET-based assay could also be used to measure the kinetics of cleavage, we incubated Atg4A or Atg4B with FRET-GATE-16 or FRET-LC3B, respectively, at different concentrations. The ratio of the fluorescence intensity (RFU) at 527 nm and 477 nm was in proportion to the substrate concentrations and the corresponding cleavage products, suggesting that the assay was suitable for kinetics analysis (Fig. 3A). The initial velocity of the cleavage was thus determined based on the change of the 527 nm/477 nm ratio and then plotted against the substrate concentrations. From the fitted curve, the key kinetic parameters, Vmax and KM, were derived (Fig. 3B–D).
Figure 3. Determination of the kinetics of Atg4A and Atg4B using the FRET-based assay. (A–C) FRET-LC3B and FRET-GATE-16 at different concentrations (25 μg/ml to 500 μg/ml) were incubated with Atg4B (2 μg/ml in A, 0.5 μg/ml in B) or Atg4A (2.5 μg/ml, C) for 30 min at 37°C. The RFUs at 527 nm and 477 nm were determined and the ratios were calculated (A). The initial velocity (V, y-axis) at each substrate concentration was defined as the increment of the substrate per second ([mM]/s), and was calculated based on RFU at the 5 min time point of the reaction for Atg4B (B) or at the 30 min time point of the reaction for Atg4A (C) The velocity was then plotted against the concentration of the substrate (S [mM], x-axis). The curves were fitted using the ligand-binding method (SigmaPlot 10.0). (D). The kinetic parameters KM and kcat/KM were derived from the fitted curves. Data represent the mean ± SD from three independent experiments. ***p < 0.001 (paired t-test, A).
The data indicated that Atg4B had a higher affinity and a higher catalytic efficiency than Atg4A toward the two substrates. GATE-16 was a better substrate than LC3B, particularly for Atg4A, which could not cleave LC3B. These findings were consistent with previous determinations using LC3B-GST and GATE-16-GST as the substrates in an SDS-PAGE-based assay.17,22 The catalytic efficiency of Atg4B toward FRET-GATE-16 (221,000 mol−1S−1, Fig. 3D) was in the same order of magnitude as that toward GATE-16-GST (107, 000 mol−1S−1)17 despite the different assay formats used for the measurement. The catalytic efficiency of Atg4B toward LC3B-GST17 and FRET-LC3B (Fig. 3D) was also quite comparable (89,600 vs 120,000 mol−1S−1). These observations are consistent with the notion that Atg4B is less selective toward the substrates and their configurations. On the other hand, it is noted that Atg4A is more selective than Atg4B in substrate preference, only being able to hydrolyze the GABARAP subfamily substrates, and is therefore likely more sensitive to the configuration of the substrate. Consistently, the catalytic efficiency of Atg4A toward FRET-GATE-16 (1,310 mol−1S−1, Fig. 3D) seemed to be particularly lower than that toward GATE-16-GST (12,800 mol−1S−1),17 showing a preference toward GATE-16-GST over FRET-GATE-16.
Measurement of Atg4 activities in cell lysates using the FRET-based assay
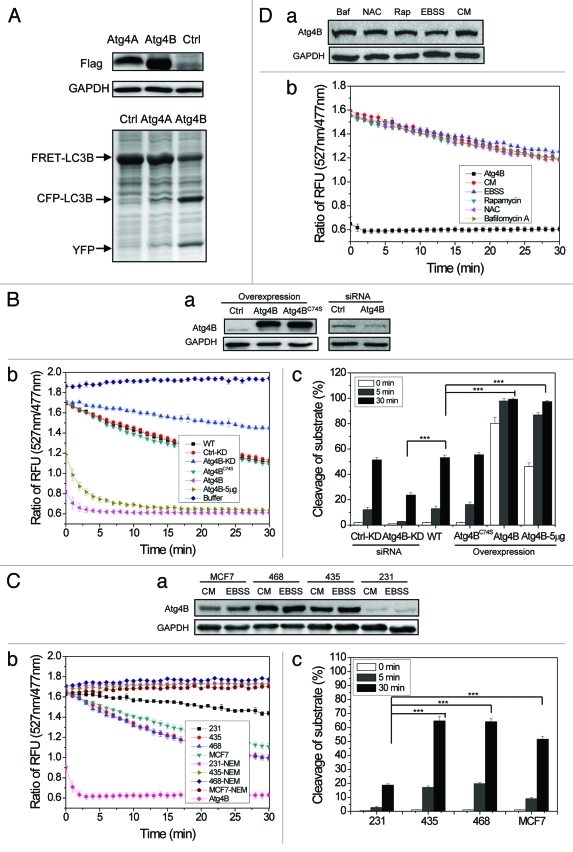
While the above studies used purified recombinant Atg4 proteins, we sought to determine whether the FRET assay would be also effective for the measurement of Atg4 activity in biological samples. We investigated this issue by examining the activity of Atg4B in the following studies. Lysates prepared from HEK-293A cells with or without various overexpressed Atg4 constructs were incubated with the FRET-LC3B substrate. While the SDS-PAGE-based assay clearly revealed the difference in the extent of FRET-LC3B cleavage with the overexpressed Atg4B (Fig. 4A), the FRET-assay provided a more quantitative measurement (Fig. 4B). Thus, while only about 10% of incubated FRET-LC3B (3 μg) was cleaved by the endogenous Atg4B in 20 μg of lysates in 5 min, nearly 100% of the same amount of FRET-LC3B could be cleaved by 20 μg of lysates from Atg4B-overexpressing cells, and nearly 90% of cleavage was reached with 5 μg of such lysates. The overexpression of neither Atg4BC74S (Fig. 4B) nor Atg4A (Fig. 4A) caused an elevation of the cleavage above the level reached by the endogenous Atg4B. Conversely, knockdown of Atg4B by about 50% led to the reduction of the cleavage by 60% in a 30 min reaction (Fig. 4B). Thus, the FRET assay could specifically and quantitatively measure the Atg4B activity in a biological sample.
Figure 4. Measurements of Atg4B activity in cell lysates using the FRET-based assay. (A) HEK-293A cells were transfected with Flag-Atg4A, Flag-Atg4B or vector for 24 h. Cells were harvested and cell lysates (25 μg) were mixed with FRET-LC3B (5 µg) for 20 min before the reaction was stopped by the addition of sample loading buffer, and separated by SDS-PAGE. The expression level of Flag-Atg4A and Flag-Atg4B was detected by immunoblot assay using an anti-Flag antibody. The cleavage was assessed after CBB staining. (B) HEK-293A cells were transfected with Flag-Atg4B, Flag-Atg4BC74S or the control vectors. Alternatively, they were transfected with a control siRNA or a siRNA against Atg4B. Cell lysates (20 μg) were prepared and subjected to immunoblot analysis (a), or incubated with FRET-LC3B (3 µg) for the cleavage assay (b). Five micrograms (indicated) or 20 µg (all others) of lysates were used. WT, control; KD, control or Atg4B siRNA-treated. The percentage of substrate cleavage (c) was calculated. (C) Four breast cancer cell lines were cultured in complete medium (CM) or EBSS for 4 h. Cell lysates (20 μg) were prepared and subjected to immunoblot analysis (a), or incubated with FRET-LC3B for the cleavage assay (b). A positive control group with purified recombinant Atg4B (2 μg) was included. A generic cysteine protease inhibitor, NEM (100 µM), was included in some reactions as indicated. The percentage of substrate cleavage (c) was calculated. (D) HEK-293A cells were incubated in EBSS, or in complete medium alone (CM) or with rapamycin (Rap, 2 µM), N-acetylcysteine (NAC, 20 mM) or bafilomycin A1 (Baf, 1 µM) for 4 h. Cell lysates were prepared and subjected to immunoblot analysis (a), or incubated with FRET-LC3B for the cleavage assay (b). A positive control group with purified recombinant Atg4B (2 μg) was included (indicated as Atg4B). Data represent the mean ± SD from three independent experiments. ***p < 0.001 (one way ANOVA, B, c; and C, c).
To determine whether the assay could discriminate Atg4B activity in cells constitutively expressing different levels of Atg4B, we analyzed a panel of 4 breast cancer lines and found that the Atg4B activity was well correlated with the expression level of Atg4B, with MDA-MB-231 being the lowest, followed by MCF7, MDA-MB-436 and MDA-MB-468 (Fig. 4C). The cleavage activity was fully suppressed by N-ethylmaleimide (NEM), a general cysteine protease inhibitor.
Considering these data, we next sought to determine whether the Atg4B expression and activity could change during autophagy. Incubation of cells in EBSS, a starvation medium, did not alter the expression of Atg4B in the breast cancer cell lines (Fig. 4C, a) and in HEK-293A cells (Fig. 4D, a). Consistently, application of other chemicals that affect autophagy, such as rapamycin, N-acetylcysteine, or bafilomycin A1, did not change the Atg4B expression level (Fig. 4D, a), or the activity (Fig. 4D, b). These data indicate that the cleavage potency of Atg4B was more dependent on the expression level than on the autophagy status. This notion, however, does not rule out the possibility that Atg4 activity could vary inside the cells following proper autophagy stimulation,26 but failed to be detected in the in vitro condition.
Configuration of the FRET-based assay for high-throughput screening (HTS)
One major advantage of the FRET assay over the SDS-PAGE-based assay is that it can be easily configured to a high-throughput format for analysis or for screening. To ascertain HTS compatibility and robustness, we examined the performance of the FRET assay in multiple replicates in 384-well plates.
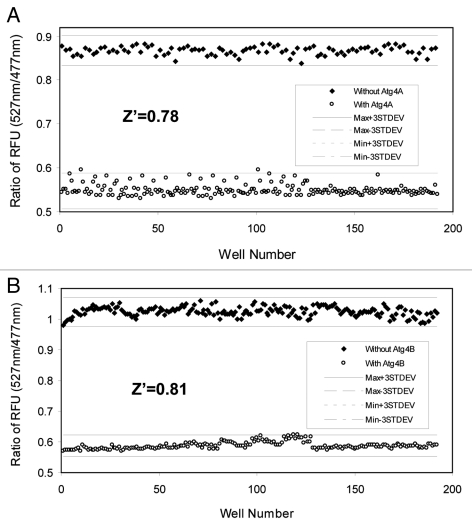
An important parameter for evaluating the reliability and robustness of the high-throughput assay is the Z’ factor.27 This factor evaluates the size of the errors in the data relative to the difference in the maximum and minimum values in the assay, which gives rise to an estimate of the assay window. The Z’ factor can assume any value ≤ 1. A Z’ factor of 1 denotes an ideal assay with complete separation of positive and negative controls. Z’ factor > 0 denotes separation between the means ± 3 SD of the positive and negative control. In HTS, Z’ factors of > 0.5 are considered excellent. We determined that the Z’ factors for the Atg4A FRET-GATE-16 assay and the Atg4B FRET-LC3B assay were 0.78 and 0.81, respectively (Fig. 5), which indicated a robust performance of these assays. Thus, the FRET-based Atg4 assay has the quality to be successfully employed in HTS.
Figure 5. Determination of the performance of the FRET-based Atg4 assay in a high-throughput format. Atg4A (10 μg/ml, A) or Atg4B (2 µg/ml, B) were mixed with 60 μg/ml of FRET-GATE-16 (A) or FRET-LC3B (B) in a total volume of 20 µl in 384-well plates. After 60 min incubation, the RFU ratio of 527 nm/477 nm was determined for each of the reactions in 192 wells (solid circle). Control reactions with no Atg4 enzymes were set up in the same way (open circle). The ranges of the maximal and minimal values ± three standard deviations were indicated and the Z’ factors for both Atg4A and Atg4B assays were calculated.
Analysis of chemical interference of Atg4 activity using the FRET-based assay
The FRET assay would allow for a better quantitative determination and comparison of various enhancing or inhibiting modulators of Atg4 enzymes. Using this assay, we first examined the effect of six representative protease inhibitors on Atg4A and Atg4B activities. These chemicals include a disulfide bond reducer, tris-2-carboxyethyl-phosphine (TCEP), a sulfhydryl group blocker, N-ethylmaleimide (NEM), a serine protease inhibitor, 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), a general cysteine protease inhibitor, E64-D, a general aspartic peptidase inhibitor, pepstatin A, and an aminopeptidase inhibitor, bestatin.
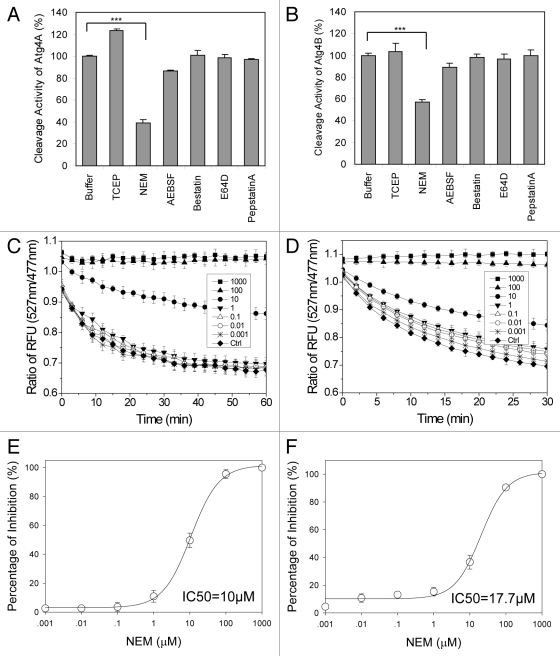
We found that only NEM, but not other chemicals at the tested concentrations, was able to inhibit the activity of Atg4A and Atg4B (Fig. 6A and B), consistent with previous findings.15,18 This indicates that while the cysteine residue of Atg4A and Atg4B is important for enzymatic activity, generic cysteine protease inhibitors, such as E64-D, are not effective in suppressing this activity. Consistently, TCEP showed an enhancing effect on the reaction, especially for Atg4A, suggesting the importance of maintaining cysteine residues in reduced status.
Figure 6. Assessment of selected protease inhibitors on Atg4 using the FRET-based assay. (A and B) Atg4A (10 µg/ml, A) or Atg4B (2 µg/ml, B) were incubated with the indicated compounds (10 μM) for 30 min at 37°C in a 384-well plate. The FRET signal was recorded after adding FRET-GATE-16 (A) or FRET-LC3B (B) (50 μg/ml) to a total volume of 50 µl. The RFU ratios of 527 nm/477 nm at the reaction time of 60 min (A) or 30 min (B) were determined. The relative Atg4 cleavage activity was expressed using the ratios normalized to that of the control (buffer only). (C and D) Atg4A (10 μg/ml, C) or Atg4B (2 µg/ml, D) were incubated with NEM at different concentrations (0–1,000 μM) for 30 min at 37°C in a 384-well plate. FRET-LC3B (50 μg/ml) was then added and the fluorescence signals at 477 nm and 527 nm were measured consecutively in the next 60 min (C) or 30 min (D) and the ratios of 527 nm/477 nm were determined. (E and F) The percentages of inhibition on the activity of Atg4A (E) or Atg4B (F) in the presence of different concentrations of NEM at the time when maximal cleavage was achieved (60 min for Atg4A, E, 30 min for Atg4B, F) were calculated. The IC50 was then determined for each enzyme-compound combination. Data represent the mean ± SD from three independent experiments. ***p < 0.001 (one way ANOVA, A and B).
To determine the potency of NEM, different concentrations of NEM were applied and the RFU ratio of 527 nm/477 nm of the cleavage reaction was reversed in a concentration-dependent manner (Fig. 6C and D). The IC50 was determined to be 10 µM for Atg4A and 17.7 µM for Atg4B (Fig. 6E and F). Thus, the FRET-based Atg4 assay can quantitatively determine the effects of small molecule modulators of Atg4A and Atg4B.
Discussion
The mammalian Atg8 homologs are a group of proteins with diverse sequences, belonging to two subfamilies, the LC3 subfamily and the GABARAP subfamily. Diversity in their function has been reported in which LC3B seems to be mostly involved in the elongation of the phagophore membrane, whereas GATE-16 is more important for autophagosome maturation at a later stage.14 The initial processing of Atg8 molecules requires the cysteine protease Atg4, which has multiple mammalian homologs. While Atg4A and Atg4B have potent enzymatic activities, Atg4C and Atg4D seem to have only minor effects on Atg8 processing, and their physiological significance is not clear.17,18,28,29 Atg4B has the widest substrate spectrum, being able to cleave mammalian Atg8 homologs of both subfamilies. Deletion of Atg4B led to significant defects in the lipidation of all Atg8 molecules.30 Interestingly, the phenotypes of Atg4B-deficient mice are limited to inner ear development and balance functions,30 suggesting that its function could be complemented by other Atg4 homologs, such as Atg4A. The latter is able to process the GABARAP subfamily of Atg8 proteins, but not the LC3 subfamily of Atg8 proteins. Thus the functional complementarity of Atg4 homologs could be based on the functional complementarity of the two Atg8 subfamilies. It also seems important that both Atg4A and Atg4B need to be affected if endogenous regulatory or exogenous intervening mechanisms are to be exerted at this level of autophagy function. Methods that allow rapid detection of Atg4A and Atg4B activities with a great sensitivity and specificity are needed to understand these mechanisms.
Several approaches to measuring Atg4 activity have been developed. The classical method is based on electrophoretic separation of cleaved substrates on SDS-PAGE, using Atg8 substrates fused with a tag C-terminal to the cleavage site.9,19,20 The fused tag allows the detection of cleavage based on either the change of the size of the substrate or the appearance of the C-terminal cleavage products. This method has been used widely with no need for special equipment. We have successfully used this method to determine the kinetic parameters of all mammalian Atg4 homologs.17 The SDS-PAGE-based method is, however, rather cumbersome for quantification of the cleavage, is less suitable for regular cellular samples than recombinant Atg4, and is certainly not adaptable for high throughput.
Subsequent assay development had been focused on the aspects of being more quantitative and sensitive, while adopting the principle of using fusion substrates. A successful example is the use of phospholipase A2 (PLA2) as the fusion partner of LC3B.21 After cleavage by Atg4B, PLA2 is separated from LC3B and regains its enzymatic activity, which is detected with a secondary fluorogenic assay. Thus the activity of PLA2 is in proportion to the level of LC3B-PLA2 cleavage, or Atg4B activity. This method seems to be very sensitive and specific, and is suitable for high throughput screening. However, one possible concern for the two-reaction design is that interference with the PLA2 reaction may be mistakenly attributed to the change in the Atg4 activities.22,31
To detect Atg4 activity in situ, two other methods have been developed. One uses a LC3B fusion construct that contains Gaussia luciferase and β-actin, which would allow the hybrid molecule to be sequestered inside the cells in the actin network.21 The cleavage by Atg4B results in the release of Gaussia luciferase, which is exported to the extracellular space where it can be measured by luminometry. This assay is also a coupled assay in which the luciferase reaction may render the assay prone to changes not related to the Atg4 activity. In addition, the introduction of the luciferase fusion construct to the cell can be labor intensive. Recently, a simplified one-reaction design has been proposed.26 This system uses FITC-labeled Atg4 substrate peptides conjugated to polymeric nanoparticles, which are highly permeable to cells. Once inside the cells, the peptides are cleaved by Atg4, releasing and dequenching the labeled fluorescent dye. The fluorescence is captured by a CCD camera and the intensity reflects the Atg4 activity. The advantage of this system is that there is no involvement of any secondary reactions and there are no complicated methods needed to introduce the substrate into cells. A different concern is the use of a 4-amino acid peptide based on the consensus sequence of several Atg8 homologs, instead of the full length proteins, as the substrate of Atg4 will likely reduce the efficiency of the cleavage reaction because of the requirement for conformational change of the enzyme triggered only by the full-length substrates.17,20,22 In general, these “in vivo” assays may be suited for analysis of Atg4 activity in situ, but the substrate may be rapidly hydrolyzed by the high basal level of Atg4 activity as soon as it is introduced into cells. These assays are also in general difficult to be configured to the high throughput format.
The assay reported in this study uses FRET-based substrates and is able to sensitively detect cleavage by changes in fluorescence intensity at two different wavelengths. Much less substrate is needed in this assay compared with the SDS-PAGE based assay. In addition, the FRET-based assay delivers time- and dose-dependent quantitative responses and has a broad dynamic range sufficient for quantifying and discriminating Atg4 activities under different conditions. The use of a full-length molecule as the substrate can ensure maximal specificity and hydrolysis efficiency and at the same times allow both the LC3 and the GABARAP subfamily substrates to be used to differentiate the activity of Atg4A and Atg4B. It would be also less prone to interference than other fluorescence/luminescence based assays as the signals are directly coupled with the Atg4 activity without the involvement of a secondary reaction.
With these properties, this assay has been successfully used to determine the kinetic properties of the recombinant Atg4A and Atg4B enzymes, to determine Atg4 activity in cell lysates, and to assess the potency of chemical inhibitors. The robust performance of the assay in high throughput format suggests that it is suitable for screening chemical modulators. Finally, although not examined in this study, the FRET-substrate construct can be transfected into cells to analyze Atg4 activity in situ. It is thus believed that this FRET-based Atg4 assay has overcome some of the key limitations of current methodology and offers an efficient way to determine Atg4 activity under a variety of conditions.
Materials and Methods
Chemicals, antibodies, bacterial stains and plasmids
All chemical reagents were obtained from Sigma-Aldrich unless otherwise stated. Anti-Flag was obtained from Sigma-Aldrich (F3165), anti-Atg4B was from Abgent (Ap1809c), anti-GAPDH was from Novus (NB300-221) and secondary antibodies conjugated with HRP were from Jackson Immunochemicals (115-035-062 and 111-035-045). The Escherichia coli strain BL21 (DE3) and the plasmid pcDNA3.1 (+) (V790-20) were from Invitrogen. The plasmid pET-28a (+)(69864) was from Novagen, and pEYFP-C1 (6004-1) and pECFP-C1 (6075-1) were from Clontech.
Protein expression and purification
The open reading frames encoding human Atg4A, Atg4B, Atg4BC74S, LC3B and GATE-16 were cloned and constructed as previously described.17 To express LC3B and GATE-16 with the N-terminal His-CFP-tag and C-terminal YFP-tag in E. coli, full-length LC3B or GATE-16 was first cloned into pET-28a (+) between the BamHI and HindIII restriction sites. The DNA fragment of ECFP and YFP were amplified from the commercial vectors pECFP-C1 and pEYFP-C1. ECFP was inserted at the N-terminal side of LC3B or GATE-16 at the NdeI and BamHI sites, whereas YFP was inserted at the C-terminal side at the HindIII and XhoI sites. This resulted in six additional nucleotides at both fusion sites. FRET-LC3BG120A was constructed using the ExSite in vitro site-directed mutagenesis system (Agilent Stratagene). All constructs were confirmed by sequencing and transformed into BL21 (DE3) for expression following IPTG (Research Products International Corp., I56000) induction. Expressed proteins were purified by affinity chromatography using PrepEase (USB, 78796) and gel filtration using a Superdex 75 column (GE Health, 17-1068-01), and verified by SDS-PAGE and Commassie Brilliant Blue (CBB) staining.
Cell culture, transfection and immunoblot assay
HEK-293A cells were grown in DMEM containing 10% fetal bovine serum and 1% antibiotics. To express Atg4 tagged with Flag in mammalian cells, the cDNA fragments encoding Atg4A, Atg4B and Atg4BC74S with the tag were introduced into the mammalian expression vector pcDNA3.1 (+). HEK-293A cells were transfected with the indicated constructs using Lipofectamine 2000 transfection reagent (Invitrogen, 11668-027) according to the manufacturer’s instruction. After 24 h incubation, transfected cells were harvested and lysed with Buffer A (150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM glycerophosphate, 1 mM Na3VO4, 1% Triton X-100, 50 mM TRIS-HCl, pH 7.5). To suppress the endogenous expression level of Atg4B, siRNA against Atg4B (SantaCruz Biotech, sc-72584) was transfected into HEK293A cells using Oligofectamine (Invitrogen).
For immunoblot assay, lysates were separated by SDS-PAGE and transferred to PVDF membranes. After the application of the appropriate antibodies, signals were developed using Immobilon Western detection kit (Millipore, WBKLS0500) according to the manufacturer’s instructions.
Analysis of the Atg4-mediated cleavage of FRET-substrates by SDS-PAGE
Purified FRET-LC3B, FRET-LC3BG120A and FRET-GATE-16 (5 µg) were mixed with 0.25 µg of purified Atg4A, Atg4B or Atg4BC74S or 25 µg of HEK-293A cell lysate in Buffer B (150 mM NaCl, 1 mM EDTA, 50 mM TRIS-HCl, pH 7.5). After incubation at 37°C for different times the reactions were stopped by the addition of sample buffer, separated by SDS-PAGE and examined by CBB staining. The full-length protein (CFP-LC3B-YFP and CFP-GATE-16-YFP) would be cleaved into CFP-LC3B or CFP-GATE-16, and YFP by the proper Atg4. The percentage of cleavage was calculated as previously described.17 Briefly, the optical density (OD) of the product protein bands is divided by the OD of all the protein bands: percentage of cleavage (%) = 100% * (ODYFP + ODCFP-LC3B/CFP-GATE-16)/ (ODYFP + ODCFP-LC3B/CFP-GATE-16 + ODFRET-LC3B/FRET-GATE-16).
Measurement of Atg4 activity using the FRET-based assay
Purified Atg4A and Atg4B proteins or cell lysate were mixed with FRET-LC3B or FRET-GATE-16 in Buffer B. The assay volumes were 400 µl (cuvette format), 200 µl (96-well format) or 20–50 µl (384-well format, respectively. After incubation at 37°C for a given time, the cleavage of the substrates was measured using a fluorescence spectrometer (Cary Eclips, Agilent, or M5, Molecular Device). The excitation wavelength was 434 nm. Emission was measured at 477 nm (CFP) and 527 nm (YFP or FRET). The RFU ratio of 527 nm/477 nm was calculated and used to determine the degree of cleavage since it decreased proportionally to the extent of substrate cleavage. The percentage of cleavage of the substrates was thus determined based on the proportion of the total RFU change, which is calculated as following: percentage of cleavage (%) at a given condition (x) = (RFUmax - RFUX)/(RFUmax - RFUmin)) *100%, in which the RFUs are the 527 nm/477 nm ratios of reactions under no cleavage (RFUmax), under highest cleavage (RFUmin), and under a given condition (RFUX).
Measurement of Atg4 kinetics using the FRET-based assay
Both FRET-LC3B and FRET-GATE-16 can be efficiently cleaved by Atg4B at a dose of 25–500 µg/ml. Atg4A or Atg4B were incubated with FRET-LC3B or FRET-GATE-16 at different concentrations in Buffer B in a total volume of 200 µl at 37°C for the designated times. The kinetics were determined as previously described32 with modifications. In brief, the initial velocity of the cleavage reaction was defined as V = [S]t/t, in which [S]t is the substrate concentration ([S]) at a given time point (t). [S]t was estimated based on the fluorescence change of the substrates at 527 nm ([S]t-527nm) as well as on the fluorescence change of the substrates at 477 nm ([S]t-477nm), so that [S]t = ([S]t-527nm + [S]t-477nm)/2. [S]t-527nm was calculated as [S]*(RFUmax-527nm − RFUt-527nm)/(RFUmax-527nm − RFUmin-527nm), whereas [S]t-477nm = [S]*(RFUt-477nm − RFUmin-477nm)/(RFUmax-477nm − RFUmin-477nm), in which [S] is the initial substrate concentration, RFUmax and RFUmin are the fluorescence intensity at the maximal and minimal levels across the entire reaction period at the corresponding wavelength, and RFUt is the fluorescence intensity at a given time (t). The initial velocity (V) was then plotted against the concentration of the substrate at the given time points ([S]t) and the curve was fitted using the ligand-binding method (SigmaPlot 10.0, Systat Software), from which the Vmax ([S]/sec) and KM (Michaelis constant, [S]) for each enzyme-substrate reaction were derived based on the Michaelis-Menten formula. The catalytic constant (kcat, sec−1) was determined by dividing Vmax by the concentration of the enzyme. The catalytic efficiency is defined as kcat/KM (mol−1Lsec−1).
High-throughput screening format of the Atg4 assay
Atg4A (10 µg/ml) or Atg4B (2 µg/ml) were first incubated with chemical compounds (10 μM) for 30 min at 37°C in 384-well plates. FRET-GATE-16 or FRET-LC3B (60–100 µg/ml) was then added to a total volume of 20–50 µl. After 30 min (for the Atg4B/FRET-LC3B reaction) or 60 min (for the Atg4A/FRET-GATE-16 reaction), the fluorescence intensity was recorded. The relative cleavage activity of Atg4A or Atg4B was calculated based on the change of 527 nm/477 nm ratio as described above. The positive control for the cleavage was without any chemicals, whereas the negative control was without the Atg4 enzyme. The performance of the screening was measured by the Z factor (Z’): Z’ = 1–3*(σp+σn)/|µp-µn|, where σ is the standard deviation and µ is the mean for positive (p) and negative (n) controls.27
The potency of inhibition by a given chemical was measured by the percentage of the inhibition of Atg4 cleavage activity, which was calculated using the formula modified from the one used for calculating the percentage of cleavage (see above): percentage of inhibition (%) = (RFUX - RFUmin)/(RFUmax – RFUmin)*100%, in which RFUX is the RFU ratio of 527 nm/477 nm in the presence of a given concentration of the chemical, RFUmin is the ratio in the absence of chemicals, and RFUmax is the ratio in the presence of a maximal inhibitory concentration of the chemical. IC50 was determined by plotting the percentage of inhibition against the concentration of the chemical, which was fitted with a nonlinear 4-parameter fitting method using SigmaPlot 10.0.
Statistical analysis
All experiments have been performed at least three times. Data shown are the mean ± SD from three experiments and were subjected to t-test or one-way ANOVA followed by Holm-Sidak’s post-hoc analysis as indicated in the figure legends. A level of p < 0.001 was considered significant.
Acknowledgment
This work is in part supported by an NIH grant to X.-M.Y. (R01CA 83817).
Glossary
Abbreviations:
- AEBSF
4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride
- CBB
Coomassie Brilliant Blue
- CFP
cyan fluorescent protein
- FRET
fluorescence resonance energy transfer
- GABARAP
γ-aminobutyric acid receptor-associated protein
- GATE-16
Golgi-associated ATPase enhancer-16
- HTS
high throughput screening
- LC3B
microtubule-associated protein 1 A/B light chain 3B
- NEM
N-ethylmaleimide
- RFU
relative fluorescence unit
- TCEP
Tris-2-carboxyethyl-phosphine
- YFP
yellow fluorescent protein
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/18777
References
- 1.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–48. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 4.Komatsu M, Kominami E, Tanaka K. Autophagy and neurodegeneration. Autophagy. 2006;2:315–7. doi: 10.4161/auto.2974. [DOI] [PubMed] [Google Scholar]
- 5.Jin S, White E. Role of autophagy in cancer: management of metabolic stress. Autophagy. 2007;3:28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ichimura Y, Imamura Y, Emoto K, Umeda M, Noda T, Ohsumi Y. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J Biol Chem. 2004;279:40584–92. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- 7.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–8. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 9.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–76. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16:2967–83. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–76. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 12.Tanida I, Komatsu M, Ueno T, Kominami E. GATE-16 and GABARAP are authentic modifiers mediated by Apg7 and Apg3. Biochem Biophys Res Commun. 2003;300:637–44. doi: 10.1016/S0006-291X(02)02907-8. [DOI] [PubMed] [Google Scholar]
- 13.Tanida I, Sou YS, Minematsu-Ikeguchi N, Ueno T, Kominami E. Atg8L/Apg8L is the fourth mammalian modifier of mammalian Atg8 conjugation mediated by human Atg4B, Atg7 and Atg3. FEBS J. 2006;273:2553–62. doi: 10.1111/j.1742-4658.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- 14.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mariño G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez-Otin C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278:3671–8. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 16.Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem. 2003;278:51841–50. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- 17.Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin XM. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem. 2011;286:7327–38. doi: 10.1074/jbc.M110.199059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scherz-Shouval R, Sagiv Y, Shorer H, Elazar Z. The COOH terminus of GATE-16, an intra-Golgi transport modulator, is cleaved by the human cysteine protease HsApg4A. J Biol Chem. 2003;278:14053–8. doi: 10.1074/jbc.M212108200. [DOI] [PubMed] [Google Scholar]
- 19.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J Biol Chem. 2005;280:40058–65. doi: 10.1074/jbc.M509158200. [DOI] [PubMed] [Google Scholar]
- 21.Ketteler R, Sun Z, Kovacs KF, He WW, Seed B. A pathway sensor for genome-wide screens of intracellular proteolytic cleavage. Genome Biol. 2008;9:R64. doi: 10.1186/gb-2008-9-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu CW, Drag M, Bekes M, Zhai D, Salvesen GS, Reed JC. Synthetic substrates for measuring activity of autophagy proteases: autophagins (Atg4) Autophagy. 2010;6:936–47. doi: 10.4161/auto.6.7.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jares-Erijman EA, Jovin TM. FRET imaging. Nat Biotechnol. 2003;21:1387–95. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 24.Kouno T, Mizuguchi M, Tanida I, Ueno T, Kanematsu T, Mori Y, et al. Solution structure of microtubule-associated protein light chain 3 and identification of its functional subdomains. J Biol Chem. 2005;280:24610–7. doi: 10.1074/jbc.M413565200. [DOI] [PubMed] [Google Scholar]
- 25.Paz Y, Elazar Z, Fass D. Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J Biol Chem. 2000;275:25445–50. doi: 10.1074/jbc.C000307200. [DOI] [PubMed] [Google Scholar]
- 26.Choi KM, Nam HY, Na JH, Kim SW, Kim SY, Kim K, et al. A monitoring method for Atg4 activation in living cells using peptide-conjugated polymeric nanoparticles. Autophagy. 2011;7:1052–1062. doi: 10.4161/auto.7.9.16451. [DOI] [PubMed] [Google Scholar]
- 27.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 28.Mariño G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–83. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 29.Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci. 2009;122:2554–66. doi: 10.1242/jcs.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mariño G, Fernandez AF, Cabrera S, Lundberg YW, Cabanillas R, Rodriguez F, et al. Autophagy is essential for mouse sense of balance. J Clin Invest. 2010;120:2331–44. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ketteler R, Seed B. Quantitation of autophagy by luciferase release assay. Autophagy. 2008;4:801–6. doi: 10.4161/auto.6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauer-Fields JL, Spicer TP, Chase PS, Cudic M, Burstein GD, Nagase H, et al. Screening of potential a disintegrin and metalloproteinase with thrombospondin motifs-4 inhibitors using a collagen model fluorescence resonance energy transfer substrate. Anal Biochem. 2008;373:43–51. doi: 10.1016/j.ab.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]