Abstract
Cationic liposome–DNA (CL–DNA) complexes are being pursued as nonviral gene delivery systems for use in applications that include clinic trials. However, to compete with viral vectors for systemic delivery in vivo, their efficiencies and pharmacokinetics need to be improved. The addition of poly (ethylene glycol)-lipids (PEGylation) prolongs circulation lifetimes of liposomes, but inhibits cellular uptake and endosomal escape of CL–DNA complexes. We show that this limits their transfection efficiency (TE) in a manner dependent on the amount of PEG-lipid, the lipid/DNA charge ratio, and the lipid membrane charge density. To improve endosomal escape of PEGylated CL–DNA complexes, we prepared an acid-labile PEG-lipid (HPEG2K-lipid, PEG MW 2000) which is designed to lose its PEG chains at the pH of late endosomes. The HPEG2K-lipid and a similar but acid-stable PEG-lipid were used to prepare PEGylated CL–DNA complexes. TLC and dynamic light scattering showed that HPEG2K-CL–DNA complexes are stable at pH 7.4 for more than 24 hours, but the PEG chains are cleaved at pH 5 within one hour, leading to complex aggregation. The acid-labile HPEG2K-CL–DNA complexes showed enhanced TE over complexes stabilized with the acid-stable PEG-lipid. Live-cell imaging showed that both types of complexes were internalized to quantitatively similar particle distributions within the first 2 hours of incubation with cells. Thus, we attribute the increased TE of the HPEG2K-CL–DNA complexes to efficient endosomal escape, enabled by the acid-labile HPEG2K-lipid which sheds its PEG chains in the low-pH environment of late endosomes, effectively switching on the electrostatic interactions that promote fusion of the membranes of complex and endosome.
1. Introduction
Gene therapy is a promising approach in medicine, which is applicable to a wide variety of diseases. Numerous clinical trials are ongoing, with the majority focusing on cancer [1–4]. However, improved vectors for nucleic acid delivery are needed for gene therapy to reach its potential. Nonviral (synthetic) vectors, such as complexes of DNA with cationic lipids or polymers, are desirable because they possess low immunogenicity, can easily be scaled up and chemically modified, and can transfer DNA of any size [5–10]. However, in comparison with viral vectors, their TE is lower, and their pharmacokinetic properties in vivo suffer from their poor colloidal stability [11–16]. Thus, development of nonviral vectors for systemic in vivo applications remains a major challenge [17].
Specific challenges for in vivo applications of CL–DNA complexes and other nonviral vectors include their rapid clearance from circulation and activation of the complement system [18, 19]. A strategy to address these challenges is surface functionalization of CL–DNA complexes by poly (ethylene glycol) (PEG)-lipids, resulting in steric stabilization. This reduces the nonspecific interactions of CL–DNA complexes and blood components and is well known to increase circulation time for liposomes [20–22]. However, the steric shielding due to PEGylation not only reduces undesired interactions, but also suppresses the electrostatic interactions required for cellular uptake [23]. In addition, it interferes with endosomal escape of complexes by inhibiting fusion of the membranes of the complex with those of the endosome [24, 25]. Both of these processes are keys steps in the transfection mechanism. Endosomal escape, in particular, is a critical step in achieving successful transfection [26, 27], and the TE-limiting step for lamellar complexes of low to intermediate membrane charge density [28, 29]. After uptake by endocytosis, the DNA cargo of complexes which are unable to escape from endosomes eventually gets degraded as endosomes mature and fuse with lysosomes.
There are multiple design strategies that can be implemented to overcome the barriers associated with the use of PEGylation, such as tethering a specific ligand to improve low binding and uptake into the targeted cells [30–32], or designing the PEG shield to be pH-sensitive to enhance endosomal escape [33]. The latter approach has been explored widely with liposomes [34–38], and has also found some recent application with complexes of cationic polymers and nucleic acids (polyplexes) [39–43]. The only (to the best of our knowledge) work reporting use of this approach in CL–DNA complexes [44] applied an orthoester-based PEG-lipid.
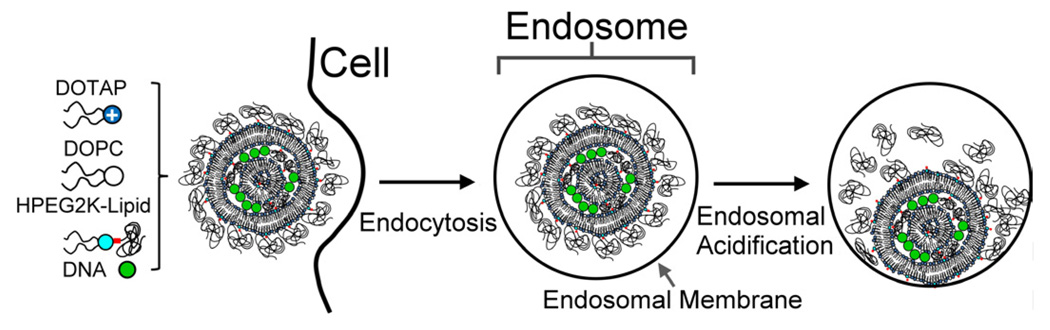
We have designed and synthesized an acylhydrazone-based acid-labile PEG-lipid (HPEG2K-lipid, PEG MW 2000) that is stable at physiological pH, but whose PEG-chain will be cleaved from the tail at lower pH, such as occurs intracellularly in maturing endosomes [45]. Thus, complexes containing HPEG2K-lipid will be sterically stabilized in the extracellular environment but lose their PEG shell in the endosome, which in turn should allow for more efficient endosomal escape of the complex because the electrostatic interactions between the membranes of complex and endosome are no longer inhibited (cf. Fig. 1). We prepared CL–DNA complexes containing HPEG2K-lipid and, for comparison, a similar but low pH-stable PEG-lipid (PEG2K-lipid), starting from mixtures of the neutral lipid 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC) with univalent cationic 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) or a custom-synthesized pentavalent cationic lipid (MVL5 [46]). The cationic lipid content was fixed at 80% mol DOTAP and 50% mol MVL5, respectively, which is in the membrane charge density (M, average charge/unit area of membrane) regime optimal for transfection [29]. We compared PEGylated complexes containing PEG2K-lipid vs. HPEG2K-lipid using dynamics light scattering (DLS, to identify their time-dependent and pH-dependent colloidal stability), live-cell imaging (to assess intracellular particle distributions), and TE measurements.
Fig. 1.
Schematic depiction of the intracellular processing of CL–DNA complexes stabilized with the low pH-sensitive HPEG2K-lipid. Complexes are taken up by the cell via endocytosis (left and middle). As the endosome matures, its contents are acidified. This cleaves the PEG-chains from the HPEG2K-lipid, which allows closer contact between the membranes of complex and endosomes. This in turn facilitates CL–DNA complex escape via membrane fusion.
2. Materials and Methods
2.1. Synthesis
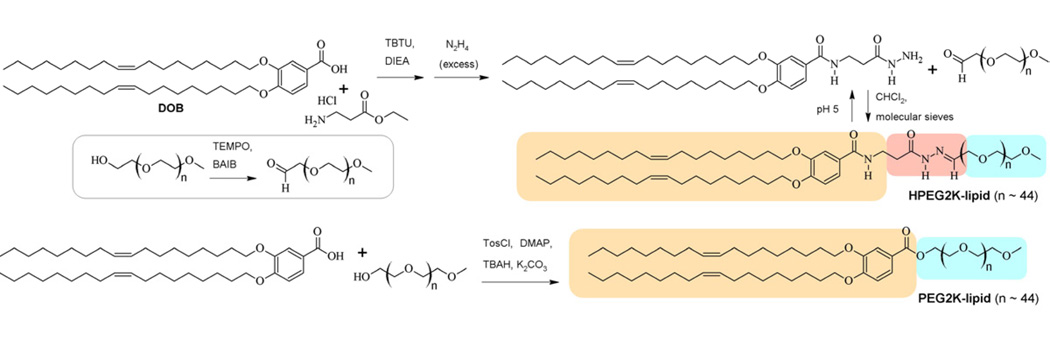
An acylhydrazone-based PEG2000-lipid and a structurally similar but low pH-stable PEG2000-lipid were synthesized according to the procedure described in the Supplementary Material. The synthesis and chemical structure of these PEG-lipids are shown in Scheme 1.
Scheme 1.
Synthesis of the acid-labile HPEG2K-lipid and the PEG2K-lipid. The lipid tails are underlain in tan, the acid-labile acylhydrazone moiety in red, and the PEG chains in blue.
2.2. Liposome Preparation
Lipofectamine 2000 was purchased from Invitrogen and used as per manufacturer’s instructions. DOTAP and DOPC were purchased as solutions in chloroform from Avanti Polar Lipids (Alabaster, AL). MVL5 was synthesized as described previously [46]. For calculation of the lipid to DNA charge ratio, full protonation (headgroup charge +5e) was assumed for MVL5. All lipids were dissolved in chloroform/methanol (9:1, v/v), and these solutions were mixed at the appropriate ratios in glass vials, dried under a stream of nitrogen, and then placed in a vacuum (rotary vane pump) for 8–12 h. To the resulting thin lipid film, sterile high resistivity (18.2 MΩ cm) water was added, and the mixture was incubated at 37 °C for at least 12 h. The final total lipid concentration was 1 mM. All aqueous lipid solutions were sonicated prior to use and stored at 4 °C.
2.3. Dynamic Light Scattering
The diameter of CL–DNA complexes was measured using a Dynapro Nanostar dynamic light scattering (DLS) instrument (Wyatt) and Dynamics 7 software. Complexes were prepared in light scattering vials (0.4 µg DNA per vial) by combining appropriate volumes of liposome (1 mg/mL) and plasmid DNA (1.4 mg/mL) stock solutions that had been diluted with Opti-MEM (Invitrogen) or buffer, to yield a final volume of 200 µL. The complexes ((H)PEG–lipid/DOTAP/DOPC = x/80/10-x, where x = 0, 5, 10) were incubated at 37 °C for the indicated time and their mean diameter measured. The pH-dependent experiments were performed at room temperature in buffers of the specified pH (pH=7.4, 6.0, 5.0, and 4.0) which contained a total of 150 mM total monovalent salt (buffer plus added monovalent salt; see Supplementary Material) to enable comparison with the results in Opti-MEM. Plots show the z-average hydrodynamic diameter as obtained from the instrument. Data points shown are the average of duplicate measurements of the same sample, with error bars showing the standard deviation.
2.4. Cell Culture and Transfection [47]
Luciferase plasmid DNA (pGL3 Control Vector, Promega) was propagated in E. coli and isolated using a Qiagen Giga Kit. Mouse fibroblast L-cells (ATCC number: CCL-1) were maintained at 37 °C in supplemented culture medium (Dulbecco’s Modified Eagle’s Medium (DMEM), containing 5% fetal bovine serum (Thermo Scientific) and 1% penicillin (Invitrogen) in a humidified atmosphere with 5% CO2 and reseeded approximately every 72 h to maintain subconfluency. For transfection, 80 000 cells/well were seeded in 24-well plates and incubated for 18–24 h prior to transfection. A solution of pGL3 plasmid DNA (4 µg/mL in Opti-MEM) was prepared from a DNA stock solution at 1.4 mg/mL. Appropriate volumes of liposome solutions (to yield the desired lipid to DNA charge ratio) were diluted with Opti-MEM. Equal volumes of liposome and DNA solutions were combined, and 200 L of this mixture (containing 0.4 µg DNA) were added per well after incubation for 20 min at room temperature. After 6 h incubation at 37 °C, the transfection medium was removed, each well was washed once with PBS, and fresh culture medium was added to each well. After another 20–24 h of incubation, cells were harvested in 150 µL of Passive Lysis Buffer (Promega), and luciferase expression was measured as per the assay manufacturer’s (Promega) instructions. A multilabel counter (Perkin-Elmer 1420 Victor3 V) was used to measure the relative light units (RLU) from the luminescence assay. Data points shown are the average of duplicate (and in some cases quadruplicate) measurements, with error bars showing the standard deviation. All experiments were repeated 2–3 times to ascertain reproducibility.
2.5. Cytotoxicity
Cytoxicity was assessed using the CellTiter 96 Aqueous One assay (Promega) for cell viability. Mouse fibroblast L-cells were seeded in 96 well plates (15 000 cells/well) in DMEM medium supplemented with 5% FBS and 1% penicillin. After 18–24 hours incubation, the cells were washed once with PBS, and subsequently 40 µL solution including medium and complexes were added (transfection concentration; 0.08 µg DNA per well). After 6 hours incubation, the complex-containing medium was replaced by a mixture of 60 µL of Opti-MEM and 20 µL of the Cell Proliferation Assay. Following 2 h of incubation, absorbance at 490 nm was measured using a scanning multi-well spectrophotometer. The experiment was performed simultaneously for all lipids and the results were normalized to control wells, which differed from the experimental wells only in that they were treated with Opti-MEM instead of PEGylated nanoparticles. Each data point represents the average of at least quadruplicate measurements, with error bars showing the standard deviation.
2.6. Optical Microscopy
Mouse fibroblast L-cells were grown to 70% confluency on poly(L-lysine)-coated glass coverslips (22 mm). The coverslips were mounted in a closed, temperature controlled chamber from Harvard Apparatus (Model # P2 and RC21-B). Images were acquired using a Nikon Diaphot 300 equipped with a Sensicam QE CCD. Brightfield images were acquired at 60-fold magnification in differential interference contrast (DIC) mode. CL–DNA complexes were prepared at transfection concentration by combining appropriate volumes solutions of fluorescently labeled liposomes (2 mM; 0.2% (of lipid weight) TRITC-DHPE (Molecular Probes)) and plasmid DNA (0.1 mg/ml stock, labeled with Cy5; diluted with Opti-MEM to the desired concentration). DNA was labeled using the Mirus Label IT Nucleic Acid Labeling Kit according to the manufacturer's protocol. The complexes were incubated for 20 min at room temperature and then added to the cells (maintained at 37 °C).
An automated Matlab routine was used to measure the uptake of particles at various timepoints (see also Fig. 6C). The routine located intracellular fluorescent particles containing Cy5 and measured the closest distance to the nuclear membrane (the DIC image was used to locate the nucleus). The cell boundary was determined using the brightfield image or the fluorescent image: early time points show a high number of fluorescent particles coating the plasma membrane which allows for clear visualization of the cell boundary in fluorescent micrographs.
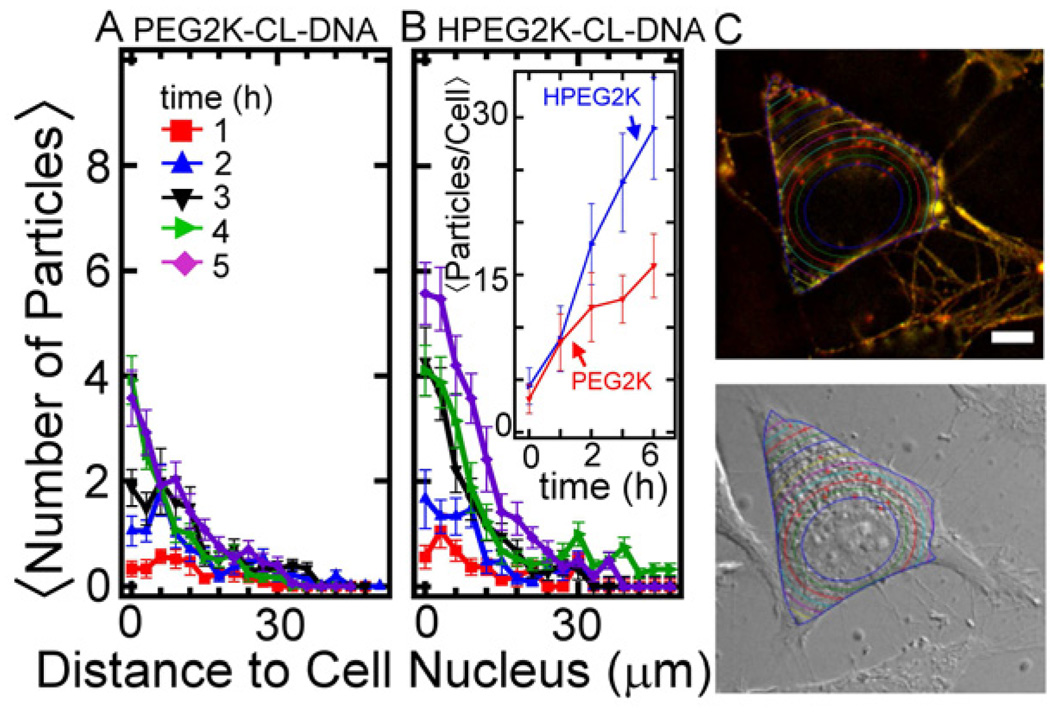
Fig. 6.
Intracellular distributions of PEGylated CL–DNA complexes (10 mol% PEG-lipid) obtained via image analysis. The number of resolved particles is plotted as a function of the distance from the nucleus for complexes containing PEG2K-lipid (A) and HPEG2K-lipid (B). Particles were only counted if they were located within the boundary of the cytoplasmic membrane. Each data point indicates the average and standard deviation from images of at least 20 cells. The inset shows the average total number of resolved particles per cell as a function of time for the two types of complexes. (C) Micrographs illustrating the automated image analysis routine. The cell nucleus was located from the DIC image, and the cell boundary from DIC and fluorescent images (cf. Fig. 5C). Colored lines equidistant from the nuclear membrane indicate how distance from nucleus was measured, and red crosshairs indicated located complexes.
3. Results and Discussion
To enable applications of nonviral vectors in systemic gene therapy in vivo, vectors with prolonged circulation times and high transfection efficiency (TE) need to be developed. Prolonged circulation may be achieved by PEGylation as shown for liposomes, but this significantly decreases TE. Causes for the drop in TE are inhibition of the electrostatic interactions between complexes and the cell surface, and interference of the PEG coat with the membrane fusion processes required for endosomal escape. One approach to counteract the latter effect is to attach PEG chains with a pH-sensitive linker which is stable at neutral pH (the pH of blood) but will be cleaved at pH 5 to 4, the pH of late endosomes. Figure 1 schematically depicts the rationale for this approach: after complexes are taken up by the cell via endocytosis (left and middle), the endosome matures, and its contents are acidified. This cleaves the PEG-chains from the HPEG2K-lipid, allowing closer contact between the membranes of complex and endosomes and thus facilitating escape of the CL–DNA complex via membrane fusion, which is mediated by electrostatic interactions. In other words, synthetic modification of the PEG-lipid enables a physicochemical route for endosomal escape as the contents of the endosome are acidified.
3.1. Lipid Design and Synthesis
To investigate this approach in detail, we designed and synthesized a new acid-labile PEG-lipid (HPEG2K-lipid) featuring an acylhydrazone bond which is prone to hydrolysis at low pH [39]. The HPEG2K-lipid was synthesized following the steps shown in Scheme 1. The lipid tail building block 3,4-dioleyloxybenzoic acid (DOB) [48, 49] was coupled with -alanine ethyl ester and the resulting ester treated with hydrazine to give acylhydrazide 1. To prepare the HPEG2K-lipid, this was reacted under anhydrous conditions with the aldehyde of PEG2000 monomethylether (mPEG2000), prepared via TEMPO oxidation [50]. In the structure of the HPEG2K-lipid in Scheme 1, the lipid tails are underlain in tan, the acid-labile acylhydrazone moiety in red, and the PEG chain in blue. TLC analysis shows that the HPEG2K-lipid is stable at pH 7.4, but quickly degrades at pH 4 to 5 (see Supplementary Material). The synthesis and structure of the acid-stable (on the time-scale of intracellular processing) PEG2K-lipid, which was prepared by esterification of DOB with mPEG2000 [48, 51], is also shown in Scheme 1.
3.2. Complex Size and Colloidal Stability
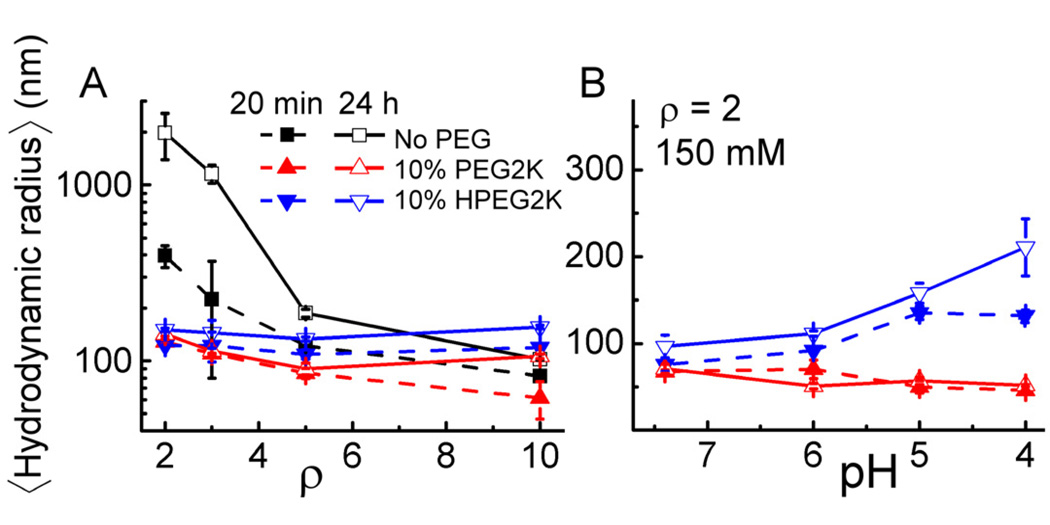
CL–DNA complexes were formed by combining DNA and aqueous lipid preparations. All complexes were prepared at a fixed mol fraction of cationic lipid. To determine the colloidal stability of PEGylated and nonPEGylated CL–DNA complexes, we measured their mean hydrodynamic radius by dynamic light scattering (DLS). Figure 2A shows the results in transfection medium (Opti-MEM; pH 7.4) for DOTAP/DOPC–DNA complexes (ΦDOTAP = 0.8) containing no PEG-lipid or 10 mol% of PEG2K-lipid or HPEG2K-lipid. The size of complexes without PEG-lipid changes drastically as a function of CL to DNA charge ratio (): the CL–DNA complexes are not stable as ρ approaches to the isoelectric point, their size increasing from a few hundred nm for newly formed complexes (20 min incubation, solid black squares) to above one µm within 24 hours incubation (open black squares); as ρ increases, the complexes maintain a well-defined size of about 100 nm because the complexes are overcharged [52] and electrostatically repel each other. All PEGylated complexes (PEG2K-lipid: red triangles; HPEG2K-lipid: blue inverse triangles) exhibit a size of around 100 nm in Opti-MEM, independent of and incubation time (Fig. 2A). The steric shielding due to PEGylation reduces particle-particle interaction, preventing aggregation even at low. Thus, PEGylated complexes will remain colloidally stable in transfection medium for more than the incubation period (6 hours) of cells with complexes, irrespective of PEG-lipid linker chemistry.
Fig. 2.
Mean hydrodynamic radius of CL–DNA complexes measured by DLS. (A) Complex size as a function of ρ at Opti-MEM (pH 7.4). (B) Size of complexes of ρ = 2 at varied pH. All CL–DNA complexes were prepared at ΦDOTAP = 0.8, i.e., with the lipid composition PEG-lipid/DOTAP/DOPC = x/80/20-x, with x = 0 or 10. Each pH buffer used to obtain the data in Fig. 2B contained a total of 150 mM monovalent salt. Error bars indicate the standard deviation of triplicate measurements.
Fig. 2B shows the sizes of PEGylated complexes (at = 2, where the size of nonPEGylated complexes is not stable over time) measured in buffers of increasing acidity at a total (buffer + added salt) monovalent salt concentration of 150 mM, which is similar to that of Opti-MEM. The size of complexes incorporating the pH-stable PEG2K-lipid (red triangles) is essentially independent of solution pH and time over the investigated time period of 24 h, with a very small decrease in size with decreasing pH. In contrast, the size of HPEG2K-lipid containing complexes (blue inverse triangles) increases with decreasing pH in a time-dependent fashion. This indicates complex aggregation due to the acid-induced dePEGylation caused by cleavage of HPEG2K-lipids, demonstrating that the acid-sensitivity of the HPEG2K-lipid can be exploited when it is incorporated into CL–DNA complexes. Importantly, the pH threshold for dePEGylation, at pH ≤ 5, corresponds to the pH of maturing endosomes.
3.3. Transfection Efficiency and Cytotoxicity
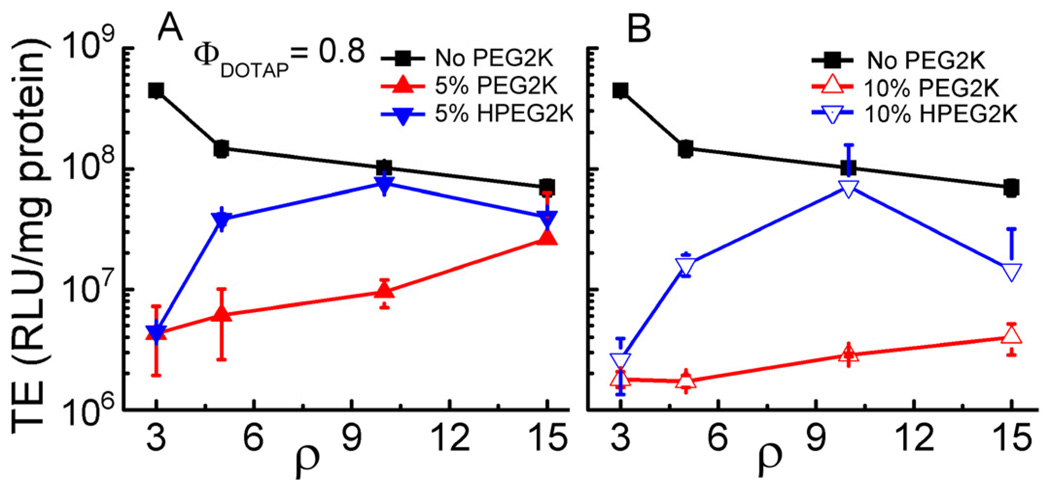
We first assessed the effect of PEGylation on transfection efficiency using the model system of DOTAP/DOPC–DNA complexes at ΦDOTAP = 0.8, i.e., with the lipid composition PEG-lipid/DOTAP/DOPC = x/80/20-x. TE was measured using a luciferase reporter gene assay in mouse L-cells. Prior work has identified ΦDOTAP = 0.8 as yielding optimal TE for DOTAP/DOPC–DNA complexes [29]. Figure 3 shows TE data for various DOTAP/DOPC–DNA complex formulations as function of ρ. In the absence of PEG-lipid (black solid squares), the maximum of TE occurs at ρ = 3, and TE then slightly decreases with increasing ρ. Addition of 5 mol% PEG2K-lipid decreases TE by two orders of magnitudes at ρ = 3, but the TE of these complexes increases with ρ to within an order of magnitude of the TE of nonPEGylated complexes at ρ = 15 (Fig. 3A, red solid triangles). The addition of 10 mol% PEG2K-lipid (Fig. 3B, red open triangles) reduced TE to levels comparable to the TE of uncomplexed DNA (≈106 RLU/mg protein). The TE of these complexes is almost independent of ρ, showing only a slight increase with ρ. Replacing the pH-stable PEG2K-lipid with the acid-labile HPEG2K-lipid has a large effect on TE (Fig. 3, blue inverse triangles). The TE of HPEG2K-containing complexes is higher for all data points and exhibits a maximum TE, at ρ = 10, approaching the TE of nonPEGylated complexes at this ρ for both 5 and 10 mol% HPEG2K-lipid.
Fig. 3.
Transfection efficiency (TE) of nonPEGylated and PEGylated DOTAP/DOPC–DNA complexes versus ρ in mouse fibroblast L-cells. The lipid compositions were (H)PEG2K-lipid/DOTAP/DOPC = x/80/20-x, where x = 0, 5, and 10. The amount of DNA was constant for all data points. Each data point represents the average and standard deviation of at least duplicate measurements.
Of note, 5 mol% PEG2000 lipid corresponds to full coverage of the bilayer with PEG chains in the mushroom conformation, while 10 mol% (the maximum amount that can be incorporated without phase separation) corresponds to PEG chains in the brush conformation, i.e., maximum sterical stabilization [53, 54]. Intriguingly, replacing PEG2K-lipid with the pH-sensitive HPEG2K-lipid only increases TE significantly at ρ > 3. The may be due to a reduction in complex stability cause by the incorporation of PEG-lipid, which is indicated by a lower total scattering intensity in DLS and fluorescence microscopy observations.
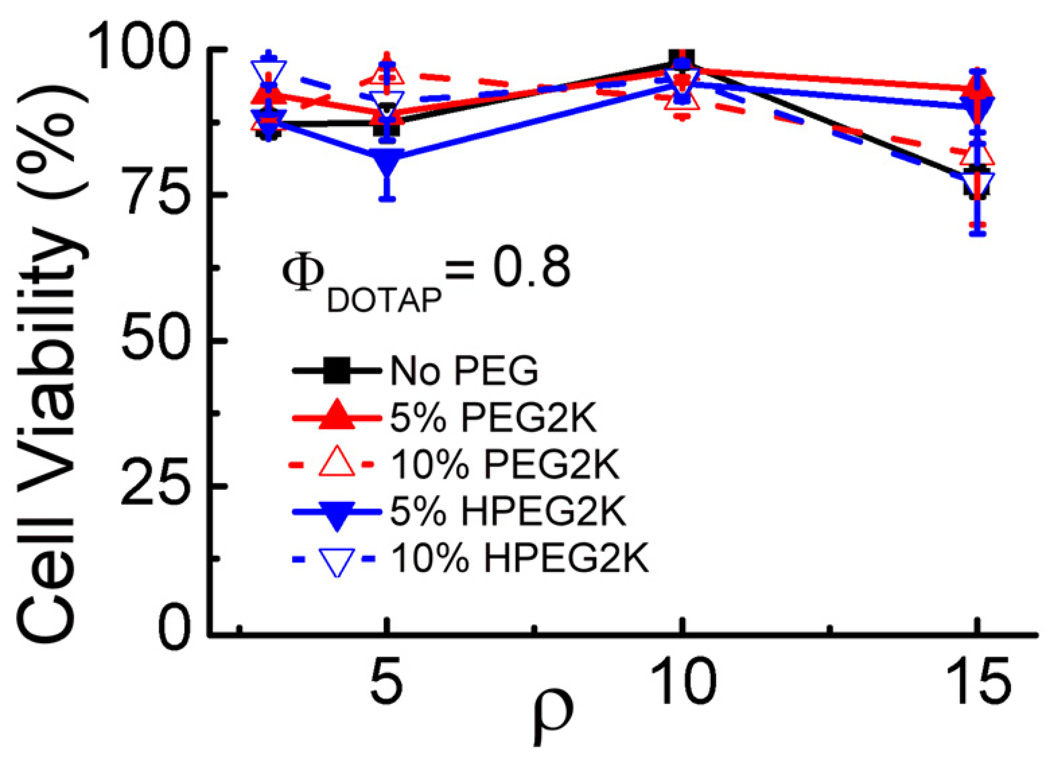
Complexes at higher CL to DNA charge ratios can exhibit lipid-caused vector toxicity [49, 55]. We thus measured cytotoxicity of the complexes used for TE measurements, employing a modified MTT assay [56]. Figure 4 shows the results as percentage of viable cells against ρ. No notable toxicity was observed for any of the investigated complexes. In particular, the toxicity of optimized HPEG-CL–DNA complexes (at ρ = 10) approaches the background value and is identical (within error) to that of nonPEGylated complexes.
Fig. 4.
Cell viability versus ρ for nonPEGylated and PEGylated DOTAP/DOPC–DNA complexes (ΦDOTAP = 0.8). Cell viability was measured by a commercial tetrazolium salt-based assay. The amount of DNA was constant for all data points. Each data point indicates the average and standard deviation from quadruplicate measurements.
3.4. Live cell imaging
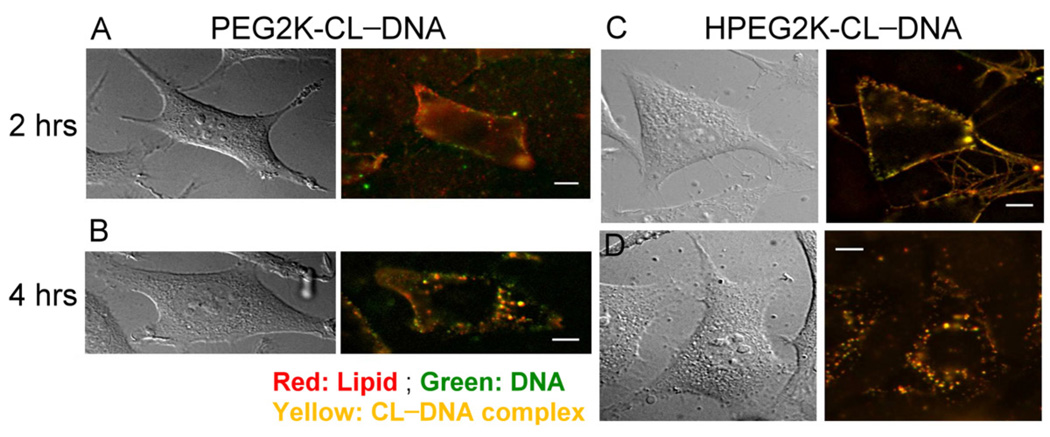
To investigate the intracellular fate of PEGylated CL–DNA complexes directly, we performed epi-fluorescence microscopy on live cells using doubly labeled complexes. Thus, complexes were prepared with Cy5-labeled DNA (green in Fig. 5) and Texas Red-DHPE as the lipid label (red in Fig. 5). Cells were treated with complexes as for transfection, and the location of the complexes was observed over time. Figure 5 shows differential interference contrast (DIC) and fluorescent micrographs of cells incubated with PEGylated (10 mol% PEG-lipid) CL–DNA complexes of = 10 (where HPEG2K-CL–DNA complexes exhibit maximum TE). At 2 hours, both PEGylated CL–DNA complexes (yellow) are bound to the plasma membrane with a small number of complexes localized within the cytoplasm (Fig. 5 A and C), independent of the PEG-lipid used. After 4 hours, fluorescent particles are seen inside of the cells in a punctuate pattern (Fig. 5 B and D). A number of complexes are still observed bound to the plasma membrane after 4 hours, suggesting a low rate constant for uptake once complexes are bound to the membrane. The imaging results were analyzed using an automated MatLab routine to quantitatively determine the intracellular spatial distribution of complexes. The resulting data is shown in Figure 6. Within the first 2 hours, PEG2K-CL–DNA and HPEG2K-CL–DNA complexes show quantitatively similar uptake and cytoplasmic distribution, suggesting the the difference in TE is due to differences in intracellular processing. However, at later time points, a larger number of HPEG-CL–DNA complexes were counted in the perinuclear region. An assessment of the actual images (cf. Fig. 6) suggests that this does not necessarily signify increased uptake. As endosomes mature and fuse (and travel to the perinuclear region, the location of the lysosomal compartment), several particles may share the same endosomal compartment and become indistinguishable by fluorescent microscopy as their separation the resolution limit of the microscope. This effect should be more pronounced for particles that remain trapped in endosmes, i.e., PEG2K-CL–DNA complexes, which indeed show larger, brighter spots in the images. Conversely, as HPEG complexes primarily remain as small, point-like particles which remain spatially separated. This is indirect evidence of endosomal escape and could explain the divergence of the total number of internalized particles for the two types of complexes at later times.
Fig. 5.
Live-cell imaging of transfection by PEGylated CL–DNA complexes: DIC and fluorescence micrographs of mouse L-cells incubated with doubly labeled PEGylated CL–DNA complexes at ΦDOTAP = 0.8 (10 mol% PEG-lipid). (A, B) Images for complexes prepared with the acid-stable PEG2K-lipid. (C, D) Images for complexes prepared with the acid-labile HPEG2K-lipid. After 2 hours of incubation, both types of complexes show are seen attached to the plasma membrane. After 4 hours, complexes containing the PEG2K-lipid appear as large, bright fluorescent spots while complexes containing the HPEG-lipid remain as small, point-like particles. As endosomes mature and fuse, more than one trapped complexes (such as complexes containing the PEG2K-lipid) may share the same endosomal compartment and become indistinguishable by fluorescent microscopy. The comparatively large fluorescent objects observed in (B) suggest that this is the case. In contrast, HPEG2K-CL–DNA complexes that escape from endosomes remain resolvable as individual particles due to the larger average spacing between them. Scale bars indicate 10 µm.
3.5. Results for the Multivalent Lipid MVL5
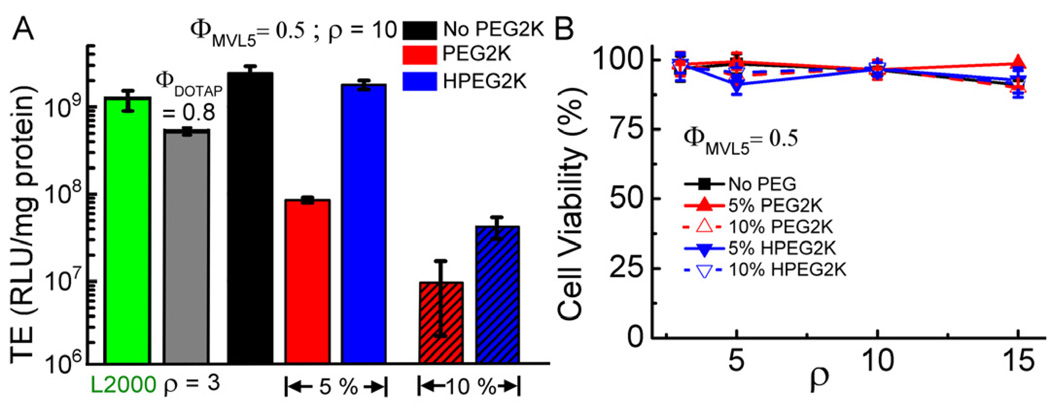
Multivalent cationic lipids efficiently transfect over a broad range of composition and are superior to univalent cationic lipids as vectors for siRNA delivery and for DNA delivery to hard-to-transfect cells [43, 55]. This prompted us to investigate the effect of PEGylation on the TE of DNA complexes of the multivalent lipid MVL5 [46]. Thus, we prepared MVL5/DOPC–DNA complexes containing 0, 5 or 10 mol% of PEG2K-lipid or HPEG2K-lipid at ΦMVL5 =0.5 and ρ =10, which corresponds to the optimal TE for nonPEGylated complexes [29]. Figure 7A shows the TE results, obtained as for DOTAP-containing complexes. Also shown is the TE of optimized DOTAP/DOPC–DNA complexes (cf. Fig. 3) and of the benchmark commercial reagent, Lipofectamine 2000. Without PEGylation, TE of MVL5/DOPC–DNA complexes is as high as that of Lipofectamine 2000 and higher than that of optimized DOTAP/DOPC–DNA complexes. Addition of 5 mol% PEG2K-lipid reduced TE, as observed for PEG2K-lipid/DOTAP/DOPC–DNA complexes (cf. Fig. 3). However, the MVL5-based complexes maintain a TE that is one order of magnitude higher than that of the DOTAP-containing complexes. Increasing the PEG2K-lipid content from 5 mol% to 10 mol% decreased TE by another order of magnitude to around 107 RLU/mg protein. Replacing PEG2K-lipid with the acid-labile HPEG2K-lipid at 5 mol% HPEG2K-lipid recovered TE to essentially the level of nonPEGylated complexes. At 10 mol% PEG-lipid, however, replacing the PEG2K-lipid with the HPEG2K-lipid only recovered a small part of the lost TE, now to a level comparable to that of similar DOTAP-containing complexes. This is further indication that addition of 10 mol% (H)PEG2K-lipid strongly reduces cell attachment and/or cell entry of complexes, in addition to inhibiting endosomal escape. The steric shielding in the brush regime effectively minimizes the complex-cell membrane interaction, cell entry/attachment becomes a main barrier that limits the recovery of TE.
Fig. 7.
(A) TE of PEGylated MVL5/DOPC–DNA complexes at ρ = 10 and controls. The lipid composition of the MVL5-based complexes was (H)PEG2K-lipid /MVL5/DOPC = x/50/50-x, with x = 0, 5, and 10. Replacing the stable PEG2K-lipid with the acid-labile HPEG2K-lipid recovers most (5 mol% PEG-lipid) or some (10 mol% PEG-lipid) of the transfection efficiency lost due to PEGylation. Each data point indicates the average and standard deviation from at least duplicate measurements. The amount of DNA is identical for all data points. (B) Cytotoxicity of PEGylated MVL5/DOPC–DNA complexes. Cell viability is plotted as a function of ρ. Each data point indicates the average and standard deviation from quadruplicate measurements.
We also assessed the cytotoxicity of PEGylated MVL5/DOPC–DNA complexes at ΦMVL5 =0.5. Figure 7B shows the percentage of viable cells as a function of. As in the case of DOTAP-containing complexes, no appreciable cytotoxicity is observed.
4. Conclusions
Using a custom-synthesized acid-labile PEG-lipid (HPEG2K-lipid) which is cleaved at the pH of maturing endosomes, we have investigated the contributions of two main barriers to transfection with PEGylated CL–DNA complexes in vitro: cellular uptake and endosomal escape. At 5 mol% PEG-lipid, steric stabilization of PEGylated CL–DNA complexes is efficient, and their TE is reduced by orders of magnitude over nonPEGylated complexes. Residual electrostatic attractions maintain cellular uptake of complexes that permits recovery of most of the TE (especially for complexes formed with multivalent MVL5) to near the level of nonPEGylated complexes upon replacing the stable PEG2K-lipid with the HPEG2K-lipid. This suggests that inhibition of endosomal escape is the main barrier for complexes containing 5 mol% PEG-lipid. At 10 mol% PEG-lipid, TE drops massively and is nearly independent of, the lipid/DNA charge ratio. Here, the only moderate improvement observed on replacing PEG2K-lipid with HPEG2K-lipid suggests that poor cellular uptake has become a major barrier. All findings are consistent with our hypothesis that the low-pH environment in late endosomes results in the removal of the PEG-coating of CL-DNA complexes prepared with the acid-labile HPEG2K, effectively switching on electrostatic forces required for optimal endosomal escape. Future work will combine selective attachment to cells via peptide or other ligands on the distal end of PEG-lipids with acid-labile PEG-lipids in the hope of achieving a synergistic effect.
Supplementary Material
Acknowledgment
This work was supported by NIH GM-59288 and NSF DMR-1101900. CLC was supported by NSRRC and Academia Sinica, Taiwan. NMR characterization was performed using the Central Facilities of the Materials Research Laboratory at UCSB which are supported by the MRSEC Program of the NSF under award no. DMR-1121053; a member of the NSF-funded Materials Research Facilities Network (www.mrfn.org). CLC thanks Dr. Yeukuang Hwu (Institute of Physics, Academia Sinica).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Edelstein ML, Abedi MR, Wixon J, Edelstein RM. Gene therapy clinical trials worldwide 1989–2004 – an overview. J Gene Med. 2004;6:597–602. doi: 10.1002/jgm.619. [DOI] [PubMed] [Google Scholar]
- 2.Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2007 – an update. J Gene Med. 2007;9:833–842. doi: 10.1002/jgm.1100. [DOI] [PubMed] [Google Scholar]
- 3.Li SD, Huang L. Gene therapy progress and prospects: non-viral gene therapy by systemic delivery. Gene Ther. 2006;13:1313–1319. doi: 10.1038/sj.gt.3302838. [DOI] [PubMed] [Google Scholar]
- 4.Ewert KK, Zidovska A, Ahmad A, Bouxsein NF, Evans HM, McAllister CS, et al. Cationic lipid–nucleic acid complexes for gene delivery and silencing: pathways and mechanisms for plasmid DNA and siRNA. Topics Curr Chem. 2010;296:191–226. doi: 10.1007/128_2010_70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L, Hung MC, Wagner E. Non-viral vectors for gene therapy (Advances in genetics, vol. 53) 2nd ed. part I. San Diego: Elsevier Academic Press; 2005. [Google Scholar]
- 6.Chesnoy S, Huang L. Structure and function of lipid–DNA complexes for gene delivery. Annu Rev Biophys Biomol Struct. 2000;29:27–47. doi: 10.1146/annurev.biophys.29.1.27. [DOI] [PubMed] [Google Scholar]
- 7.Byk G, Dubertret C, Escriou V, Frederic M, Jaslin G, Rangara R, et al. Synthesis, activity, and structure-activity relationship studies of novel cationic lipids for DNA transfer. J Med Chem. 1998;41:224–235. doi: 10.1021/jm9704964. [DOI] [PubMed] [Google Scholar]
- 8.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 9.Wagner E. Strategies to improve DNA polyplexes for in vivo gene transfer: will “artificial viruses” be the answer? Pharm Res. 2004;21:8–14. doi: 10.1023/b:pham.0000012146.04068.56. [DOI] [PubMed] [Google Scholar]
- 10.Henry CM. Gene Delivery-without Viruses. Chem Eng News. 2001;79:35–41. [Google Scholar]
- 11.Ewert KK, Slack NL, Ahmad A, Evans HM, Lin AJ, Samuel CE, et al. Cationic lipid–DNA complexes for gene therapy: understanding the relationship between complex structure and gene delivery pathways at the molecular level. Curr Med Chem. 2004;11:133–149. doi: 10.2174/0929867043456160. [DOI] [PubMed] [Google Scholar]
- 12.Ewert KK, Evans H, Ahmad A, Slack L, Lin A, Martin-Herranz A, et al. Lipoplex structures and their distinct cellular pathways. In: Huang L, Hung MC, Wagner E, editors. Non-viral vectors for gene therapy (Advances in genetics, vol. 53) 2nd ed. part I. San Diego: Elsevier Academic Press; 2005. pp. 119–155. [Google Scholar]
- 13.Ewert KK, Ahmad A, Evans H, Safinya CR. Cationic lipid–DNA complexes for non-viral gene therapy: relating supramolecular structures to cellular pathways. Expert Opin Biol Ther. 2005;5:33–53. doi: 10.1517/14712598.5.1.33. [DOI] [PubMed] [Google Scholar]
- 14.Haensler J, Szoka FC., Jr Polyamidoamine cascade polymers mediate efficient transfection of cells in culture. Bioconjug Chem. 1993;4:372–379. doi: 10.1021/bc00023a012. [DOI] [PubMed] [Google Scholar]
- 15.Tang MX, Redemann CT, Szoka FC., Jr In vitro gene delivery by degraded polyamidoamine dendrimers. Bioconjug Chem. 1996;7:703–714. doi: 10.1021/bc9600630. [DOI] [PubMed] [Google Scholar]
- 16.Erbacher P, Bettinger T, Belguise-Valladier P, Zou S, Coll JL, Behr JP, et al. Transfection and physical properties of various saccharide, poly(ethylene glycol), and antibody-derivatized polyethylenimines (PEI) J Gene Med. 1999;1:210–222. doi: 10.1002/(SICI)1521-2254(199905/06)1:3<210::AID-JGM30>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 17.Morille M, Passirani C, Vonarbourg A, Clavreul A, Benoit JP. Progress in developing cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials. 2008;29:3477–3496. doi: 10.1016/j.biomaterials.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 18.Hofland HE, Shephard L, Sullivan SM. Formation of stable cationic lipid/DNA complexes for gene transfer. Proc Natl Acad Sci U S A. 1996;93:7305–7309. doi: 10.1073/pnas.93.14.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plank C, Mechtler K, Szoka FC, Jr, Wagner E. Activation of the complement system by synthetic DNA complexes: a potential barrier for intravenous gene delivery. Hum Gene Ther. 1996;7:1437–1446. doi: 10.1089/hum.1996.7.12-1437. [DOI] [PubMed] [Google Scholar]
- 20.Nicolazzi C, Mignet N, de la Figuera N, Cadet M, Ibad RT, Seguin J, et al. Anionic polyethyleneglycol lipids added to cationic lipoplexes increase their plasmatic circulation time. J Control Release. 2003;88:429–443. doi: 10.1016/s0168-3659(03)00067-1. [DOI] [PubMed] [Google Scholar]
- 21.Ambegia E, Ansell S, Cullis P, Heyesa J, Palmera L, MacLachlan I. Stabilized plasmid–lipid particles containing PEG-diacylglycerols exhibit extended circulation lifetimes and tumor selective gene expression. Biochim Biophys Acta. 2005;1669:155–163. doi: 10.1016/j.bbamem.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Tam P, Monck M, Lee D, Ludkovski O, Leng EC, Clow K, et al. Stabilized plasmid–lipid particles for systemic gene therapy. Gene Ther. 2000;7:1867–1874. doi: 10.1038/sj.gt.3301308. [DOI] [PubMed] [Google Scholar]
- 23.Mislick KA, Baldeschwieler JD. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc Natl Acad Sci U S A. 1996;93:12349–12354. doi: 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holland JW, Hui C, Cullis PR, Madden TD. Poly(ethylene glycol)–lipid conjugates regulate the calcium-induced fusion of liposomes composed of phosphatidylethanolamine and phosphatidylserine. Biochemistry. 1996;35:2618–2624. doi: 10.1021/bi952000v. [DOI] [PubMed] [Google Scholar]
- 25.Mishar S, Webster P, Davis ME. PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles. Eur J Cell Biol. 2004;83:97–111. doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- 26.Xu Y, Szoka FC., Jr Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35:5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 27.Khalil KI, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev. 2006;58:32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 28.Lin AJ, Slack NL, Ahmad A, George CX, Samuel CE, Safinya CR. Three-dimensional imaging of lipid gene-carriers: membrane charge density controls universal transfection behavior in lamellar cationic liposome–DNA complexes. Biophys J. 2003;84:3307–3316. doi: 10.1016/S0006-3495(03)70055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmad A, Evans HM, Ewert KK, George CX, Samuel CE, Safinya CR. New multivalent cationic lipids reveal bell curve for transfection efficiency versus membrane charge density: lipid–DNA complexes for gene delivery. J Gene Med. 2005;7:739–748. doi: 10.1002/jgm.717. [DOI] [PubMed] [Google Scholar]
- 30.Kim WJ, Yockman JW, Lee M, Jeong JH, Kim YH, Kim SW. Soluble Flt-1 gene delivery using PEI-g-PEG-RGD conjugate for anti-angiogenesis. J Control Release. 2005;106:224–234. doi: 10.1016/j.jconrel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 31.Li W, Szoka FC., Jr Lipid-based nanoparticles for nucleic acid delivery. Pharm Res. 2007;24:438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- 32.Yu WJ, Pirollo KF, Rait A, Yu B, Xiang LM, Huang WQ, et al. A sterically stabilized immunolipoplex for systemic administration of therapeutic gene. Gene Ther. 2004;11:1434–1440. doi: 10.1038/sj.gt.3302304. [DOI] [PubMed] [Google Scholar]
- 33.Romberg B, Hennink WE, Storm G. Sheddable coatings for long-circulating nanoparticles. Pharm Res. 2008;25:55–71. doi: 10.1007/s11095-007-9348-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin J, Shum P, Thompson DH. Acid-triggered release via dePEGylation of DOPE liposomes containing acid-labile vinyl ether PEG-lipids. J Control Release. 2003;91:187–200. doi: 10.1016/s0168-3659(03)00232-3. [DOI] [PubMed] [Google Scholar]
- 35.Guo X, Szoka FC., Jr Steric stabilization of fusogenic liposomes by a low-pH sensitive PEG–diortho ester–lipid conjugate. Bioconjug Chem. 2001;12:291–300. doi: 10.1021/bc000110v. [DOI] [PubMed] [Google Scholar]
- 36.Auguste DT, Armes SP, Brzezinska KR, Deming TJ, Kohn J, Prud’homme RK. pH triggered release of protective poly(ethylene glycol)-b-polycation copolymers from liposomes. Biomaterials. 2006;27:2599–2608. doi: 10.1016/j.biomaterials.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 37.Gerasimov OV, Boomer JA, Qualls MM, Thompson DH. Cytosolic drug delivery using pH- and light-sensitive liposomes. Adv Drug Deliv Rev. 1999;38:317–338. doi: 10.1016/s0169-409x(99)00035-6. [DOI] [PubMed] [Google Scholar]
- 38.Drummond DC, Zignani M, Leroux JC. Current status of pH-sensitive liposomes in drug delivery. Prog Lipid Res. 2000;39:409–460. doi: 10.1016/s0163-7827(00)00011-4. [DOI] [PubMed] [Google Scholar]
- 39.Walker GF, Fella C, Pelisek J, Fahrmeir J, Boeckle S, Ogris M, et al. Toward synthetic viruses: endosomal pH-triggered deshielding of targeted polyplexes greatly enhances gene transfer in vitro and in vivo. Mol Ther. 2005;11:418–425. doi: 10.1016/j.ymthe.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 40.Knorr V, Allmendinger L, Walker GF, Paintner FF, Wagner E. An acetal-based PEGylation reagent for pH-sensitive shielding of DNA polyplexes. Bioconjug Chem. 2007;18:1218–1225. doi: 10.1021/bc060327a. [DOI] [PubMed] [Google Scholar]
- 41.Fella C, Walker GF, Ogris M, Wagner E. Amine-reactive pyridylhydrazone-based PEG reagents for pH-reversible PEI polyplex shielding. Eur J Pharm Sci. 2008;34:309–320. doi: 10.1016/j.ejps.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 42.Nie Y, Gu ZW, Wagner E. Pyridylhydrazone-based PEGylation for pH-reversible lipopolyplex shielding. Biomaterials. 2011;32:858–869. doi: 10.1016/j.biomaterials.2010.09.032. [DOI] [PubMed] [Google Scholar]
- 43.Xu Z, Gu W, Chen L, Gao Y, Zhang Z, Li Y. A smart nanoassembly consisting of acid-labile vinyl ether PEG-DOPE and protamine for gene delivery: preparation and in vitro transfection. Biomacromolecules. 2008;9:3119–3126. doi: 10.1021/bm800706f. [DOI] [PubMed] [Google Scholar]
- 44.Li W, Huang Z, MacKay JA, Grube S, Szoka FC., Jr Low-pH-sensitive poly(ethylene glycol) (PEG)-stabilized plasmid nanolipoparticles: effects of PEG chain length, lipid composition and assembly conditions on gene delivery. J Gene Med. 2005;7:67–79. doi: 10.1002/jgm.634. [DOI] [PubMed] [Google Scholar]
- 45.Asokan A, Cho MJ. Exploitation of intracellular pH gradients in the cellular delivery of macromolecules. J Pharm Sci. 2002;91:903–913. doi: 10.1002/jps.10095. [DOI] [PubMed] [Google Scholar]
- 46.Ewert KK, Ahmad A, Evans HM, Schmidt HW, Safinya CR. Efficient synthesis and cell-transfection properties of a new multivalent cationic lipid for nonviral gene delivery. J Med Chem. 2002;45:5023–5029. doi: 10.1021/jm020233w. [DOI] [PubMed] [Google Scholar]
- 47.Ewert KK, Ahmad A, Bouxsein NF, Evans HM, Safinya CR. Non-viral gene delivery with cationic liposome-DNA complexes. In: Le Doux J, editor. Gene therapy protocols (Methods in molecular biology, vol. 433) 3rd ed. Totowa, NJ: Humana Press; 2008. pp. 159–175. [DOI] [PubMed] [Google Scholar]
- 48.Schulze U, Schmidt HW, Safinya CR. Synthesis of novel cationic poly(ethylene glycol) containing lipids. Bioconjug Chem. 1999;10:548–552. doi: 10.1021/bc9801068. [DOI] [PubMed] [Google Scholar]
- 49.Shirazi RS, Ewert KK, Leal C, Majzoub RN, Bouxsein NF, Safinya CR. Synthesis and characterization of degradable multivalent cationic lipids with disulfide-bond spacers for gene delivery. Biochim Biophys Acta. 2011;1808:2156–2166. doi: 10.1016/j.bbamem.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masson C, Scherman D, Bessodes M. 2,2,6,6-Tetramethyl-1-piperidinyl-oxyl/[bis(acetoxy)-iodo]benzene-mediated oxidation: a versatile and convenient route to poly(ethylene glycol) aldehyde or carboxylic acid derivatives. J Polym Sci A. 2001;39:4022–4024. [Google Scholar]
- 51.Percec V, Tomazos D, Heck J, Blackwell H, Ungar G. Self-assembly of taper-shaped monoesters of oligo(ethylene oxide) with 3,4,5-tris(n-dodecan-1-yloxy) benzoic acid and of their polymethacrylates into tubular supramolecular architectures displaying a columnar hexagonal mesophase. J Chem Soc Perkin Trans. 1994;2:31–44. [Google Scholar]
- 52.Koltover I, Salditt T, Safinya CR. Phase diagram, stability, and overcharging of lamellar cationic lipid–DNA self-assembled complexes. Biophys J. 1999;77:915–924. doi: 10.1016/S0006-3495(99)76942-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garbuzenko O, Barenholz Y, Priev A. Effect of grafted PEG on liposome size and on compressibility and packing of lipid bilayer. Chem Phys Lipids. 2005;135:117–129. doi: 10.1016/j.chemphyslip.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 54.Kenworthy AK, Hristova K, Needham D, McIntosh TJ. Range and magnitude of the steric pressure between bilayers containing phospholipids with covalently attached poly(ethylene glycol) Biophys J. 1995;68:1921–1936. doi: 10.1016/S0006-3495(95)80369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bouxsein NF, McAllister CS, Ewert KK, Samuel CE, Safinya CR. Structure and gene silencing activities of monovalent and pentavalent cationic lipid vectors complexed with siRNA. Biochemistry. 2007;46:4785–4792. doi: 10.1021/bi062138l. [DOI] [PubMed] [Google Scholar]
- 56.Cory AH, Owen TC, Barltrop JA, Cory JG. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 1991;3:207–212. doi: 10.3727/095535491820873191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.