Abstract
Insulin-like growth factor II (IGF-II) mRNA binding protein 3 (IMP3) is emerging as a useful indicator of the progression and outcome of several cancers. IMP3 expression is associated with triple-negative breast carcinomas (TNBCs), which are aggressive tumors associated with poor outcome. In this study, we addressed the hypothesis that signaling pathways, which are characteristic of TNBCs, impact the expression of IMP3 and that IMP3 contributes to the function of TNBCs. The data obtained reveal that IMP3 expression is repressed specifically by estrogen receptor β (ERβ) and its ligand 3βA-diol but not by ERα. EGF receptor (EGFR) signaling and consequent activation of the MAP kinase pathway induce IMP3 transcription and expression. Interestingly, we discovered that the EGFR promoter contains an imperfect estrogen response element and that ERβ represses EGFR transcription. These data support a mechanism in which ERβ inhibits IMP3 expression indirectly by repressing the EGFR. This mechanism relates to the biology of TNBC, which is characterized by diminished ERβ and increased EGFR expression. We also demonstrate that IMP3 contributes to the migration and invasion of breast carcinoma cells. Given that IMP3 is an mRNA binding protein, we determined that it binds several key mRNAs that could contribute to migration and invasion including CD164 (endolyn) and MMP9. Moreover, expression of these mRNAs is repressed by ERβ and enhanced by EGFR signaling, consistent with our proposed mechanism for the regulation of IMP3 expression in breast cancer cells. Our findings show that IMP3 is an effector of EGFR-mediated migration and invasion and they provide the first indication of how this important mRNA binding protein is regulated in cancer.
Keywords: Breast cancer, IMP3, ERβ, EGFR
Introduction
IMP3 is a member of a family of insulin-like growth factor II (IGF-II) mRNA binding proteins (IMPs) consisting of IMP1, IMP2 and IMP3 (1). IMPs play important roles in RNA trafficking, stabilization, localization and cell migration, especially during early stages of both human and mouse embryogenesis (2). IMPs are expressed in developing epithelium, placenta and muscle but they are undetectable in normal adult tissues (3). Importantly, however, IMP3 is re-expressed in several malignant tissues including pancreatic, lung, renal cell, ovarian, endometrial and cervical cancers (4–9). This phenomenon has been exploited for the prognostic assessment of specific cancers. In particular, IMP3 is an accurate predictor of renal-cell carcinoma metastasis and prognosis (4), and similar trends are emerging for other cancers (10–12). We are interested in breast cancer in this context, especially in light of the finding that IMP3 is expressed preferentially in triple negative breast cancers (10). Triple negative breast cancer (TNBC) is a molecular subtype of breast cancer characterized by the absence of estrogen receptor-α (ERα), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER2) amplification. Clinically, TNBCs are usually of high histological grade, poorly differentiated and more aggressive compared to other subtypes of breast cancer (13). In fact, treatment of TNBC patients remains a challenge because of the lack of targeted therapeutic options and the high metastatic potential of TNBC cells. These observations indicate that IMP3 could prove useful for the clinical management of TNBC but much remains to be learned about its regulation and function in these tumors.
In this study, we addressed the hypothesis that signaling pathways, which are characteristic of TNBCs, impact the expression of IMP3 and that IMP3 contributes to the function of TNBCs. A salient feature of TNBC is the absence of or diminished ER signaling. More specifically, TNBCs are ERα negative and they express low levels of ERβ (14). A reasonable possibility based on these observations is that ER signaling represses IMP3. Interestingly, our data reveal that ERβ and its ligand 3βA-diol (5α-Androstane-3β, 17β-diol) repress IMP3 expression but that ERα does not contribute to this repression. Further analysis demonstrated that EGFR signaling induces IMP3 expression and that ERβ represses EGFR transcription, revealing a mechanism in which ERβ inhibits IMP3 expression indirectly by repressing the EGFR. This finding is relevant to the biology of TNBC because these tumors are characterized by over-expression of the EGFR and concomitant EGFR signaling (15). Our observation that IMP3 contributes to the migration and invasion of TNBC cells provides a functional role for this IMP in breast cancer and it suggests that IMP3 can be an effector of EGFR-mediated migration and invasion.
Results
Estrogen receptor-β suppresses IMP3 expression
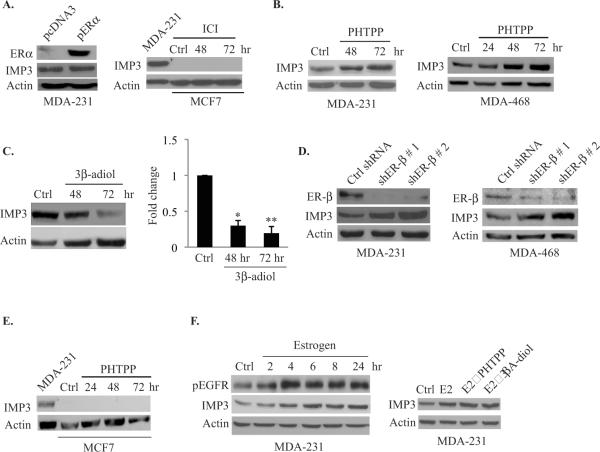
To assess the possible role of ERs in regulating IMP3 expression, we used MDA-MB-231 and MDA-MB-468 cells, which are triple negative breast carcinoma cell lines that express IMP3 but lack ERα and they express low levels of ERβ. Exogenous expression of ERα in MDA-MB-231 cells did not alter IMP3 expression (Fig. 1A). Also, inhibition of endogenous ERα activity by ICI-182780 (a selective ERα antagonist) was unable to induce IMP3 expression in MCF7 cells, which express ERα and ERβ but not IMP3 (Fig. 1A). Based on these results, we focused our attention on ERβ. PHTPP (4-[2-Phenyl-5, 7-bis (trifluoromethyl) pyrazolo [1,5-a] pyrimidin-3-yl] phenol) is a specific ERβ antagonist and it caused a significant increase in IMP3 expression in both MDA-MB-231 and MDA-MB-468 cells (Fig. 1B). This finding was substantiated using 3βA-diol, an androgen metabolite that is a specific ligand of ERβ (16). 3βA-diol decreased IMP3 expression in MDA-MB-231 cells indicating that ligand-dependent activation of ERβ represses IMP3 (Fig. 1C). Also, depletion of ERβ using shRNAs increased IMP3 expression in both MDA-MB-231 and MDA-MB-468 cells significantly (Fig. 1D). Interestingly, PHTPP did not induce IMP3 expression in MCF7 cells (Fig. 1E), indicating that the ERβ-mediated repression of IMP3 may be restricted to triple negative breast cancer cells.
Figure 1. ERβ suppresses IMP3 protein expression.
(A, left) ERα was expressed in MDA-MB-231 cells by transfecting them with an expression vector, pcDNA3/ERα (pERα). (A, right) MCF7 cells were treated with either DMSO or the ERα antagonist ICI (1 μM) and analyzed for IMP3 expression by immunoboltting. MDA-MB-231 cells were used as positive control for IMP3 expression. (B) & (E) Immunoblots show IMP3 expression in MDA-MB-231, MDA-MB-468 and MCF7 (E) cells treated with the ERβ antagonist PHTPP (10 μM). (C) MDA-MB-231 cells were treated with 3βA-diol (10 μM). Graphs on the right side of each blot represent densitometry analysis (Image J software) of IMP3 expression. (D) Blots show the effect of ERβ depletion on IMP3 expression in MDA-MB-231 and MDA-MB-468 cells. ERβ expression was depleted by transfecting cells with plasmids expressing two different shRNAs. A plasmid expressing GFP-shRNA was used as control. (F, left) Immunoblot shows the effect of estrogen (E2, 10 nM) on IMP3 protein expression in MDA-MB-231 cells. (F, right) Effect of PHTPP and 3βA-diol (24 hr) on IMP3 expression in estrogen treated MDA-MB-231 cells. The expression of IMP3, ERα and ERβ was assessed 48 hrs post-transfection. DMSO was used as control for estrogen, PHTPP and 3βA-diol treatment.
We observed that estrogen (E2) increased IMP3 expression in MDA-MB-231 cells (Fig. 1F) Although this result seems unexpected, we also found that estrogen increased EGFR phosphorylation in these cells (Fig. 1F) and that the ability of estrogen to increase EGFR phosphorylation was not affected by either inhibition (PHTPP) or stimulation (3βA-diol) of ERβ function (Fig. 1F). These findings are consistent with the report that estrogen exerts its non-genomic action through EGFR signaling (17).
EGFR signaling positively regulate IMP3 expression
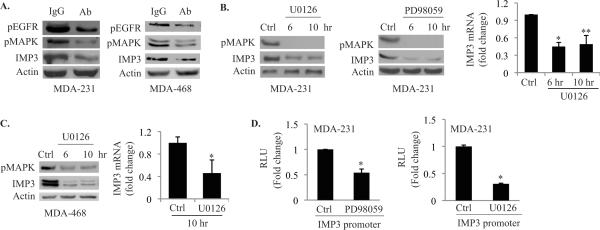
Given that high EGFR expression is associated with TNBC (15), we assessed the possibility that EGFR signaling regulates IMP3 expression. Treatment of both MDA-MB-231 and MDA-MB-468 cells with an EGFR function blocking Ab reduced IMP3 expression significantly compared to IgG- treated cells (Fig. 2A). This Ab also diminished EGFR phosphorylation and MAP kinase activation, which is a bona fide downstream effector of the EGFR signaling pathway (18) (Fig. 2A). To substantiate the involvement of EGFR signaling pathway in regulating IMP3 expression, we inhibited MEK1/2 (upstream component of MAPK) using two different inhibitors (PD98059 and U0126). As shown in Fig. 2B, IMP3 mRNA and protein expression is reduced significantly upon treatment with these inhibitors. Similar results were obtained using MDA-MB-468 cells (Fig. 2C). We also assayed the activity of the IMP3 promoter in the presence of the MEK1/2 inhibitors using a reporter construct containing the human IMP3 proximal promoter. As shown in Fig. 2D, inhibition of MEK1/2 using either U0126 or PD98059 decreased luciferase activity considerably. Collectively, our data indicate that an EGFR/MEK/MAPK pathway regulates IMP3 expression.
Figure 2. EGFR signaling positively regulates IMP3 expression.
(A) Immunoblots show the effect of blocking EGFR on IMP3 expression using a specific Ab (2 μg /mL) in MDA-MB-231 (left) and MDA-MB-468 (right) cells. Rabbit IgG was used as control. (B) & (C) MDA-MB-231 and MDA-MB-468 cells were treated with MEK1/2 inhibitors u0126 (10 μM) and PD98059 (50 μM) for different time points as indicated in the figure, and the expression of IMP3, as well as pMAPK, was analyzed by immunoblotting. IMP3 mRNA from cells treated with U0126 for 10 hr was assessed by qPCR. (D) MDA-MB-231 cells were transfected with a luciferase reporter construct containing 2.872 kb IMP3 promoter (wild type) in presence or absence of U0126 and PD98059, and luciferase activity was measured 24 h post-transfection. Data represent the mean of three independent experiments. p value (*) < 0.05.
Estrogen receptor-β suppresses EGFR expression
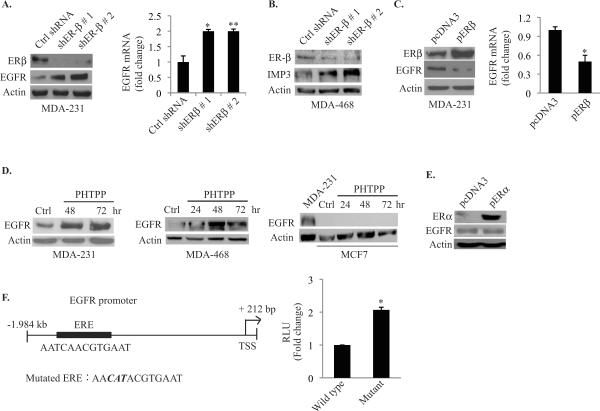
The contrasting data we obtained with ERβ and EGFR regulation of IMP3 expression raised the possibility that these receptors function in a common pathway. This possibility was supported by our observation that depletion of ERβ expression in both MDA-MB-231 and MDA-MB-468 cells increased EGFR mRNA and protein expression significantly compared to control cells expressing GFP shRNA (Fig. 3A & 3B). Similarly, over-expression of ERβ reduced EGFR protein and mRNA (Fig. 3C). To substantiate our hypothesis, we inhibited ERβ function using the selective antagonist PHTPP and observed increased EGFR protein expression (Fig. 3D). In contrast, PHTPP did not induce EGFR expression in MCF7 cells (Fig. 3D). Interestingly, restoration of ERα expression in MDA-MB-231 cells did not affect EGFR expression (Fig. 3E), consistent with our finding that ERα does not contribute to the regulation of IMP3.
Figure 3. ERβ suppresses EGFR expression.
(A) & (B) ERβ was transiently depleted in MDAMB-231 and MDA-MB-468 cells using two different shRNAs. EGFR protein and mRNA (MDA-MB-231) expression was evaluated by immunoblotting and qPCR, respectively. (C) ERβ was over-expressed in MDA-MB-231 cells using an expression construct (pERβ), and EGFR protein and mRNA expression was evaluated by immunoblotting and qPCR, respectively. (D) MDA-MB-231, MDA-MB-468 and MCF7 cells were treated with the ERβ antagonist PHTPP (10 μM) and EGFR protein expression was analyzed by immunoblotting. (E) ERα was expressed in MDA-MB-231 cells using an expression construct (pERα) and EGFR protein was assessed by immunoblotting. (F) Luciferase reporter constructs containing the wild-type 2.2 kb EGFR promoter and the mutant (mutated ERE) were transfected into MDA-MB-231 cells and luciferase activity was measured 24 h post-transfection. Data represent the mean of three independent experiments. The empty vector pcDNA3 was used as control. p value (*) < 0.05.
The EGFR promoter contains an imperfect estrogen response element (ERE) at −1.949 kb (AATCAACGTGAAT) suggestive of the possibility that ERβ may repress EGFR transcription. This imperfect ERE was identified based on its homology with the imperfect ERE present in the human vascular endothelial growth factor (VEGF) promoter which is 5'-AATCAGACTGACT-3' (19). The putative ERE present in the EGFR promoter differs from that in the VEGF promoter by a single nucleotide `A' as shown in italics, whereas the imperfect ERE present in the VEGF promoter differs from the perfect ERE (GGTCAnnnTGACC) (20) by three nucleotides (underlined). This observation suggested that the core ERE sequence TCAnnnTGA is critical for the sequence to be functional. To assess the contribution of this sequence to EGFR regulation, we generated reporter constructs containing the 2.2 kb wild type EGFR promoter or this construct containing a mutation in the core sequence (TCAACGTGA to CATACGTGA). The data obtained with these constructs revealed that mutation of the imperfect ERE increased luciferase activity by ~2-fold suggesting that this sequence has a repressive role in EGFR transcription (Fig. 3F).
IMP3 increases migration and invasion of breast cancer cells
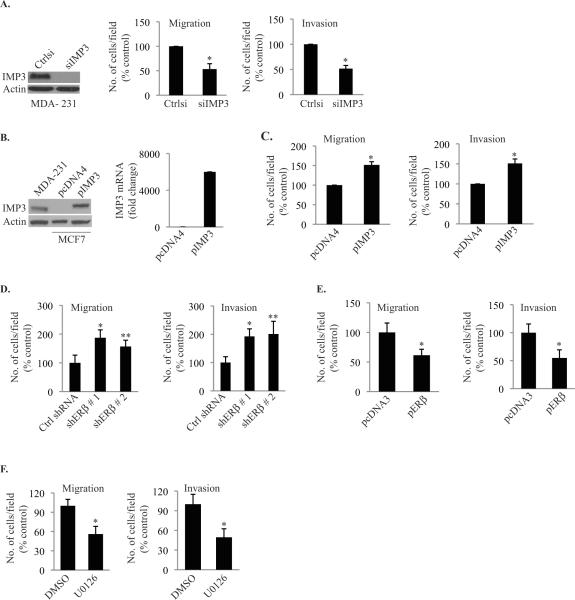
Given that triple negative breast cancers are more aggressive, possess high metastatic potential and over-express IMP3 (10, 13), it is reasonable to hypothesize that IMP3 contributes to their aggressive behavior. To address this hypothesis, we depleted IMP3 expression in MDA-MB-231 cells using siRNA and assessed their ability to migrate and invade using transwell assays. As shown in Fig. 4A, depletion of IMP3 reduced migration and invasion significantly. Moreover, expression of IMP3 in MCF7 cells, a non-triple negative and non-invasive cell line increased their migration and invasion (Fig. 4B & 4C). A functional link between ERβ and IMP3 is supported by our finding that ERβ expression represses migration and invasion as evidenced by modulation of its expression in MDA-MB-231 cells (Fig. 4D & 4E). Moreover, a functional link between EGFR signaling and IMP3 is supported by the observation that U0126, a MEK inhibitor, reduced migration and invasion significantly (Fig. 4F).
Figure 4. IMP3 enhances the migration and invasion of breast cancer cells.
(A) IMP3 was transiently depleted in MDA-MB-231 cells using siRNA (20 nM). At 72 hrs post-transfection, IMP3 protein was evaluated by immunoblotting, and migration and invasion assays were performed for another 4 h. (B) & (C) IMP3 was expressed transiently in MCF7 cells by transfecting with an expression construct (pIMP3) and IMP3 protein and mRNA were determined 48 h post-transfection by immunoblotting and qPCR, respectively. The same IMP3-expressing cells were subjected to migration and invasion assays for another 24 hrs. The empty vector pcDNA4/His/Max was used as control. (D) & (E) Migration and invasion assays were carried out with ERβ-depleted cells, as well as cells over-expressing ERβ. (F) MDA-MB-231 cells were treated with the MEK 1/2 inhibitor U0126 and assayed for migration and invasion. Data are representative of three independent experiments. p value (*) < 0.05.
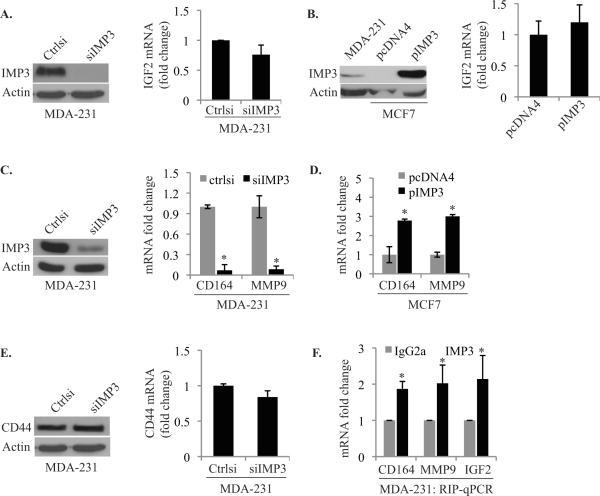
The fact that IMP3 is an mRNA binding protein suggests that it contributes to migration and invasion by regulating specific mRNAs. The obvious target for IMP3 is IGF-II but depletion of IMP3 in MDA-MB-231 cells or expression in MCF-7 cells had no effect on IGF-II mRNA expression (Fig. 5A & 5B). Several putative mRNA targets of IMP3 were recently identified based upon PAR-CLIP (Photoactivatable-Ribonucleoside-Enhanced-Crosslinking and Immunoprecipitation) (21) that could contribute to cell motility and invasion. These targets include CD164 (endolyn), RhoA, CCNG1 and CCND2, which are known to play a role in carcinogenesis (22–25). We assayed these candidate mRNAs by qPCR in IMP3-depleted MDA-MB- 231 cells and in MCF7 cells engineered to express IMP3. Although no significant change in RhoA, CCNG1 and CCND2 mRNA expression was observed (data not shown), CD164 mRNA expression was significantly decreased in IMP3-depleted MDA-MB-231 cells and it was increased in IMP3-expressing MCF7 cells (Fig. 5C & 5D). We also assayed the hyaluronan receptor CD44 expression because it is known to contribute to tumor progression and there is evidence that it can be regulated by IMP3 in HeLa cells (26). Our data, however, did not support IMP3- mediated regulation of CD44 mRNA expression in breast carcinoma cells (Fig. 5E). CD44 functions as docking site for the matrix metalloproteinase MMP9, which contributes to invasion (27). Although MMP9 had not identified as an IMP3 target, however, we observed that MMP9 mRNA expression is regulated by IMP3 (Fig. 5C & 5D). We performed Ribo-Immunoprecipitation-qPCR (RIP-qPCR) analysis to establish that IMP3 indeed binds with IGF-II, MMP9 and CD164 mRNA (Fig. 5F).
Figure 5. IMP3 binds and stabilizes CD164 and MMP9 mRNA.
(A) & (B) IGF-II mRNA expression was quantified using qPCR in IMP3-depleted MDA-MB-231 cells and in MCF7 cells engineered to express IMP3. The blot in (A) shows IMP3 depletion in MDA-MB-231 cells and in (B) IMP3 expression in MCF-7 cells. (C) & (D) CD164 and MMP9 mRNA expression was quantified using qPCR in IMP3-depleted MDA-MB-231 cells and MCF-7 cells expressing IMP3. (E) CD44 protein and mRNA expression was assessed in IMP3-depleted MDA-MB-231 cells. (F) IMP3-associated RNAs in cytoplasmic extracts of MDA-MB-231 cells were isolated using an IMP3 antibody (25 μg). Non-immune rabbit IgG was used as negative control. Expression of CD164, MMP9 and IGF-II mRNAs was quantified using qPCR. Data are representative of three independent experiments. p value (*) < 0.05.
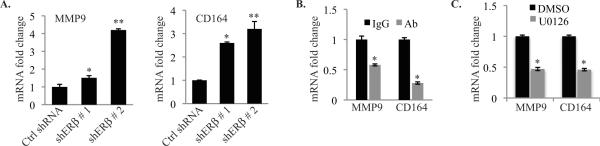
Based on our findings, IMP3 mRNA targets that we identified should be regulated by ERβ and EGFR. Indeed, depletion of ERβ increased the expression of MMP9 and CD164 mRNAs significantly in MDA-MB-231 cells (Fig. 6A). In contrast, Ab-mediated inhibition of the EGFR decreased the expression of these mRNAs (Fig. 6B). We also observed a significant reduction in the expression of these mRNAs in cells treated with MEK1/2 inhibitor U0126 (Fig. 6C).
Figure 6. CD164 and MMP9 mRNAs are suppressed by ERβ and increased by EGFR signaling.
(A–C) Expression of CD164 and MMP9 mRNAs was quantified by qPCR in MDAMB-231 cells in which ERβ had been depleted or over-expressed, or had been treated with either an EGFR antibody or the MEK 1/2 inhibitor u0126. Each experiment was repeated at least three times. p value (*) < 0.05.
Discussion
The data obtained in this study provide insight into the mechanisms that regulate the expression of IMP3 in breast carcinoma cells and that are associated with the function of this mRNA binding protein in migration and invasion. Our data revealed that IMP3 expression is repressed by ERβ but not ERα, and that it is stimulated by EGFR signaling. More specifically, we demonstrate that ERβ represses transcription of the EGFR and that EGFR signaling regulates IMP3 expression directly. An important and novel implication of this study is that IMP3 is an effector of EGFR-driven migration and invasion. Our findings have important implications for the biology and therapy of TNBC, which is characterized by elevated expression of EGFR and IMP3 (10, 28), and reduced expression of ERβ (14).
Our results highlight a role for ERβ in regulating the functions of breast carcinoma cells that is distinct from ERα. It is surprising that a role for ERα in regulating IMP3 expression was not revealed by our data because loss of ERα is a defining characteristic of TNBC (29). Establishing a distinct function for ERβ is significant because much less is known about ERβ than ERα in breast cancer biology. In addition to its ability to repress IMP3, ERβ has been shown to inhibit breast cancer cell proliferation and invasion by suppressing cell cycle genes including c-Myc, cyclin D1 and cyclin A (30). The report that ERβ expression correlates inversely with tumor grade (14) supports our conclusion that ERβ represses IMP3 because TNBC is usually high grade. Our data also indicate that there is ligand specificity in the regulation of IMP3 expression. Specifically, we demonstrated that 3βA-diol functions as an ERβ-specific ligand that represses IMP3 expression. Estrogen, in contrast, actually increased IMP3 expression in TNBC cells. Given that these cells are ERα-negative, we conclude that this effect of estrogen is non-genomic and it may be explained by its effect on EGFR signaling (17). This conclusion is supported by our observation that estrogen increased EGFR phosphorylation in ERα-negative cells independently of ERβ function.
An unexpected and novel finding in this study is that ERβ represses EGFR expression and signaling, possibly through its ability to repress EGFR transcription. Our identification of an imperfect ERE sequence in the EGFR promoter and demonstration that this sequence represses EGFR transcription supports this hypothesis. Moreover, there is precedence in the literature that ERβ can repress transcription by recruiting co-repressors such as NCoR (31). There is also indirect evidence that expression of ERβ in malignant pleural mesothelioma (MME) cells impedes EGFR signaling (32). An important issue that arises from these observations is the extent to which diminished ERβ expression in TNBC contributes to the elevated expression of EGFR that is characteristic of this breast cancer sub-type. There is evidence that EGFR gene amplification is more common in TNBCs than non-TNBC (33), but the possibility that a reduction in ERβ expression functions in concert with gene amplification should be considered.
The relationship between EGFR and IMP3 that we uncovered has important implications for the biology and therapy of breast cancer. EGFR signaling has profound effects on the proliferation, survival and invasion of tumor cells (15). Enhanced expression of EGFR in primary breast tumors predicts poor prognosis and renders them more aggressive (34–37). In the context of our findings, EGFR-driven migration and invasion contribute to tumor progression and metastasis, and the mechanisms involved have been the focus of numerous studies (38–40). We conclude from our findings that IMP3 can be an effector of EGFR-driven migration and invasion, and suggest that this contribution of IMP3 needs to be integrated with other mechanisms of EGFR-mediated migration and invasion. From a clinical perspective, the EGFR is considered to be a viable target for therapy of TNBC. Although small molecule EGFR inhibitors including gefitinib (Iressa) and the monoclonal antibody cetuximab are in clinical trials for this aggressive sub-type of breast cancer (41), none of these drugs has shown efficacy to date. We postulate based on our data that small molecule inhibitors of IMP3 would augment anti-EGFR therapy for the treatment of TNBC.
The overarching issue raised by our data is how IMP3 contributes to the aggressive behavior of breast cancers. In fact, this issue is important for several cancers in which IMP3 expression has been correlated with progression and poor outcome (4, 7, 10). A role for IMP3 in invasion was foreshadowed by the finding that IMPs (specifically IMP1 and IMP3) induce invadopodia formation by regulating the expression of CD44 in HeLa cells (26). Although our data did not reveal that CD44 expression is regulated by IMP3 in breast cancer, the induction of invadopodia formation by IMPs is consistent with our data. Recent studies have implicated IMP3 in the migration of HeLa cells (Lu et al, 2011) and in the aggressive behavior of glioblastoma cells including invasion (Suvasini 2011). In addition to a role in migration and invasion, IMP3 has been implicated in proliferation, anchorage-dependent cell growth and survival (42, 43). However, depletion of IMP3 expression in breast carcinoma cells did not affect cell cycle and anchorage-dependent growth (data not shown).
Given that IMP3 is an RNA binding protein, the most feasible hypothesis to account for its role in migration and invasion is that it affects the expression or localization of specific mRNAs that are critical for invasion and progression. Our finding that IMP3 binds and stabilizes CD164 and MMP9 mRNAs provides potential insight into this hypothesis. CD164, a cell surface receptor for sialomucin, has been implicated in prostate cancer metastasis (22) and MMP9, a type IV collagenase, contributes to carcinogenesis by several mechanisms including degradation of the basement membrane (44) and promoting the release of growth factors from the extracellular matrix (45). Moreover, MMP9 expression is correlated with poor prognosis in node negative breast cancer (46). Another relevant observation in our study is that IMP3 binds IGF-II and CD44 mRNAs but it does not alter their expression. This finding is distinct from other reports that have shown that IMP3 can activate the translation of IGF-II in leukemia cells (47) and glioblastoma cells (Suvasini et al., 2011), and regulate the expression of CD44 in Hela cells (26). The possibility that IMP3 regulates the localization or other aspects of these mRNAs merits investigation because both IGF-II and CD44 are important in the biology of breast cancer (48, 49).
In summary, our data provide the first insight into mechanisms that regulate the expression of an mRNA binding protein that is associated with TNBC and that is prognostic for progression and outcome in other cancers. These regulatory mechanisms are intimately linked to the biology of TNBC and they also reveal a novel mechanism of EGFR-driven migration and invasion. Our data also suggest that targeted IMP3 therapy could augment growth factor receptor (EGFR) inhibition in the treatment of TNBC.
Materials and Methods
Cells, reagents and treatments
The breast cancer cell lines MDA-MB-231, MDA-MB-468 (mentioned as MDA-231 and MDA-468 in the figures) and MCF7 were obtained from American Type Culture Collection (ATCC) and maintained in DMEM (low glucose) containing 10% fetal bovine serum, 1% streptomycin and penicillin at 37°C in an incubator supplied with 5% CO2. IMP3 specific siRNA was purchased from Santa Cruz Biotechnology, Inc. A non-targeting pool was used as negative control. Dharmafect-4 was used for siRNA delivery and was purchased from Thermo Scientific. Lipofectamine-2000 was used for regular DNA transfection and purchased from Invitrogen. The control plasmid vector pcDNA3 was purchased from Invitrogen and pcDNA4/His/Max was a gift from Dr. Michael Green, UMass Medical School. The lentiviruses (PLKO.1) containing ERβ shRNAs or shGFP (Open Biosystems, clone ID: TRCN0000003326, TRCN0000003327 and RHS4459) were generated in HEK293T cells and infected in MDA-MB-231 cells following standard protocols described elsewhere (50). The MEK1/2 inhibitors U0126 and PD98059 were purchased from LC Laboratories. ICI was purchased from Sigma-Aldrich. The IMP3 antibody was obtained from DAKO, Germany. ERα, EGFR and pEGFR (Tyr-1263) Abs were purchased from Santa Cruz Biotechnology Inc. pMAPK (p42/44) and pAkt (ser473) Abs were obtained from Cell Signaling Technology. The ERβ antibody was purchased from Gen Tex Inc. The IGF-II and CD44 antibodies were purchased from Abcam. Estrogen (E2), PHTPP and 3βA-diol were purchased from Tocris Biosciences and dissolved in DMSO. For 3βA-diol treatment, MDA-MB-231 cells were cultured in DMEM (low glucose) supplemented with 5 % charcoal stripped fetal bovine serum. For treatment with all other reagents, cells were cultured in normal medium (mentioned above) and serum-starved for 24 h prior to treatment.
Immunoblotting
Whole cell extracts were prepared using RIPA buffer containing EDTA and EGTA (Boston Bioproducts). A protease and phosphatase inhibitor cocktail was added separately (Roche Applied Biosciences). Extracts (40–50 μg protein) were blotted with the appropriate primary Abs and then incubated with mouse/ or rabbit IgG horse radish peroxidase conjugated secondary antibody. ECL kit (Pierce Biotechnology) was used to develop the blots.
RNA isolation and real time PCR analysis
Total RNA was isolated from cultured cells using Trizol reagent (Invitrogen) following the manufacturer's protocol. cDNA was synthesized using superscript-II reverse transcriptase (Invitrogen). mRNAs were quantified by real time PCR analysis (ABI Prism, Applied Biosystems) using Power Syber Green PCR master mix (Applied Biosystems). Quantification was performed using ΔΔCt method and GAPDH was used as reference gene. The following primer pairs were used for real time PCR analysis:
IMP3-forward: 5'-CCGCAGTTTGAGCAATCAGAA-3'
IMP3-reverse: 5'-CGAGAAAGCTGCTTGATGTGC-3'
EGFR-forward: 5'-GCCCCCACTGCGTCAAGACC-3'
EGFR-reverse: 5'-ACCTGGCCCAGTGCATCCGT-3'
ERβ-forward: 5'-AAGGTTAGTGGGAACCGTTG-3'
ERβ-reverse: 5'-ACATCCTTCACACGACCAGA-3'
MMP9-forward: 5'-TTTGACAGCGACAAGAAGTGG-3'
MMP9-reverse: 5'-AGGGCGAGGACCATAGAGG-3'
CD164-forward: 5'-GAGTGCTGTAGGATTAATTGGAAAAT-3'
CD164-reverse: 5'-GGGAGGAATGGAATTCTGC-3'
IGFII-forward: 5'-CCGAAACAGGCTACTCTCCT-3'
IGFII-reverse: 5'-AGGGTGTTTAAAGCCAATCG-3'
CD44-forward: 5'-GCCCTTCCATAGCCTAATCC-3'
CD44-reverse: 5'-CTTTGGTGTCTCCCAGAAGC-3'
Migration and invasion assays
Assays were performed using transwell chambers (8 μm polycarbonate membrane, Costar, Corning Inc) that had been coated with either collagen I or Matrigel (BD Biosciences) for migration and invasion, respectively, as described previously (51). Assays were quantified by counting the number of stained nuclei in 5 independent fields in each transwell.
RIP-qPCR
Identification of endogenous mRNA targets of IMP3 was performed by RIP (riboimmunoprecipitation) assay as described previously with minor modifications (47). Briefly MDA-MB-231 cells (~2 × 107) were harvested and extracted for 15 min on ice in 250 μl of ice-cold lysis buffer (100 mm KCl, 5 mm MgCl2, 10 mm HEPES (pH 7.0), 0.5% Nonidet P-40, 10 μm dithiothreitol) supplemented with RNase and protease inhibitors (1 ml of lysis buffer contains 5.25 μl of 40 units/ml RNase OUT (Invitrogen), 2 μl of 0.2% vanadyl ribonucleoside complexes (New England Biolabs), 50 μl of complete protease inhibitor mixture (Roche Applied Science). Extracts were cleared by centrifugation for 15 min at 13,000 rpm and supernatant was transferred to a fresh 1.5-ml tube. To pre-clear the cytoplasmic extracts, 25 μg of non-immune rabbit IgG (Sigma) was added to the supernatant and kept on ice for 45 min, then incubated with 50 μl of a 50% (v/v) suspension of protein G-Sepharose beads (Biovision, SF, California, USA) for 3 h at 4 °C with rotation. This was centrifuged at 13,000 rpm and the supernatant was recovered (pre-cleared lysate). For immunoprecipitation, the pre-cleared extract was incubated with 100 μl of a 50% suspension of protein G-Sepharose beads (Sigma) pre-coated with the same amount of either non-immune rabbit IgG (Sigma) or anti-human IMP3 antibody (25 μg) in 800 μl of NT-2 buffer (150 mm NaCl, 1 mm MgCl2, 50 mm Tris-HCl (pH 7.4), 0.05% Nonidet P-40) containing RNase inhibitor and protease inhibitors for over-night at 4 °C with rotation. Beads were washed 10 times using ice-cold NT-2 buffer, digested with 20 units of RNase-free DNase I (Promega) in 100 of μl of NT-2 buffer for 20 min at 30 °C, washed with NT-2 buffer, and further digested with 0.5 mg/ml protease K (Ambion) in 100 μl of NT-2 buffer containing 0.1% SDS at 55 °C for 30 min. RNA was extracted with Trizol (Invitrogen). Glycogen (Roche Applied Science) was added to facilitate precipitation of RNA. Real time PCR was performed with equal amount of RNA to detect IGF-II, MMP9 and CD164 mRNAs using previously described primers.
IMP3 and EGFR promoter analysis
The human IMP3 promoter (2.872 kb) was PCR amplified from human genomic DNA. The PCR amplified fragments were confirmed by restriction mapping and cloned at the NheI-Hind III sites into the pGL3 basic vector (Promega). Similarly, the EGFR promoter (2.2 kb) was cloned and ERE (AATCAACGTGAAT) was mutated to (AACATACGTGAAT) using site-directed mutagenesis (QuikChange XL Site-Directed Mutagenesis Kit, Agilent Technologies). Primer pairs used:
- IMP3 promoter:
- Forward: 5'-TATGATGCTAGCACTTGAAGTGCTAGTGCAAGACAACT
- Reverse: 5'-CAGTAGAAGCTTTCCACCAGTCTTCCTAAGTCTTAGGA
- EGFR promoter:
- Forward: 5'-TAGAAGACGCGTCAGGTACTAGCCAAGGACTACA
- Reverse: 5'-AATCGTCTCGAGGATCAATACTGGACGGAGTCAG
Luciferase reporter assays
MDA-MB-231 cells were transfected with the desired luciferase reporter constructs (express firefly luciferase) along with another construct expresses renilla luciferase to normalize the transfection efficiency. Relative Light Units (RLU) was calculated as the ratio of firefly luciferase to renilla luciferase activity (normalized luciferase activity). The protocol used for transfection and measurement of luciferase activity has been described previously (52). Luciferase activity was measured in Beckman Coultier, DTX 880 luminomiter.
Construction of IMP3, ERα and ERβ expression vectors
Human IMP3 (NM_006547) cDNA was PCR amplified using total RNA isolated from MDA-MB 231 cells and cloned into pcDNA4/His/Max vector at EcoRI/XhoI sites under CMV promoter. PCR amplified cDNA was confirmed by restriction mapping and the cloned expression vector was confirmed by sequence analysis. The ER-α and ER-β expression constructs were obtained from Shuk-Mei Ho (University of Cincinnati).
IMP3 Primers:
Forward: 5'-GCCATAGAATTCATGAACAAACTGTATATCGGAAACC-3'
Reverse: 5'-TAAGTACTCGAGTTACTTCCGTCTTGACTGAGGT-3'
Statistical analyses
The data are shown as the ± S. E. p values (*) were determined using student's t test and p ≤ 0.05 was considered as significant.
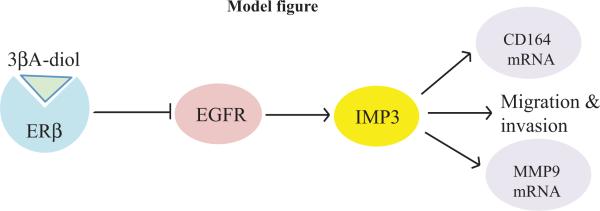
Figure 7. Proposed model for how ERβ regulates IMP3 in triple negative breast cancer cells.
The interaction of ERβ with its ligand 3βA-diol suppresses EGFR expression. EGFR signaling induces IMP3 expression. IMP3 facilitates the migration and invasion of triple negative breast carcinoma cells by a mechanism that may involve its ability to specific mRNAs such as CD164 and MMP9.
Acknowledgements
NIH Grants CA80789 and 89209 supported this work. We thank Dr. Xiofang Yang for her input into this project, and Drs. Paul Mak and Hira Lal Goel for their valuable suggestions and technical assistance. We also thank Bryan Pursell for his technical support.
Support: This work was supported by NIH Grants CA80789 and 89209.
Footnotes
Conflict of Interest The authors declare no conflict of interest.
References
- 1.Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Molecular and Cellular Biology. 1999;19(2):1262–70. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mueller-Pillasch F, Pohl B, Wilda M, Lacher U, Beil M, Wallrapp C, et al. Expression of the highly conserved RNA binding protein KOC in embryogenesis. Mechanisms of Development. 1999;88(1):95–9. doi: 10.1016/s0925-4773(99)00160-4. [DOI] [PubMed] [Google Scholar]
- 3.Mueller-Pillasch F, Lacher U, Wallrapp C, Micha A, Zimmerhackl F, Hameister H, et al. Cloning of a gene highly overexpressed in cancer coding for a novel KH-domain containing protein. Oncogene. 1997;14(22):2729–33. doi: 10.1038/sj.onc.1201110. [DOI] [PubMed] [Google Scholar]
- 4.Jiang Z, Chu PG, Woda BA, Rock KL, Liu Q, Hsieh CC, et al. Analysis of RNA-binding protein IMP3 to predict metastasis and prognosis of renal-cell carcinoma: a retrospective study. Lancet Oncology. 2006;7(7):556–64. doi: 10.1016/S1470-2045(06)70732-X. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Rock KL, Woda BA, Jiang Z, Fraire AE, Dresser K. IMP3 is a novel biomarker for adenocarcinoma in situ of the uterine cervix: an immunohistochemical study in comparison with p16(INK4a) expression. Modern Pathology. 2007;20(2):242–7. doi: 10.1038/modpathol.3800735. [DOI] [PubMed] [Google Scholar]
- 6.Lu D, Yang X, Jiang NY, Woda BA, Liu Q, Dresser K, et al. IMP3, a New Biomarker to Predict Progression of Cervical Intraepithelial Neoplasia Into Invasive Cancer. The American Journal of Surgical Pathology. 2011;35(11):1638–45. doi: 10.1097/PAS.0b013e31823272d4. [DOI] [PubMed] [Google Scholar]
- 7.Simon R, Bourne PA, Yang Q, Spaulding BO, di Sant'Agnese PA, Wang HL, et al. Extrapulmonary small cell carcinomas express K homology domain containing protein overexpressed in cancer, but carcinoid tumors do not. Human Pathology. 2007;38(8):1178–83. doi: 10.1016/j.humpath.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Yantiss RK, Woda BA, Fanger GR, Kalos M, Whalen GF, Tada H, et al. KOC (K homology domain containing protein overexpressed in cancer): a novel molecular marker that distinguishes between benign and malignant lesions of the pancreas. The American Journal of Surgical Pathology. 2005;29(2):188–95. doi: 10.1097/01.pas.0000149688.98333.54. [DOI] [PubMed] [Google Scholar]
- 9.Zheng W, Yi X, Fadare O, Liang SX, Martel M, Schwartz PE, et al. The oncofetal protein IMP3: a novel biomarker for endometrial serous carcinoma. The American Journal of Surgical Pathology. 2008;32(2):304–15. doi: 10.1097/PAS.0b013e3181483ff8. [DOI] [PubMed] [Google Scholar]
- 10.Walter O, Prasad M, Lu S, Quinlan RM, Edmiston KL, Khan A. IMP3 is a novel biomarker for triple negative invasive mammary carcinoma associated with a more aggressive phenotype. Human Pathology. 2009;40(11):1528–33. doi: 10.1016/j.humpath.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Cha J, Kim J, Kim KY, Kim HJ, Nam W, et al. Insulin-like growth factor II mRNA-binding protein 3: a novel prognostic biomarker for oral squamous cell carcinoma. Head & Neck. 2011;33(3):368–74. doi: 10.1002/hed.21457. [DOI] [PubMed] [Google Scholar]
- 12.Li D, Yan D, Tang H, Zhou C, Fan J, Li S, et al. IMP3 is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Annals of Surgical Oncology. 2009;16(12):3499–506. doi: 10.1245/s10434-009-0648-5. [DOI] [PubMed] [Google Scholar]
- 13.Irvin WJ, Jr., Carey LA. What is triple-negative breast cancer? Eur J Cancer. 2008;44(18):2799–805. doi: 10.1016/j.ejca.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 14.Marotti JD, Collins LC, Hu R, Tamimi RM. Estrogen receptor-beta expression in invasive breast cancer in relation to molecular phenotype: results from the Nurses' Health Study. Modern Pathology. 2010;23(2):197–204. doi: 10.1038/modpathol.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foley J, Nickerson NK, Nam S, Allen KT, Gilmore JL, Nephew KP, et al. EGFR signaling in breast cancer: bad to the bone. Seminars in Cell & Developmental Biology. 2010;21(9):951–60. doi: 10.1016/j.semcdb.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerini V, Sau D, Scaccianoce E, Rusmini P, Ciana P, Maggi A, et al. The androgen derivative 5alpha-androstane-3beta,17beta-diol inhibits prostate cancer cell migration through activation of the estrogen receptor beta subtype. Cancer Research. 2005;65(12):5445–53. doi: 10.1158/0008-5472.CAN-04-1941. [DOI] [PubMed] [Google Scholar]
- 17.Zhang XT, Kang LG, Ding L, Vranic S, Gatalica Z, Wang ZY. A positive feedback loop of ER-alpha36/EGFR promotes malignant growth of ER-negative breast cancer cells. Oncogene. 2011;30(7):770–80. doi: 10.1038/onc.2010.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signalling. Experimental Cell Research. 2003;284(1):31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 19.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Research. 2001;29(14):2905–19. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein-Hitpass L, Schorpp M, Wagner U, Ryffel GU. An estrogen-responsive element derived from the 5' flanking region of the Xenopus vitellogenin A2 gene functions in transfected human cells. Cell. 1986;46(7):1053–61. doi: 10.1016/0092-8674(86)90705-1. [DOI] [PubMed] [Google Scholar]
- 21.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Havens AM, Jung Y, Sun YX, Wang J, Shah RB, Buhring HJ, et al. The role of sialomucin CD164 (MGC-24v or endolyn) in prostate cancer metastasis. BMC Cancer. 2006;6:195. doi: 10.1186/1471-2407-6-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellenbroek SI, Collard JG. Rho GTPases: functions and association with cancer. Clinical & Experimental Metastasis. 2007;24(8):657–72. doi: 10.1007/s10585-007-9119-1. [DOI] [PubMed] [Google Scholar]
- 24.Reimer CL, Borras AM, Kurdistani SK, Garreau JR, Chung M, Aaronson SA, et al. Altered regulation of cyclin G in human breast cancer and its specific localization at replication foci in response to DNA damage in p53+/+ cells. The Journal of Biological Chemistry. 1999;274(16):11022–9. doi: 10.1074/jbc.274.16.11022. [DOI] [PubMed] [Google Scholar]
- 25.Sicinski P, Donaher JL, Geng Y, Parker SB, Gardner H, Park MY, et al. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature. 1996;384(6608):470–4. doi: 10.1038/384470a0. [DOI] [PubMed] [Google Scholar]
- 26.Vikesaa J, Hansen TV, Jonson L, Borup R, Wewer UM, Christiansen J, et al. RNA-binding IMPs promote cell adhesion and invadopodia formation. The EMBO Journal. 2006;25(7):1456–68. doi: 10.1038/sj.emboj.7601039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Q, Stamenkovic I. Localization of matrix metalloproteinase 9 to the cell surface provides a mechanism for CD44-mediated tumor invasion. Genes & Development. 1999;13(1):35–48. doi: 10.1101/gad.13.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van de Rijn M, Perou CM, Tibshirani R, Haas P, Kallioniemi O, Kononen J, et al. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. The American Journal of Pathology. 2002;161(6):1991–6. doi: 10.1016/S0002-9440(10)64476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korsching E, Jeffrey SS, Meinerz W, Decker T, Boecker W, Buerger H. Basal carcinoma of the breast revisited: an old entity with new interpretations. Journal of Clinical Pathology. 2008;61(5):553–60. doi: 10.1136/jcp.2008.055475. [DOI] [PubMed] [Google Scholar]
- 30.Paruthiyil S, Parmar H, Kerekatte V, Cunha GR, Firestone GL, Leitman DC. Estrogen receptor beta inhibits human breast cancer cell proliferation and tumor formation by causing a G2 cell cycle arrest. Cancer Research. 2004;64(1):423–8. doi: 10.1158/0008-5472.can-03-2446. [DOI] [PubMed] [Google Scholar]
- 31.Webb P, Valentine C, Nguyen P, Price RH, Jr., Marimuthu A, West BL, et al. ERbeta Binds N-CoR in the Presence of Estrogens via an LXXLL-like Motif in the N-CoR C-terminus. Nuclear Receptor. 2003;1(1):4. doi: 10.1186/1478-1336-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinton G, Thomas W, Bellini P, Manente AG, Favoni RE, Harvey BJ, et al. Estrogen receptor beta exerts tumor repressive functions in human malignant pleural mesothelioma via EGFR inactivation and affects response to gefitinib. PloS One. 2010;5(11):e14110. doi: 10.1371/journal.pone.0014110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryden L, Jirstrom K, Haglund M, Stal O, Ferno M. Epidermal growth factor receptor and vascular endothelial growth factor receptor 2 are specific biomarkers in triple-negative breast cancer. Results from a controlled randomized trial with long-term follow-up. Breast Cancer Research and Treatment. 2010;120(2):491–8. doi: 10.1007/s10549-010-0758-6. [DOI] [PubMed] [Google Scholar]
- 34.Garcia Castro C, Ravina M, Castro V, Salido EC. Expression of epidermal growth factor receptor (proto-oncogene c-erbB-1) and estrogen receptor in human breast carcinoma. An immunocytochemical study of 70 cases. Archives of Gynecology and Obstetrics. 1993;252(4):169–77. doi: 10.1007/BF02426354. [DOI] [PubMed] [Google Scholar]
- 35.Martinazzi M, Crivelli F, Zampatti C, Martinazzi S. Epidermal growth factor receptor immunohistochemistry in different histological types of infiltrating breast carcinoma. Journal of Clinical Pathology. 1993;46(11):1009–10. doi: 10.1136/jcp.46.11.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newby JC, A'Hern RP, Leek RD, Smith IE, Harris AL, Dowsett M. Immunohistochemical assay for epidermal growth factor receptor on paraffin-embedded sections: validation against ligand-binding assay and clinical relevance in breast cancer. British journal of cancer. 1995;71(6):1237–42. doi: 10.1038/bjc.1995.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heldin CH, Westermark B. Growth factors: mechanism of action and relation to oncogenes. Cell. 1984;37(1):9–20. doi: 10.1016/0092-8674(84)90296-4. [DOI] [PubMed] [Google Scholar]
- 38.Xu L, Nilsson MB, Saintigny P, Cascone T, Herynk MH, Du Z, et al. Epidermal growth factor receptor regulates MET levels and invasiveness through hypoxia-inducible factor-1alpha in non-small cell lung cancer cells. Oncogene. 2010;29(18):2616–27. doi: 10.1038/onc.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Meter TE, Broaddus WC, Rooprai HK, Pilkington GJ, Fillmore HL. Induction of membrane-type-1 matrix metalloproteinase by epidermal growth factor-mediated signaling in gliomas. Neuro-oncology. 2004;6(3):188–99. doi: 10.1215/S1152851703000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z, Xu X, Bai L, Chen W, Lin Y. Epidermal growth factor receptor-mediated tissue transglutaminase overexpression couples acquired tumor necrosis factor-related apoptosis-inducing ligand resistance and migration through c-FLIP and MMP-9 proteins in lung cancer cells. The Journal of Biological Chemistry. 2011;286(24):21164–72. doi: 10.1074/jbc.M110.207571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agrawal A, Gutteridge E, Gee JM, Nicholson RI, Robertson JF. Overview of tyrosine kinase inhibitors in clinical breast cancer. Endocrine-Related Cancer. 2005;12(Suppl 1):S135–44. doi: 10.1677/erc.1.01059. [DOI] [PubMed] [Google Scholar]
- 42.Suvasini R, Shruti B, Thota B, Shinde SV, Friedmann-Morvinski D, Nawaz Z, et al. Insulin growth factor-2 binding protein 3 (IGF2BP3) is a glioblastoma-specific marker that activates phosphatidylinositol 3-kinase/mitogen-activated protein kinase (PI3K/MAPK) pathways by modulating IGF-2. The Journal of Biological Chemistry. 2011;286(29):25882–90. doi: 10.1074/jbc.M110.178012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao B, Hu Y, Brewer G. RNA-binding Protein Insulin-like Growth Factor mRNA-binding Protein 3 (IMP-3) Promotes Cell Survival via Insulin-like Growth Factor II Signaling after Ionizing Radiation. The Journal of Biological Chemistry. 2011;286(36):31145–52. doi: 10.1074/jbc.M111.263913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng ZS, Cohen AM, Guillem JG. Loss of basement membrane type IV collagen is associated with increased expression of metalloproteinases 2 and 9 (MMP-2 and MMP-9) during human colorectal tumorigenesis. Carcinogenesis. 1999;20(5):749–55. doi: 10.1093/carcin/20.5.749. [DOI] [PubMed] [Google Scholar]
- 45.Belotti D, Paganoni P, Manenti L, Garofalo A, Marchini S, Taraboletti G, et al. Matrix metalloproteinases (MMP9 and MMP2) induce the release of vascular endothelial growth factor (VEGF) by ovarian carcinoma cells: implications for ascites formation. Cancer Research. 2003;63(17):5224–9. [PubMed] [Google Scholar]
- 46.Scorilas A, Karameris A, Arnogiannaki N, Ardavanis A, Bassilopoulos P, Trangas T, et al. Overexpression of matrix-metalloproteinase-9 in human breast cancer: a potential favourable indicator in node-negative patients. British Journal of Cancer. 2001;84(11):1488–96. doi: 10.1054/bjoc.2001.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao B, Hu Y, Herrick DJ, Brewer G. The RNA-binding protein IMP-3 is a translational activator of insulin-like growth factor II leader-3 mRNA during proliferation of human K562 leukemia cells. The Journal of Biological Chemistry. 2005;280(18):18517–24. doi: 10.1074/jbc.M500270200. [DOI] [PubMed] [Google Scholar]
- 48.Ellis MJ, Jenkins S, Hanfelt J, Redington ME, Taylor M, Leek R, et al. Insulin-like growth factors in human breast cancer. Breast Cancer Research and Treatment. 1998;52(1–3):175–84. doi: 10.1023/a:1006127621512. [DOI] [PubMed] [Google Scholar]
- 49.Bourguignon LY, Wong G, Earle C, Krueger K, Spevak CC. Hyaluronan-CD44 interaction promotes c-Src-mediated twist signaling, microRNA-10b expression, and RhoA/RhoC up-regulation, leading to Rho-kinase-associated cytoskeleton activation and breast tumor cell invasion. The Journal of Biological Chemistry. 2010;285(47):36721–35. doi: 10.1074/jbc.M110.162305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, Zhang X, Shen P, Loggie BW, Chang Y, Deuel TF. Identification, cloning, and expression of human estrogen receptor-alpha36, a novel variant of human estrogen receptor-alpha66. Biochemical and Biophysical Research Communications. 2005;336(4):1023–7. doi: 10.1016/j.bbrc.2005.08.226. [DOI] [PubMed] [Google Scholar]
- 51.Shaw LM, Rabinovitz I, Wang HH, Toker A, Mercurio AM. Activation of phosphoinositide 3-OH kinase by the alpha6beta4 integrin promotes carcinoma invasion. Cell. 1997;91(7):949–60. doi: 10.1016/s0092-8674(00)80486-9. [DOI] [PubMed] [Google Scholar]
- 52.Mak P, Leung YK, Tang WY, Harwood C, Ho SM. Apigenin suppresses cancer cell growth through ERbeta. Neoplasia. 2006;8(11):896–904. doi: 10.1593/neo.06538. [DOI] [PMC free article] [PubMed] [Google Scholar]