Abstract
In the adult mammalian CNS, chondroitin sulfate proteoglycans (CSPGs) and myelin–associated inhibitors (MAIs) stabilize neuronal structure and restrict compensatory sprouting following injury. The Nogo receptor family members NgR1 and NgR2 bind to MAIs and have been implicated in neuronal inhibition. Here we show that NgR1 and NgR3 bind with high–affinity to the glycosaminoglycan moiety of proteoglycans and participate in CSPG inhibition in cultured neurons. Nogo receptor triple mutants (NgR123−/−), but not single mutants, show enhanced axonal regeneration following retro–orbital optic nerve crush injury. The combined loss of NgR1 and NgR3 (NgR13−/−), but not NgR1 and NgR2 (NgR12−/−), is sufficient to mimic the NgR123−/− regeneration phenotype. Regeneration in NgR13−/− mice is further enhanced by simultaneous ablation of RPTPσ, a known CSPG receptor. Collectively, these results identify NgR1 and NgR3 as novel CSPG receptors, demonstrate functional redundancy among CSPG receptors, and provide unexpected evidence for shared mechanisms of MAI and CSPG inhibition.
In the adult mammalian central nervous system (CNS), structural neuronal plasticity is restricted by a number of extrinsic (environmental) and cell–intrinsic growth inhibitory mechanisms 1,2. While such mechanisms are believed to be important for stabilization of intricate networks of neuronal connectivity in CNS health, they also limit adaptive neuronal growth and sprouting following brain or spinal cord injury (SCI). Spontaneous repair following severe CNS injury is incomplete and commonly associated with permanent neurological deficits. Thus, a detailed understanding of the mechanisms that block neuronal growth and repair is of great interest, both biologically and clinically.
A large number of CNS inhibitory cues has been identified 2–4. In experimental animal models of SCI, acute blockage of myelin-associated inhibitors (MAIs) 5,6 or enzymatic degradation of chondroitin sulfate proteoglycans (CSPGs) with chondroitinase ABC (Ch’aseABC) 7–9 promotes neuronal sprouting and correlates with improved behavioral outcomes.
The best characterized MAIs are the reticulon family member Nogo, myelin associated glycoprotein (MAG), and oligodendrocyte myelin glycoprotein (OMgp) 2. Three isoforms of Nogo have been identified, all of which contain a 66 amino acid loop (Nogo66) that signals neuronal inhibition. Mechanistic studies identified the Nogo66 receptor–1 (NgR1) and paired immunoglobulin (Ig)–like receptor B (PirB) as functional receptors for MAIs 10,11. NgR1 is comprised of 8.5 leucine–rich repeats (LRRs), flanked by N–terminal (NT–) and C–terminal (CT–) LRR capping domains. The NT–LRR–CT cluster of NgR1 is fused to a ~ 100 amino acid residue stalk region and connected to the plasma membrane by a glycosylphosphatidylinositol (GPI) anchor 11. NgR1 and its close relative NgR2 show overlapping, yet distinct binding preferences toward MAIs. Nogo66 and OMgp bind selectively to NgR1 2, while MAG associates with NgR1 and NgR2 12. The related molecule NgR3 is poorly characterized, and thus far no functional NgR3 ligand(s) have been identified. In vitro, loss of NgR1 renders neurons more resistant to Nogo66–, MAG–, and OMgp–induced growth cone collapse but not to longitudinal neurite outgrowth inhibition on substrate bound inhibitors 13–15. MAIs activate RhoA, RockII, and conventional isoforms of protein kinase C (PKC) to destabilize the neuronal cytoskeleton 16,17. Similar to NgR1, PirB supports binding of Nogo66, MAG, and OMgp. In culture, functional ablation of PirB promotes neurite outgrowth on substrate–bound MAIs and crude CNS myelin. Interestingly, the combined perturbation of PirB and NgR1 signaling leads to a further release of neurite outgrowth inhibition on crude CNS myelin but not on recombinant Nogo66 or MAG 10.
CSPGs are a diverse class of extracellular matrix molecules that influence axonal growth and guidance of developing neurons 18. Following injury to the adult CNS, CSPG expression is upregulated and abundant in reactive astrocytes associated with glial scar tissue 4,19,20. CSPGs are comprised of a protein core with covalently attached glycosaminoglycan (GAG) side chains. GAG chains are large, unbranched polymers composed of ~20–200 repeating disaccharide units. Chondroitin sulfate (CS)–GAGs contain alternating units of N–acetyl–galactosamine and glucuronic acid. Most commonly, the hydroxyl groups at position 4 (CS–A) or position 6 (CS–C) of N–acetyl–galactosamine are sulfated. In CS–B, iduronic acid replaces glucuronic acid in the CS disaccharide unit. In CS–D and CS–E, two sulfate groups per disaccharide unit are present. CSPG inhibition is largely abrogated by bacterial chondroitinase–ABC (Ch’aseABC), indicating that CS–GAGs are important for neuronal growth inhibition 7,8,21,22.
Similar to MAIs, CSPG–mediated inhibition depends on activation of RhoA and conventional PKCs 16,17,23. Mechanistic studies recently identified the receptor protein tyrosine phosphatase sigma (RPTPσ) as a high–affinity receptor for CSPGs 24. RPTPσ is a member of the leukocyte common antigen–related protein (LAR) family that also includes LAR and RPTPδ. RPTPσ binds to CS–GAG chains and the structurally related heparan sulfate (HS)–GAG chains via its first Ig like domain 24,25. The association of RPTPσ with CS– and HS–GAGs critically depends on the presence of an evolutionarily conserved cluster of basic amino acid residues. Functional ablation of RPTPσ enhances neurite outgrowth in the presence of CSPGs in vitro, and following CNS injury promotes growth of sensory afferents 24, corticospinal tract axons 26, and retinal ganglion cell axons 27. The incomplete release of CSPG inhibition in RPTPσ –deficient neurons suggests the existence of additional mechanisms of CSPG inhibition. Here we report on the identification of the Nogo receptor (NgR) family members NgR1 and NgR3 as novel CSPG receptors.
Results
NgRs participate in prototypic MAI independent inhibition
To determine the role of Nogo receptor family members in CNS myelin inhibition, we generated NgR1, NgR2, NgR3 triple null mice (NgR123−/−) (Supplementary Fig. 1). NgR123−/− mice are born at Mendelian ratios, viable into adulthood, fertile, and indistinguishable from wildtype (WT) littermate controls at the gross anatomical level. When plated on crude CNS myelin, postnatal day 7 (P7) cerebellar granule neurons (CGNs), but not dorsal root ganglion (DRG) neurons, of NgR123−/− mice show a significant (P< 0.001, one way ANOVA, Tukey’s post–hoc), yet incomplete release of growth inhibition (Fig. 1 and Supplementary Fig. 2). Compared to CGNs isolated from WT, NgR1−/−, NgR12−/−, or NgR3−/− mice, CGNs from NgR123−/− mutants grow significantly longer neurites on myelin. Remarkably, in two different types of neurons, CGNs and DRGs, the combined loss of NgR1 and NgR2 does not result in enhanced neurite growth on crude CNS myelin (Fig. 1 and Supplementary Fig. 2). Because only NgR1 and NgR3, but not NgR2, are expressed in P7 CGNs 12,15, this suggests that NgR3 participates in myelin inhibition. This is somewhat surprising, as NgR3 does not associate with recombinant Nogo, MAG or OMgp 12.
Figure 1. Loss of all three Nogo Receptors results in enhanced growth on CNS myelin.
(a) Postnatal day 7 (P7) cerebellar granule neurons (CGNs) from WT, NgR1−/−, NgR12−/−, and NgR3−/− pups are strongly inhibited when plated on crude CNS myelin substrate (40μg/ml). In marked contrast, CGNs from NgR triple mutant (NgR123−/−) mice grow longer neurites on CNS myelin. On CNS myelin isolated from NogoABC;MAG;OMgp triple null (NMO−/−) mice (40μg/ml), CGNs from WT, NgR1−/−, NgR12−/−, and NgR3−/− pups show enhanced neurite outgrowth. A further release of inhibition is observed when NgR123−/− neurons are plated on NMO−/− CNS myelin. On a BSA control substrate, neurite length of all five genotypes is comparable. (b) Quantification of neurite length. At least 300 neurites of TuJ1–labeled cells were counted per condition (n=9 independent experiments). Light gray bars (BSA); black bars (WT myelin); dark gray bars (NMO myelin). Results are presented as mean ±SEMs. ** P< 0.001 (one way ANOVA, Tukey’s post–hoc). Scale bar, 50μm.
To directly test whether NgR3 participates in neurite outgrowth inhibition of endogenously expressed Nogo, MAG, or OMgp, experiments were repeated with CNS myelin isolated from Nogo, MAG, OMgp triple mutant mice (NMO−/−) 28. Consistent with previous reports 28,29, NMO−/− myelin is less inhibitory than WT myelin (Fig. 1 and Supplementary Fig. 2). Importantly, on NMO−/− myelin, CGNs from NgR123−/− mice continue to extend longer neurites (P< 0.001, one way ANOVA, Tukey’s post–hoc) than CGNs from WT, NgR1−/−, NgR12−/−, or NgR3−/− mice (Fig. 1). This observation indicates that NgR3 participates in Nogo–, MAG– and OMgp–independent growth inhibition. Because loss of NgR1 or NgR3 alone is not sufficient to promote growth on myelin, this suggests some degree of functional redundancy among these two receptors.
NgR1 and NgR3, but not NgR2, associate with neural GAGs
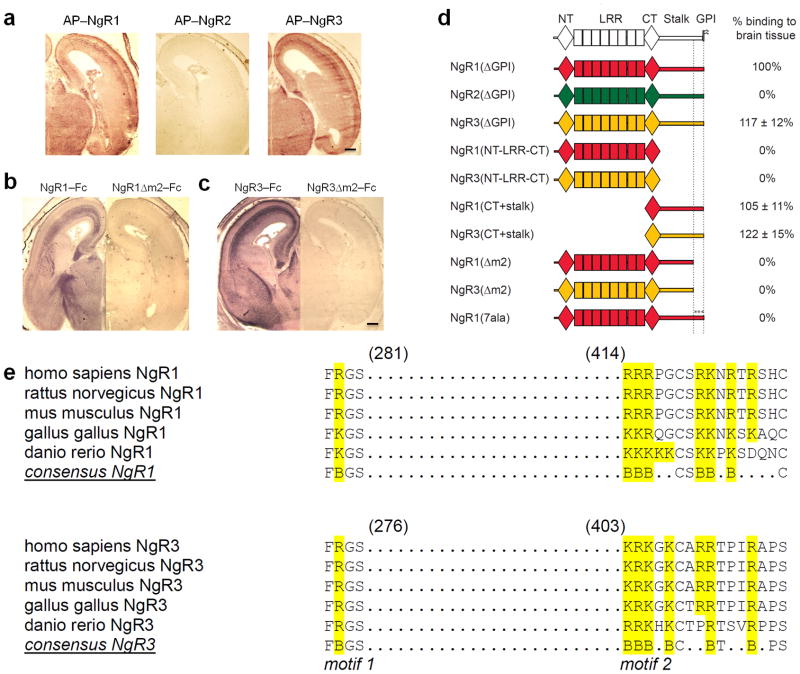
To identify candidate NgR3 ligand(s), we generated alkaline phosphatase (AP)–tagged receptor fusion proteins and assayed binding to rat brain tissue sections. Prior to the onset of CNS myelination, NgR1 and NgR3, but not NgR2, bind strongly to numerous fiber tracts in the brain and spinal cord (Fig. 2a). After myelination, a more uniform binding pattern is observed with a much less pronounced labeling of fiber tracts (data not shown). Importantly, brain sections of NMO−/− and NgR123−/− triple mutants, p75−/− single mutants, and mice lacking select gangliosides show no substantial reduction in soluble receptor binding (Supplementary Fig. 3). In COS–7 cells, components of the NgR1 holoreceptor complex, including p75, TROY, and Lingo–1 30, fail to support NgR3 binding (Supplementary Fig. 4). This suggests that binding of NgR3 to brain is not mediated by previously identified components of the NgR1 complex. Receptor deletion studies further revealed that the LRRs are not required for binding, and identified two discontinuous and evolutionarily conserved sequence motifs, in both NgR1 and NgR3, that are necessary for binding to brain (Fig. 2b–e and data not shown). Motif 1 (m1) is located in the CT capping domain, and interestingly, overlaps with the FRG motif previously shown to participate in sialic acid–dependent binding of the ganglioside GT1b to NgR1 31. Motif 2 (m2), separated from m1 by approximately 130 amino acid residues, is located near the juxtamembrane region of the NgR1 and NgR3 stalk domain. Motif m2 in NgR1 and NgR3 is comprised of a highly conserved cluster of basic amino acid residues, deletion of which completely abolishes binding to brain (Fig. 2b–e). Furthermore, a soluble form of NgR1 in which the basic residues of m2 are replaced by seven alanines [NgR1(7ala)–Fc] no longer binds to brain tissue (Fig. 2d).
Figure 2. NgR1 and NgR3, but not NgR2, contain two discontinuous and evolutionarily conserved sequence motifs necessary for binding to brain tissue.
(a) Coronal sections of E18 rat brain showing the binding pattern of AP–NgR1 and AP–NgR3. No binding is observed for AP–NgR2. (b–c) Binding of (b) NgR1–Fc and (c) NgR3–Fc to E18 brain sections is abolished upon deletion of a cluster of basic residues (motif m2) in the stalk region. (d) Schematic of receptor deletion constructs and their relative binding to E18 rat brain tissue compared to soluble NgR1 [NgR1(ΔGPI)]. Soluble NgR1 (red) and NgR3 (yellow), but not NgR2 (green), bind strongly to brain tissue sections. The LRRs of NgR1, previously shown to participate in myelin inhibitor binding, are dispensable for binding to neural tissue. Deletion of a cluster of basic amino acid residues in the C–terminal region of the NgR1 and NgR3 stalk (motif m2), or replacement of these residues with alanines [NgR1(7ala)], completely abolishes binding. (e) Sequence alignment of binding motifs m1 and m2 of NgR1 and NgR3. In the LRR–CT domain, residues F278 and R279 in NgR1 and residues F273 and R274 in NgR3 (motif m1) are important for GAG binding. Motif m2 is comprised of a cluster of basic amino acids, including residues 414–426 in NgR1 and residues 403–415 in NgR3. The conserved residues are bolded and the basic residues are in blue. Scale bar, 40μm.
To assess whether the association of NgR1 and NgR3 with neural tissue is the result of a protein–protein interaction, brain sections were subjected to heat or protease treatment. Remarkably, binding was largely resistant to either treatment (Supplementary Fig. 3), suggesting a possible interaction with neural glycan(s). Pretreatment of brain tissue sections with various glycosidases revealed sensitivity to heparinase and Ch’aseABC. Moreover, in the presence of heparin, binding was completely abolished (Fig. 3a and Supplementary Fig. 3). Together these studies suggest that NgR1 and NgR3 associate with neural glycosaminoglycans.
Figure 3. NgR1 and NgR3 interact directly with specific GAGs.
(a) Binding of soluble AP–NgR1 to P1 rat brain tissue sections is sensitive to heparinase (Hep’ase) and chondroitinase–ABC (Ch’aseABC) treatment (1unit/ml). (b) Binding of soluble AP–NgR1 and AP–NgR3 to E18 rat brain tissue sections in the presence of PBS (vehicle control), 50μg/ml monosulfated CS–GAGs (CS–A, CS–B, or CS–C), or 50μg/ml disulfated CS–GAGs (CS–D or CS–E). Only CS–B, CS–D, and CS–E compete effectively with NgR1 and NgR3 for binding to brain tissue sections. (c) ELISA binding studies revealed a direct association of soluble NgR1 and RPTPσ(1–3) with specific GAGs (insets show Scatchard plot analysis). The calculated Kds for CS–B, CS–D, CS–E, and heparin are 3.2, 1.3, 1.0, and 1.2nM (NgR1) and 1.4, 3.0, 0.1, and 0.1nM [RPTPσ(1–3)], respectively. (d) WT P7 rat CGNs are strongly inhibited when plated on substrate–bound CSPGs. Long neurites are seen on a BSA control substrate. In the presence of NgR1–Fc or RPTPσ(1–3)–Fc, but not NgR1(7ala)–Fc or IgG1 control, CSPG inhibition is abolished. On a BSA substrate, soluble receptors do not influence neurite outgrowth. Quantification of neurite length is shown as a percentage of the IgG1–BSA control (100%). At least 300 neurites of TuJ1–labeled cells were counted per condition (n=4 independent experiments). Gray bars (BSA); black bars (CSPGs). Results are presented as mean ±SEMs. ** P< 0.001 (one way ANOVA, Tukey’s post–hoc), n.s.= not significant. Scale bar: a, 20μm; b, 40μm; d, 50μm.
NgR1 and NgR3 complex with select CS–GAGs
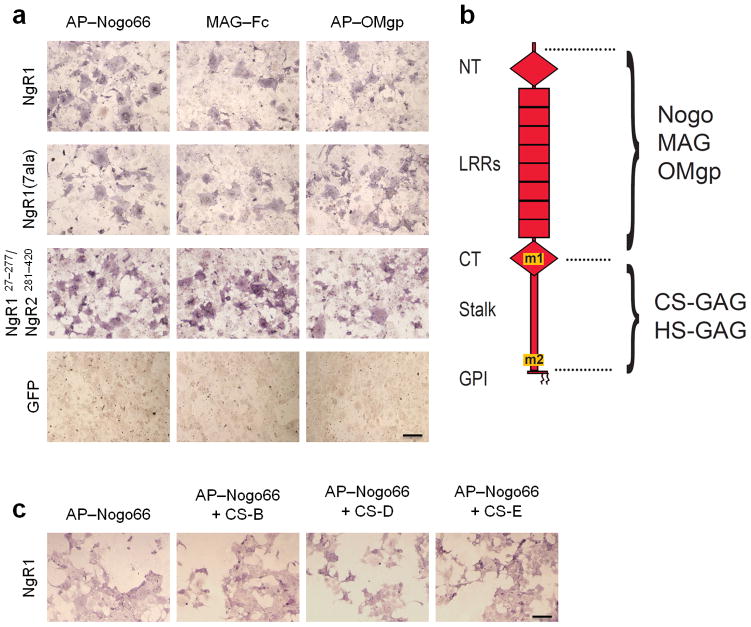
To examine the specificity of the GAG association, AP–NgR1 and AP–NgR3 fusion proteins were preincubated with various types of CS–GAGs. Strikingly, CS–B, CS–D and CS–E, but not CS–A or CS–C, effectively compete with soluble NgR1 and NgR3 for binding to brain sections (Fig. 3b). To test whether NgR1 and NgR3 bind to purified GAGs directly, we developed a sandwich ELISA, in which biotinylated GAGs were adsorbed to streptavidin–coated microtiter plates and then incubated with soluble AP–tagged NgRs (Supplementary Fig. 5a). Consistent with binding experiments to rat brain tissue, NgR1 and NgR3 bind robustly to heparin and purified CS–GAGs, indicating that these receptors bind GAGs directly (Fig. 3c and Supplementary Fig. 5b). NgR1 and NgR3 bind with high specificity and selectivity to different types of monosulfated and disulfated GAGs. Strong binding was observed to monosulfated CS–B, and disulfated CS–D and CS–E. The dissociation constants for these interactions are in the low nanomolar range (Fig. 3c and Supplementary Fig. 5b). No interactions with CS–A or CS–C were detected. This argues against a nonspecific interaction with negatively–charged compounds and underscores the selectivity of the NgR1– and NgR3–GAG associations.
Notably, the first three Ig like domains of RPTPσ [RPTPσ(1–3)] show very similar GAG–binding profiles (Fig. 3c). At increasing doses, RPTPσ(1–3)–Fc effectively competes with NgR1 for binding to CS–E, indicating that these two receptors complex with, at least in part, overlapping CS–GAG epitopes (Supplementary Fig. 5c). Functional studies with primary neurons show that soluble RPTPσ(1–3)–Fc and NgR1–Fc block the growth inhibitory activity of CSPGs toward P7 CGNs in vitro. The neutralizing effects of NgR1–Fc critically depend on the presence of the GAG–binding motif m2, as soluble NgR1(7ala)–Fc fails to block CSPG inhibition (Fig. 3d).
The CSPG and MAI binding sites on NgR1 are dissociable
To further characterize the relation of CSPG and MAI binding sites on NgR1, we generated a chimeric receptor construct in which the GAG–binding portion of NgR1 (amino acid residues 278–445) was replaced by the corresponding, non–GAG–binding sequence of NgR2 (amino acid residues 281–420). Nogo66, MAG, and OMgp bind strongly to this chimeric NgR1/NgR2 receptor, indicating that the GAG–binding sequences of NgR1 are not necessary for MAI binding (Fig. 4a). A soluble form of this same chimeric receptor fails to bind to rat brain tissue sections or to GAGs directly (Supplementary Fig. 5d–e). This suggests that MAIs and CSPGs bind to distinct and dissociable sites on NgR1 (Fig. 4b). Moreover, the presence of CS–B, CS–D, or CS–E does not substantially influence binding of AP–Nogo66 to NgR1 (Fig. 4c).
Figure 4. Binding sites for MAIs and CSPGs on NgR1 are distinct and dissociable.
(a) In transiently transfected COS–7 cells, wildtype NgR1 and the GAG–binding–deficient mutant [NgR1(7ala)] show very similar binding of the MAIs AP–Nogo66, MAG–Fc, and AP–OMgp. The NgR1 fragment F278–E445 (AP–NgR1Δ15CT+stalk) is sufficient for high–affinity binding to neural GAGs. When residues 278–445 in NgR1 are replaced with the corresponding sequences of NgR2, resulting in construct NgR1C27–K277/NgR2V281–L420, binding of MAIs is not diminished compared to wildtype NgR1. Ligand binding to GFP–transfected–COS 7 cells is shown as a negative control. (b) Schematic of NgR1, showing the regions necessary for binding of MAIs and GAGs. The GAG–binding motifs m1 and m2 of NgR1 are distinct and dissociable from the Nogo–, MAG–, and OMgp–binding sequences. (c) In the presence of bath applied CS–B, CS–D, or CS–E GAGs (1mg/ml), binding of AP–Nogo66 to NgR1–expressing COS–7 cells is not altered. Scale bar, 20μm.
Neuronal Nogo receptors participate in CSPG inhibition
To determine whether loss of NgRs leads to disinhibition of neurite growth on substrate bound CSPGs, CGNs from NgR single and compound mutants were analyzed (Fig. 5). Loss of NgR1 or NgR3 alone, or the combined loss of NgR1 and NgR2 (NgR12−/−), is not sufficient to attenuate CSPG inhibition. Loss of all three NgRs (NgR123−/−), however, results in significant (P< 0.001, one way ANOVA, Tukey’s post–hoc), yet incomplete release of CSPG inhibition. Furthermore, release of inhibition for CGNs isolated from NgR123−/− and RPTPσ−/− pups is comparable (Fig. 5). Dose–response experiments show that when challenged with high concentrations of CSPGs, NgR123−/− and RPTPσ−/− neurons are strongly inhibited and lose their growth advantage over WT neurons (Supplementary Fig. 6a). This suggests that these receptor systems share some degree of functional redundancy. At high doses of CSPGs, loss of NgRs may be compensated by RPTPσ and vice versa. NgRs are not abundantly expressed in P7 DRGs 15 and NgR123−/− DRG neurons are not disinhibited on CSPG substrate. In a parallel experiment with DRG neurons from RPTPσ−/− mice, neurite length is increased on CSPGs (Supplementary Fig. 6b). Collectively, these studies show that NgR1 and NgR3 bind CS–GAGs directly and participate in CSPG–mediated neurite outgrowth inhibition in a neuronal cell–type specific manner.
Figure 5. Nogo receptors mediate CSPG inhibition.
(a) In vitro, WT P7 CGNs are strongly inhibited when plated on substrate–bound CSPGs (10μg/ml). Studies with CGNs from NgR1−/−, NgR12−/−, and NgR3−/− mutants revealed no significant release of CSPG inhibition. Loss of all three NgRs (NgR123−/−) or RPTPσ alone (RPTPσ−/−) leads to a significant, yet incomplete release of CSPG inhibition. On a control substrate (BSA), neurite length of all six genotypes is comparable. (b) Quantification of neurite length. Gray bars (BSA); black bars (CSPGs). At least 300 neurites of TuJ1–labeled cells were counted per condition (n=8 independent experiments). Results are presented as mean ±SEMs. ** P< 0.001 (one way ANOVA, Tukey’s post–hoc), n.s.= not significant. Scale bar, 70μm.
NgR1 and NgR3 associate in a ligand dependent manner
NgRs are GPI–anchored proteins, and thus depend on interactions with transmembrane receptor components to signal growth inhibition across the neuronal plasma membrane. To assess whether NgR1 and NgR3 employ shared signaling mechanisms, we assayed binding of NgR3–Fc to the previously identified NgR1 receptor components p75, TROY, and Lingo–1 in COS–7 cells. We observed no binding of soluble NgR3 to p75, TROY, Lingo–1, or NgR1 (Supplementary Fig. 4). There are conflicting results on whether NgR1 and NgR3 interact 32,33. We therefore revisited this issue and found that NgR1 and NgR3 are part of the same immune complex when co–expressed in HEK293T cells. The NgR1–NgR3 association is ligand dependent and is only observed in the presence of exogenously–applied CSPGs (Supplementary Fig. 4), suggesting that the two receptors may be part of the same receptor complex. In this same assay, no association of NgR1 with NgR2 is observed, neither in the presence nor the absence of CSPGs. Next we examined whether NgR1, NgR3, and p75 may be part of the same receptor complex. In HEK293T cells co–transfected with NgR1, NgR3, and p75, anti–NgR1–pull down experiments revealed that the three receptors are present in the same immune complex if cells are treated with CSPGs (Supplementary Fig. 4).
To directly test whether p75 is important for CSPG mediated neurite outgrowth inhibition, P7 CGNs from p75−/− mice were plated on substrate bound CSPGs. Loss of p75 does not result in a significant release of CSPG inhibition (Supplementary Fig. 4). Together these experiments show that p75, NgR1, and NgR3 do interact in the presence of CSPGs; however, p75 is not necessary for CSPG–mediated outgrowth inhibition. Our studies confirm and expand on previous work showing that versican V2 mediates neurite outgrowth inhibition in CGNs and DRG neurons in a p75–independent manner 17.
CSPGs in the injured CNS support binding of NgR1 and NgR3
Similar to other CNS fiber tracts, severed retinal ganglion cell (RGC) axons in the rodent optic nerve fail to show spontaneous long distance axonal regeneration. Retro–orbital crush injury to the optic nerve results in a global upregulation of CSPGs along the nerve 34. Importantly, injured but not control optic nerve sections support strong binding of soluble NgR1–Fc and NgR3–Fc, and the GAG–binding motif m2 of NgR1 and NgR3 is necessary for this binding (Fig. 6). Moreover, binding of soluble receptors is largely abrogated by pretreatment of injured optic nerve sections with Ch’aseABC. Residual binding of NgR1–Fc is likely due to association with endogenous MAIs (Fig. 6). Together these studies suggest that CSPGs are endogenous ligands for neuronal NgR1 and NgR3.
Figure 6. Binding of soluble NgR1 Fc and NgR3 Fc to optic nerve is enhanced by injury.
(a) Longitudinal sections of uninjured and injured adult mouse optic nerve (7 days following retro–orbital nerve crush) were incubated with NgR1–Fc, NgR3–Fc, NgR1(7ala)–Fc, and NgR3Δm2–Fc. Soluble NgR1–Fc and NgR1(7ala)–Fc, but not NgR3–Fc or NgR3Δm2–Fc, bind weakly to uninjured optic nerve sections. Following injury, binding of NgR1–Fc and NgR3–Fc is strongly increased and depends on the presence of the proteoglycan binding motif m2, as no increase was observed for NgR1(7ala)–Fc and NgR3 m2–Fc. Following crush injury to the optic nerve, GFAP immunoreactivity is upregulated along the entire nerve. Treatment of injured optic nerve sections with 1unit/ml chondroitinase ABC–(Ch’ase) strongly reduces NgR1–Fc and NgR3–Fc binding, suggesting that endogenous CSPGs participate in the binding of NgR1 and NgR3. (b) Quantification of binding to optic nerve sections. All binding is shown as a fold increase relative to AP–Fc. At least 20 sections were counted per condition (n=4 independent experiments). Light gray bars (Uninjured); black bars (Injured); dark gray bars (Injured + Ch’ase). Results are presented as mean ±SEMs. ** P< 0.001 (one way ANOVA, Tukey’s post–hoc), n.s.= not significant. Scale bar, 30μm.
Regeneration is enhanced in NgR123−/− and NgR13−/− mice
In the adult mouse retina, NgR1, NgR2, and NgR3 are all strongly expressed in RGCs (Fig. 7a). Retinal stratification (Fig. 7b) and optic nerve myelination (Fig. 7c) in NgR123−/− mice appear normal. To assess RGC axon targeting to the superior colliculus, the suprachiasmatic nucleus, and the lateral geniculate nucleus, the right eye of adult WT and NgR123−/− mice was injected with Alexa 594–conjugated Cholera Toxin β (CTB–red) tracer, and the left eye with Alexa 488–conjugated Cholera Toxin β (CTB–green) tracer. No defects in RGC axon central projections or target innervation were observed (Fig. 7d–f). Thus, germline ablation of all three NgRs does not appear to compromise retinal stratification, optic nerve myelination, or RGC axonal pathfinding.
Figure 7. Retinal stratification, optic nerve myelination, and RGC central projections appear normal in NgR123−/− mice.
(a) Sections of adult wildtype (WT) and Nogo receptor triple mutant (NgR123−/−) mouse retina were subjected to in situ hybridization with digoxigenin–labeled cRNA probes specific for NgR1, NgR2, and NgR3 transcripts. All three receptors are strongly expressed in the ganglion cell layer (arrow) and the inner nuclear layer, but are absent from the outer nuclear layer of the retina. No signal was detected on parallel–processed sections of NgR123−/− retina. (b) Hoechst 33342 nuclear staining, as well as anti–calbindin and anti–calretinin immunolabeling, of adult WT and NgR123−/− retina did not reveal any noticeable differences in retinal organization among the two genotypes. (c) Toluidine blue labeling of epon–embedded adult WT and NgR123−/− optic nerve cross sections reveals a comparable number of axons and degree of myelinated fibers. (d–f) The fidelity of RGC central projections in six–week–old WT and NgR123−/− mice was assessed by anterograde fiber tracing. Five days after injection of Alexa 594–conjugated Cholera Toxin β into the right eye and Alexa 488–conjugated Cholera Toxin β into the left eye, mice were sacrificed, perfused, and brain sections analyzed by fluorescence microscopy. Right eye (red) and left eye (green) RGC projections to the (d) superior colliculus, (e) suprachiasmatic nucleus and (f) lateral geniculate nucleus in NgR123−/− mice are indistinguishable from age–matched WT controls. Scale bar: a, b, 80μm; c, 5μm; d, 100μm; e, f, 60μm.
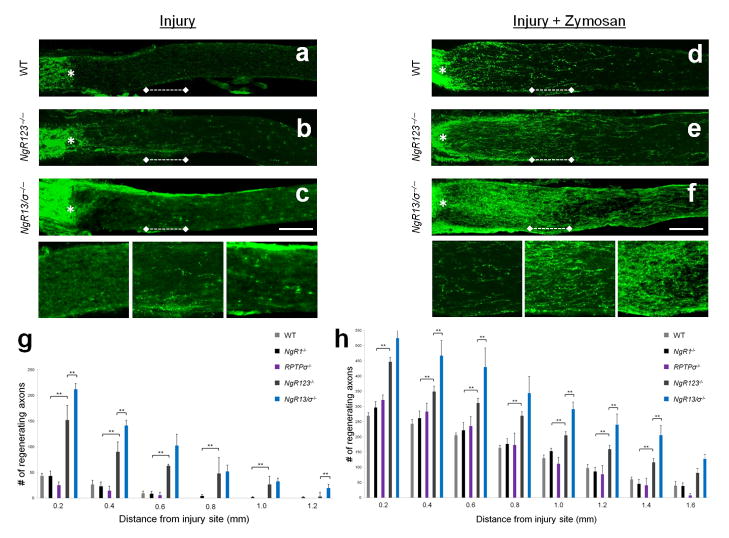
To assess whether NgRs contribute to the regenerative failure of injured CNS axons, we performed retro–orbital optic nerve crush injury in Nogo receptor single and compound mutant mice. Compared to injured wildtype controls, NgR123−/− mice show a modest but significant (P< 0.001, one way ANOVA, Tukey’s post–hoc) increase in RGC axon regeneration (Fig. 8). At two weeks post–injury, significantly more GAP43+ fibers are observed at 0.2–1.0 mm distal to the injury site in NgR123−/− mice compared to WT mice. Because NgR1 and NgR2 are known to associate with MAIs, the NgR123−/− regeneration phenotype may be a reflection of (i) decreased Nogo, MAG and OMgp inhibition, (ii) decreased CSPG inhibition, or (iii) a combination thereof. To address this issue, we directly compared regeneration of NgR1−/−, NgR2−/−, and NgR3−/− single mutants, as well as NgR12−/− and NgR13−/− double mutants, to NgR123−/− triple mutants. Loss of NgR1, NgR2, or NgR3 alone, or the combined loss of NgR1 and NgR2 (NgR12−/−), does not result in significantly enhanced RGC axon regeneration compared to WT mice (Fig. 8, Supplementary Fig. 7, and Supplementary Table). However, NgR13−/− mice show a similar degree of axon regeneration as NgR123−/− mice. This suggests a novel role for NgR3 in signaling neuronal growth inhibition. When coupled with neurite outgrowth studies in vitro, showing that NgR1 and NgR3 operate as functionally redundant CSPG receptors, this suggests that the optic nerve regeneration in NgR13−/− and NgR123−/− mice is at least in part a reflection of decreased CSPG inhibition.
Figure 8. NgR123−/− and NgR13/RPTPσ−/− compound mutants show enhanced fiber regeneration following crush injury to the optic nerve.
Two weeks following injury, regenerating axons in optic nerve sections were visualized by anti–GAP43 immunolabeling. The injury site is marked with an asterisk. (a) WT mice show very limited regenerative axonal growth following injury. (b) In NgR123−/− mice, many GAP43+ fibers grow beyond the lesion site. (c) In NgR13/RPTPσ−/− (NgR13/σ−/−) mice, a further increase of GAP43+ fiber growth is observed. (a′), (b′), and (c′) depict higher magnification images of the region 0.5 to 0.75mm distal to the lesion site [dotted line in images (a), (b), and (c), respectively]. (g) Quantification of the number of GAP43+ axons at 0.2 to 1.2mm distal to the lesion site. Light gray bars (WT, n=6); black bars (NgR1−/−, n=7); purple bars (RPTPσ−/−, n=5); dark gray bars (NgR123−/−, n=8); blue bars (NgR13/σ−/−, n=4). (d) Intraocular injection of Zymosan enhances regenerative axonal growth in WT mice. A further increase is observed in (e) NgR123−/− mice, which is further enhanced in (f) NgR13/σ−/−, mice. (d′), (e′), and (f′) depict higher magnification images of the region 0.5 to 0.75mm distal to the lesion site [dotted line in images (d), (e), and (f), respectively]. (h) Quantification of the number of GAP43+ axons at 0.2 to 1.6mm distal to the lesion site in Zymosan–injected mice. Light gray bars (WT + Zymosan, n=6); black bars (NgR1−/− + Zymosan, n=6); purple bars (RPTPσ−/− + Zymosan, n=4); dark gray bars (NgR123−/− + Zymosan, n=8); blue bars (NgR13/σ−/− + Zymosan, n=3). Results are presented as mean ±SEMs. ** P< 0.05 (one way ANOVA, Tukey’s post–hoc). Scale bar, 200μm.
As RPTPσ is expressed in adult RGCs 27, we examined whether the combined loss of NgR1 and NgR3 on an RPTPσ−/− background (NgR13/RPTPσ−/−) results in a further increase of regenerating axons. Few regenerating axons were observed in RPTPσ−/− single mutants, with no significant difference compared to WT controls. Compared to NgR13−/− double mutants, NgR13/RPTPσ−/− triple mutants show a further increase in the number of regenerating axons (P< 0.001, one way ANOVA, Tukey’s post–hoc), suggesting a genetic interaction among these receptors (Fig. 8, Supplementary Fig. 7, and Supplementary Table).
An advantage of optic nerve regeneration studies is that the growth potential of RGCs can be sensitized by intraocular (i.o.) injection of the yeast cell wall extract Zymosan, resulting in the release of RGC survival and growth–promoting factors, including oncomodulin 35, CNTF, and LIF 36. WT mice that receive i.o. Zymosan show greatly enhanced regeneration of RGC axons, exceeding the regeneration observed in non–Zymosan–treated NgR123−/− and NgR13/RPTPσ−/− mice (Fig. 8). Importantly, NgR123−/− mice that receive i.o. Zymosan show significantly more (P< 0.05, one way ANOVA, Tukey’s post–hoc) regenerating axons than WT, NgR1−/−, NgR2−/−, NgR3−/−, or RPTPσ−/− single mutants, as well as NgR12−/− double mutants, subjected to i.o. Zymosan. NgR13−/− and NgR123−/− mice with i.o. Zymosan show a similar regeneration phenotype. At several distances from the injury site, NgR13/RPTPσ−/− triple mutants with i.o. Zymosan show a further increase in the number of regenerating axons compared to NgR123−/− mice with i.o. Zymosan (P< 0.05, one way ANOVA, Tukey’s post–hoc) (Fig. 8, Supplementary Fig. 7, and Supplementary Table).
In mice, optic nerve injury leads to the death of ~ 70% of RGCs by two weeks post–injury (Supplementary Fig. 8). The enhanced regeneration observed in NgR123−/− mice is not a result of increased RGC survival, as similar numbers of injury induced–RGC death were observed in WT and NgR123−/− triple mutants. Intraocular Zymosan administration partially protects RGCs from axotomy–induced cell death; however, the protective effect of Zymosan is similar in WT and NgR123−/− mice (Supplementary Fig. 8). Consistent with the view that a decrease in RGC death is not sufficient to promote axonal regeneration, p53–deficient RGCs are more resistant to injury–induced cell death but fail to show enhanced regeneration 1. To assess whether i.o. Zymosan influences expression of NgRs or RPTPσ in RGCs, we performed in situ hybridization at 3 and 7 days post–Zymosan injection, but did not observe any obvious changes (Supplementary Fig. 9).
Discussion
The main finding of the present study is the identification of two novel CSPG receptors. We show that NgR1 and NgR3 bind directly and with high affinity to select types of CS–GAGs and operate as functionally redundant CSPG receptors. Loss of NgR family members individually is not sufficient to overcome CSPG inhibition; however, the combined loss of NgR1 and NgR3 leads to a significant release of CSPG inhibition. In NgR123−/− triple mutants, severed RGC axons show enhanced regenerative growth. Interestingly, NgR13−/−, but not NgR12−/− double mutants, phenocopy the optic nerve regeneration phenotype of NgR123−/− mice. A further enhancement of axon regeneration is observed in NgR13/RPTPσ triple mutants, revealing a genetic interaction among NgR family members and the previously identified CSPG receptor RPTPσ. Collectively, our studies provide unexpected evidence for shared receptor mechanisms for “prototypic myelin inhibitors” and CSPGs, two major classes of growth inhibitory molecules abundant in the adult mammalian CNS.
NgR1 and NgR3 bind with high selectivity to CS GAGs
CSPG inhibition depends on the presence of CS–GAG chains; we therefore explored the molecular basis of Nogo receptor–GAG interactions. We identified two sequence motifs in each receptor, both of which are necessary for GAG binding. Motif 1 is located in the LRR–CT capping domain and identical to the GT1b binding motif identified in NgR1 31. Motif 2 is located in the distal stalk region juxtaposed to the GPI anchor.
Remarkably, NgR1 and NgR3 show exquisite specificity toward select types of CS–GAGs. Binding to monosulfated CS–B, but not CS–A or CS–C, is very robust. In addition, the disulfated GAGs CS–D and CS–E bind strongly to NgR1 and NgR3. Identical binding preferences were observed for RPTPσ. Competition of soluble NgR1 and RPTPσ(1–3) ectodomain for CS–E binding suggests that two very different protein modules complex with at least partially overlapping CS–GAG structures. Dose–response experiments show that loss of all NgRs or RPTPσ is sufficient to attenuate inhibition of neurite outgrowth at low and intermediate, but not at high doses of CSPGs. A very recent study identified the receptor tyrosine phosphatase LAR as a CSPG receptor 37. Together, these findings reveal a significant degree of functional redundancy among CSPG receptor mechanisms and suggest that antagonism of multiple NgR and LAR family members will be required to fully overcome CSPG inhibition.
Additive effects of manipulating extrinsic and intrinsic pathways
The relatively modest regeneration phenotype observed in NgR123−/− and NgR13/RPTPσ−/− mice at two weeks post–injury is consistent with previous studies showing that expression of a dominant negative form of NgR1 in RGCs 38 or blocking of RhoA with C3 transferase 39 is not sufficient to promote substantial regeneration of severed optic nerve axons. In a similar vein, removal of one or several MAIs results in inconsistent and often poor regeneration in spinal cord injured mice 28,29. Collectively, mouse genetic studies suggest that germline ablation of multiple growth inhibitory ligands or receptors is not sufficient to promote robust and long–distance regeneration in different fiber tracts of the injured adult CNS.
A significant impact of environmental inhibitory signals on limiting axon regeneration was revealed, however, when genetic manipulations were combined with activation of RGC intrinsic growth programs. On an NgR13−/−, NgR123−/−, or NgR13/RPTPσ−/− background, i.o. Zymosan injection results in significantly enhanced axonal growth distal to the injury site compared to WT, NgR12−/−, or RPTPσ−/− mutant mice with i.o. Zymosan. While the additive effects of simultaneous release of growth inhibitory mechanisms and activation of intrinsic growth programs have been reported 38,40,41, our data show that in growth enabled RGCs, members of the NgR family and LAR family collaborate to negatively impact the number and length of regenerating axons following CNS injury.
NgR3 participates in neuronal growth inhibition
Enhanced axon regeneration observed in the optic nerve of NgR123−/− mice is mimicked by NgR13−/−, but not NgR12−/− double mutants. This suggests that on an NgR1−/− background, NgR3, but not NgR2, contributes to the regenerative failure of severed RGC axons. As NgR3 does not directly associate with Nogo, MAG, or OMgp, but supports CSPG binding and participates in CSPG inhibition in vitro, our findings suggest that NgR3–CSPG–mediated growth inhibition contributes to the regenerative failure of CNS axons in vivo.
While CSPGs are the first ligands identified for NgR3, they also bind to NgR1, further underscoring the high promiscuity of NgR1. CSPGs are found in crude CNS myelin preparations 17,42 and present in the CNS myelin used for this study (data now shown). Similar to the enhanced neurite outgrowth of NgR123−/− neurons on CNS myelin (Fig. 1), the enhanced growth of neurons functionally depleted of NgR1 and PirB 10 may be, at least in part, a reflection of decreased MAI and CSPG inhibition.
Implications for experience dependent neural plasticity
While it has been known for some time that MAIs and CSPGs share similar downstream signaling pathways 16,17, the level at which MAI and CSPG signaling cascades converge to regulate neuronal cytoskeletal dynamics has not yet been determined. Here we identify NgR1 and NgR3 as novel and functionally redundant CSPG receptors. We provide evidence that Nogo, MAG, OMgp, and CSPGs share receptor components and perhaps signal through related receptor complexes to block neuronal plasticity, sprouting, and axonal regeneration. In support of this idea, the myelin inhibitor Nogo–A shares structural and sequential similarities with neurocan, an inhibitory CSPG implicated in blocking neuronal regeneration 43, suggesting a common origin for two seemingly unrelated inhibitors of growth. The newly discovered connection between CSPGs and NgRs is not only relevant for neuronal repair, but may also provide a mechanistic explanation for why two seemingly unrelated manipulations, such as Ch’aseABC infusion into the mature visual cortex and germline ablation of NgR1 or Nogo result in enhanced ocular dominance plasticity following monocular deprivation 21,44. Mounting evidence suggests that mechanisms that limit neuronal growth and plasticity following CNS injury and disease resemble those that negatively regulate neuronal growth and synaptic structure under physiological conditions 45,46.
The identification of NgRs as shared receptors for MAIs and CSPGs provides new insights into how a diverse group of inhibitory cues regulates neuronal structure and function under physiological conditions and following injury. We propose that Nogo receptors are part of a multicomponent receptor system that serves as a signaling platform to initiate pathways that limit neuronal growth and increase structural stability of synapses. When combined with recent findings that NgR1and its ligands Nogo and OMgp influence synaptic transmission 47, experience–dependent network refinement 44, and spatial memory 48, the present findings expand the function of these molecules beyond neural repair, and shed light on a vital part of the neuronal machinery that limits growth and plasticity in CNS health and disease.
Methods
Transgenic mice
All animal handling and surgical procedures were performed in compliance with local and national animal care guidelines and approved by the University of Michigan Committee on Use and Care of Animals (UCUCA). NogoABC−/−;MAG−/−;OMgp−/−, RPTPσ−/−, NgR1−/−, NgR2−/−, and p75NTR−/− mice have been described 15,24,28. NgR3−/− germline mutants were generated by Lexicon Genetics and kindly provided by M. Greenberg (Harvard Medical School). NgR1 and NgR2 conditional mutants have been described elsewhere 31. NgR3 conditional knockout mice were generated by flanking exon2 with loxP sites (Supplementary Fig. 1). To generate germline deletion mutants, conditional knockouts were crossed with protamine–cre transgenic mice and then intercrossed with each other, or onto an RPTPσ−/− background, to generate double and triple mutants.
To confirm that NgR123−/− mice are null for NgR1, NgR2, and NgR3, brain extracts of adult WT and NgR123−/− mice were analyzed by western blotting. To enrich for NgRs, brain membranes were isolated, lysed in RIPA buffer (Sigma) containing protease inhibitor cocktail (Sigma), and affinity precipitated with agarose Concanavalin A beads (Vector Laboratories) overnight at 4°C. Bound glycoproteins were subjected to SDS–PAGE, blotted onto nitrocellulose membranes (Thermo Fisher Scientific), and probed with polyclonal anti–NgR1, anti–NgR2, anti–NgR3 or anti–β–actin.
Neurite outgrowth assays
To assay myelin inhibition, 96–well plates were coated with poly–D–lysine hydrobromide (50μg/ml; Sigma) overnight, rinsed in water, air dried, and then incubated with a 5μl spot of BSA (40μg/ml) or CNS myelin (40μg/ml) prepared from wildtype or NogoABC−/−;MAG−/−;OMgp−/− (NMO−/−) mice 28. Proteins were adsorbed to poly–D–lysine for 3 hours at 37°C. Wells were then rinsed and incubated with laminin (10μg/ml; Sigma) for 1 hour at 37°C. P7 CGNs and DRGs were prepared as described previously 12, and cultured for 24 hours before fixation with 4% paraformaldehyde, blocking in 1% horse serum and 0.1% Triton X–100 in PBS, and staining with anti–class III β–tubulin (TuJ1; Promega). To visualize the spotted myelin, wells were also stained with anti–myelin basic protein (Sigma). Alexa Fluor–conjugated secondary antibodies (Invitrogen) were used for fluorescent labeling.
To assay CSPG inhibition, 5μl spots (1, 2, 10, 100μg/ml) of either a mixture of large, extracellular chondroitin sulfate proteoglycans isolated from embryonic chicken brain (Millipore) or BSA were adsorbed on 96–well plates for 3 hours at 37°C before coating with poly–D–lysine hydrobromide and laminin. After 24 hours in culture, neurite length of CGNs or DRGs was determined as described above. To visualize the spotted CSPGs, wells were also stained with anti–chondroitin sulfate (CS56; Sigma). For some experiments, receptor fusion proteins (10μg/ml) or the ROCK inhibitor Y–27632 (10μM) were added to the wells at the time of CGN plating.
Construction of fusion proteins
AP– and Fc–tagged fusion proteins were constructed by standard PCR cloning using the Tth–DNA polymerase (Applied Biosystems). Constructs for AP–Fc, AP–NgR1(ΔGPI), AP–NgR2(ΔGPI), AP–NgR3(ΔGPI), AP–Nogo66, AP–OMgp, and MAG–Fc have been described previously 12,18. Additional constructs included AP–NgR1NT–LRR–CT(Ala24–V311), AP–NgR2NT–LRR–CT(Ser30–Thr314), AP–NgR3NT–LRR–CT(Ser22–Pro307), AP–NgR1CT+stalk (Val263–Glu445), AP–NgR2CT+stalk (Ala264–Ser397), and AP–NgR3CT+stalk (Asp258–Val413). Soluble Fc fusion proteins contain a PAM–myc signal sequence and a C–terminal Fc region of human IgG1. Constructs were assembled in the expression vector pcDNA3.0 as described 12 and include NgR1–Fc(Cys27–Gly448), NgR1Δm2–Fc(Cys27–Gly412), NgR1(7ala)–Fc (residues 414–429 RRRPGCSRKNRTRSHC of NgR1 were replaced by AAAPGCSAATSTASHC – the location of an internal Spe1 restriction site introduced for PCR construction is underlined), NgR2–Fc(Cys31–Gly399), NgR3–Fc(Pro25–Val420), NgR3Δm2–Fc(Pro25–Met397), NgR1(Cys27–Lys277)/NgR2(Val281–Gly399)–Fc, and RPTPσ(Ig1–3)–Fc(Glu30–Val315).
Binding assays
COS–7 cells grown in 24–well plates were transiently transfected (Lipofectamine 2000) with plasmid DNA encoding p75NTR, TROY, Lingo–1, L–MAG, NgR1, NgR3, NgR1(7ala), chimeric NgR1(Cys27–Lys277)/NgR2(Val281–Leu420), or GFP. Ligand receptor binding studies were carried out and developed as described previously 12. For some COS–7 cell binding assays, CS–GAGs, at a concentration of 1mg/ml, were added to the wells at the time of ligand incubation.
To monitor binding of soluble Nogo receptors to brain tissue sections in situ, binding studies with embryonic (E18) and neonatal (P1–P3) rat or mouse brains were carried out as described previously 12,18. Additional binding studies included longitudinal optic nerve sections of adult mice. Consecutive optic nerve sections were stained with anti–GFAP (Millipore).
To assess whether the association of AP–NgR1CT+stalk or AP–NgR3CT+stalk to brain tissue sections is mediated by protein–protein interactions, brain sections were preincubated at 75°C for 3 hours or treated with trypsin (10 units for 30 minutes at 37°C) prior to incubation with AP–fusion proteins. To explore whether glycoconjugates participate in binding of soluble Nogo receptors to brain tissue, sections were enzymatically treated with glycosidases according to manufacturer’s instructions prior to adding the AP–fusion proteins. Enzymes included N–acetylglucosaminidase, Vibrio cholerae neuraminidase, heparinase III (Flavobacterium heparinium), chondroitinase–ABC (all from Calbiochem), glycopeptidase F (New England Biolabs), and endoneuraminidase N (kindly provided by U. Rutishauser). Some injured optic nerve sections were incubated with chondroitinase–ABC (1unit/ml in Tris–acetate buffer – pH 8.0 – with 0.02% BSA) for 3 hours at 37°C prior to addition of receptor fusion proteins. To assay whether specific types of GAG side chains block binding of soluble NgRs to brain tissue, competition binding experiments were carried out in the presence of heparin or different types of CS–GAG chains (50μg/ml, Seikagaku Glycobiology).
To determine whether NgRs or RPTPσ bind directly to GAG chains, enzyme–linked immunosorbent assays (ELISAs) were used as described previously 49. Briefly, GAG chains were biotinylated with EDC and EZ–Link Sulfo–NHS–LC–LC–Biotin (Thermo Scientific) and adsorbed for 15 minutes to ELISA plates (Immulon4, NUNC) precoated with streptavidin (5μg/ml; Invitrogen). Plates were blocked (5% BSA), rinsed with HBS, and incubated with various amounts of fusion proteins (diluted with 5% BSA) for 2 hours at 22°C. Following five washes with HBS, bound AP activity was monitored with a BluePhos Microwell Substrate Kit (KPL). For competitive binding experiments, immobilized GAG chains were preincubated with various amounts of RPTPσ(Ig1–3)–Fc for 16 hours at 4°C, and then incubated with AP–NgR1 (1nM) for 2 hours at 22°C. Bound AP activity was measured as described above.
Immunoprecipitation
HEK293T cells (in 10 cm culture dishes) were transfected with various combinations of p75NTR, NgR1, NgR1–myc, NgR2–myc, and NgR3–myc expression constructs. After 48 hours, the cells were incubated in lysis buffer containing the following: 20mM Tris–HCl (pH 7.5), 150mM NaCl, 5mM EDTA, 1% NP–40, and protease inhibitor mixture. For some assays, cells were incubated for 30 minutes with 100μg/ml of CSPG mixture prior to lysis. Cell lysates were tumbled overnight at 4°C in the presence of either anti–p75 (Promega) or anti–NgR1, and precipitated with Protein G Plus/Protein A–Agarose (Calbiochem) after incubation at 4°C for 2 hours. Precipitated beads were rinsed three times with lysis buffer, and bound proteins were eluted with 2X SDS sample buffer. Precipitates were analyzed by immunoblotting, using anti–NgR1, anti–p75 (Promega), anti myc (Cell Signaling), or anti–β–actin (Sigma).
Optic nerve surgery
Adult mice (6–8 weeks of age) of either sex were anesthetized with an intraperitoneal injection of Ketamine (100mg/kg; Fort Dodge Animal Health) and Xylazine (10mg/kg; Akorn, Inc.). The optic nerve was exposed through an incision in the conjunctiva and compressed for 10 seconds with angle jeweler’s forceps (Dumont #5, Fine Science Tools) at approximately 1mm behind the eyeball. Care was taken not to damage or rupture the ophthalmic artery. For intraocular (i.o.) injection of Zymosan, 5μl of a suspension (12.5μg/μl in sterile PBS; Sigma) was injected manually using a Hamilton syringe with a 30 gauge removable needle. Following optic nerve surgery, the operated eye was rinsed with sterile PBS and ophthalmic ointment was applied (Butler AHS). All surgeries were performed under aseptic conditions. Fourteen days after optic nerve injury, mice were given a lethal dose of anesthesia and perfused through the heart with PBS followed by ice–cold 4% paraformaldehyde.
Histochemical studies
In situ hybridization of mouse retina with cRNA probes specific for NgR1, NgR2, NgR3, and RPTPσ was carried out as described previously 12,27. For immunohistochemical procedures, cryosections of adult retina were stained with anti–calbindin (Swant, 1:2,500 dilution) or anti calretinin (Swant, 1:2,500 dilution), and then counterstained with Hoechst 33342 (1:30,000 dilution). To assess axon density and myelination, optic nerves were embedded in epon and stained with Toluidine blue. To assess retinal ganglion cell death at various time points following optic nerve injury, retinal sections were stained with anti–class III–β–tubulin (TuJ1), and in some instances, with anti–active caspase–3 (Promega).
For intraocular injections of anterograde tracer, 6–week–old mice received bilateral injections (2μl) of 1μg/μl Alexa 488– and Alexa 594–conjugated Cholera Toxin β (Invitrogen) in the left and right eye, respectively. Five days post–injection, mice were perfused transcardially, and their brains were dissected, post–fixed in 4 % paraformaldehyde overnight, and cryoprotected in 30% sucrose overnight. Brain tissue was embedded in OCT Tissue–Tek Medium (Sakura Finetek) and coronal sections (50μm thickness) were imaged.
To visualize regenerating axons in the injured optic nerve, eyes with optic nerves attached were dissected, post–fixed, and cryoprotected. Optic nerves were embedded and longitudinal sections (14μm thickness) were stained with anti–GAP–43. The appropriate Alexa Fluor–conjugated secondary antibody (Invitrogen) was then used for fluorescent labeling.
Statistical analysis
For quantification of neurite outgrowth, UTHSCSA ImageTool for Windows was used, and processes equal or longer to approximately one cell body diameter were measured. For each condition, at least 150 neurites were quantified, and the mean and SEM of neurite length for each genotype was determined from multiple, independent experiments. For quantification of retinal ganglion cell death, the density of TuJ1–positive cells in the ganglion cell layer per field of view (at least 10 sections, 3 independent experiments per condition) was counted. For quantification of activated caspase–3–positive retinal ganglion cells, the number of cells labeled for activated caspase–3 was calculated as a percentage of the total number of cells (TuJ1–positive) per field of view (at least 10 sections, 3 independent experiments per condition). Quantification of optic nerve binding assays and in situ hybridization (at least 20 sections, 4 independent experiments per condition) was performed as previously described 50, using Microsuite Five (Olympus) quantification software. All data were analyzed using one–way analysis of variance followed by Tukey’s post–hoc comparisons. All statistics were performed using SigmaStat 3.0 for Windows (Systat Software).
To assess regenerative axonal growth, the number of GAP–43–positive axons at prespecified distances from the injury site was counted in at least three sections per nerve. These numbers were converted into the number of regenerating axons per nerve at various distances as described previously 38. All data were analyzed using one–way analysis of variance followed by Tukey’s pos–hoc comparisons. All statistics were performed using GraphPad Prism 5.00 (GraphPad Software). Our finding that loss of all three NgRs elicits significant retinal ganglion cell regeneration is based on two independently generated data sets produced by two independent surgeons (K.T.B. and Y.K). Both data sets were analyzed separately and lead to the same conclusions (Supplementary Table). In addition, no significant differences in axon regeneration following injury (with or without intraocular Zymosan injection) were observed between mice on three different genetic backgrounds (129, C57BL/6, BALB/c) (data not shown).
Supplementary Material
Acknowledgments
This work was supported by the Neuroscience Training Grant T32EY017878 and the University of Michigan Rackham Merit Fellowship (T.L.D.), the Cellular and Molecular Biology Training Grant T32GM007315 (K.T.B. and Y.A.M.), the National Research Service Award Ruth Kirschstein Fellowship F31NS061589 (S.J.R.), the New York State Spinal Cord Injury Research Program, the Dr. Miriam and Sheldon G. Adelson Medical Foundation on Neural Repair and Rehabilitation, the US Department of Veterans Affairs (1I01RX000229–01), the National Institute of Neurological Disorders and Stroke R56NS047333 (R.J.G.) and the National Eye Institute (L.I.B.). We thank Michel Tremblay for RPTPσ−/− mice; Michael Greenberg for NgR3−/− mice; Brian Pierchala for p75NTR−/− mice; Brian Bates, David Howland, and Mary L. Mercado for their assistance in the generation and initial analysis of NgR123−/− triple mutant mice; Urs Rutishauser for Endo–N; David Figge and Yuko Yasui for assistance in ELISA binding assays; Yuqin Yin for training in optic nerve surgery; Yuntao Duan for generation of the RPTPσ(Ig1–3)–Fc construct; Justin Barbieri for technical assistance; and Margaret M. Zaleska for project administration.
Footnotes
Author Contributions:
R.J.G. conceived the study; T.L.D., L.I.B., H.M.G., and R.J.G. designed the experiments; T.L.D., K.T.B., Y.A.M., Y.K., S.J.R., C.D.L., Y.K., and R.J.G. performed experiments; T.L.D., K.T.B., and Y.K. contributed to data analysis and figure preparation; K.L.A., A.W., C.G.G., and B.Z. generated and provided mice or reagents for the study; and T.L.D. and R.J.G. wrote the manuscript. All authors read and agreed on the final version of the manuscript.
Competing Financial Interests:
The authors declare no competing financial interests.
References
- 1.Park KK, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu BP, Cafferty WB, Budel SO, Strittmatter SM. Extracellular regulators of axonal growth in the adult central nervous system. Philos Trans R Soc Lond B Biol Sci. 2006;361:1593–1610. doi: 10.1098/rstb.2006.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winzeler AM, et al. The lipid sulfatide is a novel myelin-associated inhibitor of CNS axon outgrowth. J Neurosci. 2011;31:6481–6492. doi: 10.1523/JNEUROSCI.3004-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci. 2004;5:146–156. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- 5.Bregman BS, et al. Recovery from spinal cord injury mediated by antibodies to neurite growth inhibitors. Nature. 1995;378:498–501. doi: 10.1038/378498a0. [DOI] [PubMed] [Google Scholar]
- 6.Li S, et al. Blockade of Nogo-66, myelin-associated glycoprotein, and oligodendrocyte myelin glycoprotein by soluble Nogo-66 receptor promotes axonal sprouting and recovery after spinal injury. J Neurosci. 2004;24:10511–10520. doi: 10.1523/JNEUROSCI.2828-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradbury EJ, et al. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature. 2002;416:636–640. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Alias G, Barkhuysen S, Buckle M, Fawcett JW. Chondroitinase ABC treatment opens a window of opportunity for task-specific rehabilitation. Nat Neurosci. 2009;12:1145–1151. doi: 10.1038/nn.2377. [DOI] [PubMed] [Google Scholar]
- 9.Massey JM, et al. Chondroitinase ABC digestion of the perineuronal net promotes functional collateral sprouting in the cuneate nucleus after cervical spinal cord injury. J Neurosci. 2006;26:4406–4414. doi: 10.1523/JNEUROSCI.5467-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atwal JK, et al. PirB is a functional receptor for myelin inhibitors of axonal regeneration. Science. 2008;322:967–970. doi: 10.1126/science.1161151. [DOI] [PubMed] [Google Scholar]
- 11.Fournier AE, GrandPre T, Strittmatter SM. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature. 2001;409:341–346. doi: 10.1038/35053072. [DOI] [PubMed] [Google Scholar]
- 12.Venkatesh K, et al. The Nogo-66 receptor homolog NgR2 is a sialic acid-dependent receptor selective for myelin-associated glycoprotein. J Neurosci. 2005;25:808–822. doi: 10.1523/JNEUROSCI.4464-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chivatakarn O, Kaneko S, He Z, Tessier-Lavigne M, Giger RJ. The Nogo-66 receptor NgR1 is required only for the acute growth cone-collapsing but not the chronic growth-inhibitory actions of myelin inhibitors. J Neurosci. 2007;27:7117–7124. doi: 10.1523/JNEUROSCI.1541-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JE, Liu BP, Park JH, Strittmatter SM. Nogo-66 receptor prevents raphespinal and rubrospinal axon regeneration and limits functional recovery from spinal cord injury. Neuron. 2004;44:439–451. doi: 10.1016/j.neuron.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 15.Zheng B, et al. Genetic deletion of the Nogo receptor does not reduce neurite inhibition in vitro or promote corticospinal tract regeneration in vivo. Proc Natl Acad Sci U S A. 2005;102:1205–1210. doi: 10.1073/pnas.0409026102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sivasankaran R, et al. PKC mediates inhibitory effects of myelin and chondroitin sulfate proteoglycans on axonal regeneration. Nat Neurosci. 2004;7:261–268. doi: 10.1038/nn1193. [DOI] [PubMed] [Google Scholar]
- 17.Schweigreiter R, et al. Versican V2 and the central inhibitory domain of Nogo-A inhibit neurite growth via p75NTR/NgR-independent pathways that converge at RhoA. Mol Cell Neurosci. 2004;27:163–174. doi: 10.1016/j.mcn.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Kantor DB, et al. Semaphorin 5A is a bifunctional axon guidance cue regulated by heparan and chondroitin sulfate proteoglycans. Neuron. 2004;44:961–975. doi: 10.1016/j.neuron.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Rhodes KE, Fawcett JW. Chondroitin sulphate proteoglycans: preventing plasticity or protecting the CNS? J Anat. 2004;204:33–48. doi: 10.1111/j.1469-7580.2004.00261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKeon RJ, Hoke A, Silver J. Injury-induced proteoglycans inhibit the potential for laminin-mediated axon growth on astrocytic scars. Exp Neurol. 1995;136:32–43. doi: 10.1006/exnr.1995.1081. [DOI] [PubMed] [Google Scholar]
- 21.Pizzorusso T, et al. Reactivation of ocular dominance plasticity in the adult visual cortex. Science. 2002;298:1248–1251. doi: 10.1126/science.1072699. [DOI] [PubMed] [Google Scholar]
- 22.Houle JD, et al. Combining an autologous peripheral nervous system “bridge” and matrix modification by chondroitinase allows robust, functional regeneration beyond a hemisection lesion of the adult rat spinal cord. J Neurosci. 2006;26:7405–7415. doi: 10.1523/JNEUROSCI.1166-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powell EM, Mercado ML, Calle-Patino Y, Geller HM. Protein kinase C mediates neurite guidance at an astrocyte boundary. Glia. 2001;33:288–297. doi: 10.1002/1098-1136(20010315)33:4<288::aid-glia1027>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 24.Shen Y, et al. PTPsigma is a receptor for chondroitin sulfate proteoglycan, an inhibitor of neural regeneration. Science. 2009;326:592–596. doi: 10.1126/science.1178310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aricescu AR, McKinnell IW, Halfter W, Stoker AW. Heparan sulfate proteoglycans are ligands for receptor protein tyrosine phosphatase sigma. Mol Cell Biol. 2002;22:1881–1892. doi: 10.1128/MCB.22.6.1881-1892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fry EJ, Chagnon MJ, Lopez-Vales R, Tremblay ML, David S. Corticospinal tract regeneration after spinal cord injury in receptor protein tyrosine phosphatase sigma deficient mice. Glia. 2010;58:423–433. doi: 10.1002/glia.20934. [DOI] [PubMed] [Google Scholar]
- 27.Sapieha PS, et al. Receptor protein tyrosine phosphatase sigma inhibits axon regrowth in the adult injured CNS. Mol Cell Neurosci. 2005;28:625–635. doi: 10.1016/j.mcn.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Lee JK, et al. Assessing spinal axon regeneration and sprouting in Nogo-, MAG-, and OMgp-deficient mice. Neuron. 2010;66:663–670. doi: 10.1016/j.neuron.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cafferty WB, Duffy P, Huebner E, Strittmatter SM. MAG and OMgp synergize with Nogo-A to restrict axonal growth and neurological recovery after spinal cord trauma. J Neurosci. 2010;30:6825–6837. doi: 10.1523/JNEUROSCI.6239-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamashita T, Fujitani M, Yamagishi S, Hata K, Mimura F. Multiple signals regulate axon regeneration through the Nogo receptor complex. Mol Neurobiol. 2005;32:105–111. doi: 10.1385/MN:32:2:105. [DOI] [PubMed] [Google Scholar]
- 31.Williams G, et al. Ganglioside inhibition of neurite outgrowth requires Nogo receptor function: identification of interaction sites and development of novel antagonists. J Biol Chem. 2008;283:16641–16652. doi: 10.1074/jbc.M802067200. [DOI] [PubMed] [Google Scholar]
- 32.Barton WA, et al. Structure and axon outgrowth inhibitor binding of the Nogo-66 receptor and related proteins. EMBO J. 2003;22:3291–3302. doi: 10.1093/emboj/cdg325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Kuang X, Zhang J. Nogo receptor 3, a paralog of Nogo-66 receptor 1 (NgR1), may function as a NgR1 co-receptor for Nogo-66. J Genet Genomics. 2011;38:515–523. doi: 10.1016/j.jgg.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Ohlsson M, Mattsson P, Svensson M. A temporal study of axonal degeneration and glial scar formation following a standardized crush injury of the optic nerve in the adult rat. Restor Neurol Neurosci. 2004;22:1–10. [PubMed] [Google Scholar]
- 35.Yin Y, et al. Oncomodulin links inflammation to optic nerve regeneration. Proc Natl Acad Sci U S A. 2009;106:19587–19592. doi: 10.1073/pnas.0907085106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leibinger M, et al. Neuroprotective and axon growth-promoting effects following inflammatory stimulation on mature retinal ganglion cells in mice depend on ciliary neurotrophic factor and leukemia inhibitory factor. J Neurosci. 2009;29:14334–14341. doi: 10.1523/JNEUROSCI.2770-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher D, et al. Leukocyte Common Antigen-Related Phosphatase Is a Functional Receptor for Chondroitin Sulfate Proteoglycan Axon Growth Inhibitors. J Neurosci. 2011;31:14051–14066. doi: 10.1523/JNEUROSCI.1737-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer D, He Z, Benowitz LI. Counteracting the Nogo receptor enhances optic nerve regeneration if retinal ganglion cells are in an active growth state. J Neurosci. 2004;24:1646–1651. doi: 10.1523/JNEUROSCI.5119-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer D, Petkova V, Thanos S, Benowitz LI. Switching mature retinal ganglion cells to a robust growth state in vivo: gene expression and synergy with RhoA inactivation. J Neurosci. 2004;24:8726–8740. doi: 10.1523/JNEUROSCI.2774-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadoya K, et al. Combined intrinsic and extrinsic neuronal mechanisms facilitate bridging axonal regeneration one year after spinal cord injury. Neuron. 2009;64:165–172. doi: 10.1016/j.neuron.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujita Y, Endo S, Takai T, Yamashita T. Myelin suppresses axon regeneration by PIR-B/SHP-mediated inhibition of Trk activity. EMBO J. 2011;30:1389–1401. doi: 10.1038/emboj.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niederost BP, Zimmermann DR, Schwab ME, Bandtlow CE. Bovine CNS myelin contains neurite growth-inhibitory activity associated with chondroitin sulfate proteoglycans. J Neurosci. 1999;19:8979–8989. doi: 10.1523/JNEUROSCI.19-20-08979.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shypitsyna A, Malaga-Trillo E, Reuter A, Stuermer CA. Origin of Nogo-A by domain shuffling in an early jawed vertebrate. Mol Biol Evol. 2010 doi: 10.1093/molbev/msq313. [DOI] [PubMed] [Google Scholar]
- 44.McGee AW, Yang Y, Fischer QS, Daw NW, Strittmatter SM. Experience-driven plasticity of visual cortex limited by myelin and Nogo receptor. Science. 2005;309:2222–2226. doi: 10.1126/science.1114362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee H, et al. Synaptic function for the Nogo-66 receptor NgR1: regulation of dendritic spine morphology and activity-dependent synaptic strength. J Neurosci. 2008;28:2753–2765. doi: 10.1523/JNEUROSCI.5586-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zagrebelsky M, Schweigreiter R, Bandtlow CE, Schwab ME, Korte M. Nogo-A stabilizes the architecture of hippocampal neurons. J Neurosci. 2010;30:13220–13234. doi: 10.1523/JNEUROSCI.1044-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raiker SJ, et al. Oligodendrocyte-myelin glycoprotein and Nogo negatively regulate activity-dependent synaptic plasticity. J Neurosci. 2010;30:12432–12445. doi: 10.1523/JNEUROSCI.0895-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karlen A, et al. Nogo receptor 1 regulates formation of lasting memories. Proc Natl Acad Sci U S A. 2009;106:20476–20481. doi: 10.1073/pnas.0905390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Briani C, Berger JS, Latov N. Antibodies to chondroitin sulfate C: a new detection assay and correlations with neurological diseases. J Neuroimmunol. 1998;84:117–121. doi: 10.1016/s0165-5728(97)00209-9. [DOI] [PubMed] [Google Scholar]
- 50.Robak LA, et al. Molecular basis of the interactions of the Nogo-66 receptor and its homolog NgR2 with myelin-associated glycoprotein: development of NgROMNI-Fc, a novel antagonist of CNS myelin inhibition. J Neurosci. 2009;29:5768–5783. doi: 10.1523/JNEUROSCI.4935-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.