Abstract
Most disease association mapping algorithms are based on hypothesis testing procedures that test one variant at a time. Those methods lose power when the disease mutations are jointly tagged by multiple variants, or when gene-gene interaction exist. Nearby variants are also correlated, for which procedures ignoring the dependence between variants will inevitably produce redundant results. With a large number of variants genotyped in current genome-wide disease association studies, simultaneous multi-variant association mapping algorithms are strongly desired. We present a novel Bayesian method for automatic detection of multi-variant joint association in genome-wide case-control studies. Our method has improved power and specificity over existing tools. We fit a joint probabilistic model to the entire data and identify disease variants simultaneously. The method dynamically accounts for the strong linkage disequilibrium (LD) between variants. As a result, only the primary disease variants will be identified, with all secondary associations due to LD effects filtered out. Our method better pinpoints the disease variants with improved resolution. The method is also computationally efficient for genome-wide studies. When applied to a real dataset of inflammatory bowel disease (IBD) containing 401,473 variants in 4,720 individuals, our method detected all previously reported IBD loci in the same data, and recovered two missed loci. We further detected two novel inter-chromosome interactions. The first is between STAT3 and PARD6G, and the second is between DLG5 and an intergenic region at 5p14. We further validated the two interactions in an independent study.
Keywords: disease association mapping, Bayesian graph, linkage disequilibrium, Markov chain Monte Carlo
1 Introduction
Genome-wide association study (GWAS) for complex diseases has become routine in recent years (WTCCC, 2007). The goal is to identify potential locations in the genome containing mutations that may affect the risk of complex diseases. Single nucleotide polymorphism (SNP) is a typical marker used in GWAS. A common approach in case control setting is to compare the SNP genotype distribution between the affected and the unaffected individuals. If a disease mutation occurs at a locus in the genome, which is typically unobserved, its nearby SNPs will demonstrate association with the disease due to linkage disequilibrium (LD). LD can be understood as dependence in statistical sense, and LD diminishes over distance. It is thus expected that SNPs genotyped in a GWAS can capture the information of unobserved disease mutations nearby. The genotyped SNPs are called tagging SNPs. Intuitively, with more SNPs genotyped in a GWAS, the more likely that a disease mutation is tagged or observed, and thus increases the power of GWAS. The problem is however complicated by the increasing number of SNPs, such as the additional computation burden and the issues created by LD among densely genotyped SNPs.
Many current association mapping approaches only focus on testing single-SNP association with the disease. Despite their simplicity, single-SNP methods inevitably lose power when interaction association exists between SNPs (Moore et al 2002; Marchini et al 2005; Zhang and Liu 2007). Single-SNP methods also perform poorly when the disease mutations are jointly captured by multiple tagging SNPs (Zhang et al 2002; Kuno et al 2004; de Bakker et al 2005), which is common due to the nature of the tagging SNP selection criteria. A few recent studies (Cirulli and Goldstein 2010) further suggested that the risk of complex traits may be attributable to rare mutations, in which case single-SNP methods will be powerless regardless of how many tagging SNPs are genotyped. Due to these reasons, advanced statistical methods for automatic detection of multi-SNP association mapping are strongly desired (Cordell 2009). Testing multi-SNP associations is an interesting and important problem, yet the task is very difficult because there are too many possible SNP combinations to be explored in the genome-wide scale. A few computationally efficient multi-SNP mapping algorithms have emerged recently, demonstrating that whole-genome multi-SNP association mapping is computationally feasible, and at the same time statistically powerful (Marchini et al 2005; Zhang and Liu 2007; Schwartz et al 2008; Wu et al 2009; Wan et al 2010; Zhang et al 2011; Liu et al 2011).
Even if one is only interested in single-SNP association, most existing methods have problems when many SNPs in strong LD are genotyped in a region. In particular, LD can create a large number of secondary disease associations around a true disease mutation. Most methods use hypothesis testing procedures (Marchini et al 2005; Wan et al 2010; Liu et al 2011) to test each SNP or SNP set separately in a step-wise manner, ignoring the LD effects. As a result, they are likely to report a large number of redundant association, with biased effect estimates, due to missing important variants in their tests. Distinct from the hypothesis testing procedures, there are also joint disease mapping approaches (Zhang and Liu 2007; Schwartz et al 2008; Wu et al 2009; Zhang et al 2011) that fit a single model to all SNPs simultaneously, and detect SNP association conditioning on all other SNPs. Although joint mapping approaches are potentially more powerful and robust than hypothesis testing approaches, many existing algorithms are computationally demanding for large-scale studies, and most methods cannot sufficiently account for the complex LD effects among dense SNPs. We show in our simulation study that insufficient modeling of LD could substantially increase the analysis complexity and compromise the disease mapping results.
In this paper, we introduce a novel Bayesian graphical method, called BEAM3, for large-scale association mapping. BEAM3 simultaneously detects single-SNP, multi-local-SNP, and long-distance SNP-SNP interaction associations. BEAM3 is particularly advantageous for analyzing a large number of SNPs with possibly strong LD. There are two major improvements of BEAM3 compared to existing methods and our previous BEAM models (Zhang and Liu 2007; Zhang et al 2011). First, multi-SNP associations and high-order interactions have exponentially growing model complexity, for which saturated models are powerless. The new method detects flexible interaction structures using graphs, which substantially reduces the multi-SNP model complexity and hence improves the power. Second, BEAM3 achieves computational efficiency for GWAS by constructing only the disease association graphs, while avoiding inference of the unknown huge and complicated dependence graph for genome-wide SNPs. The latter is typical in practice but is only applicable to much smaller studies. Third, the method uses a flexible graph structure to implicitly account for the unknown LD among SNPs, such that only the primary disease associations will be reported. All secondary associations due to LD effects will be filtered out. BEAM3 therefore produces cleaner results with greatly improved mapping sensitivity and specificity, which will greatly simplify the downstream analysis. Our new method is well suited in a Bayesian framework. Various disease SNPs and interactions are automatically evaluated using regularized probabilities, with minimum input requirement from the user. Biological expert information can also be easily incorporated into our model as priors.
With recent advance in high-throughput sequencing technologies (Metzker et al 2010), it becomes cost-effective to genotype millions of SNPs in GWAS. It is thus strongly desirable to develop advanced statistical methods, with unified and flexible single-SNP and multi-SNP models, to improve the power of detecting subtle disease associations and interactions. We performed large-scale simulation study to demonstrate the superior performance of BEAM3 compared to several existing methods, including penalized regression methods, machine learning methods, and our previous Bayesian partition methods. We further applied BEAM3 to a GWAS dataset of inflammatory bowel disease (IBD) from WTCCC (2007). We successfully identified all previously reported IBD loci from this data and recovered two missed loci. We also identified two local joint-tagging association at known IBD loci, and two inter-chromosome interactions. We validated the two inter-chromosome interactions in an independent study.
2 Material and Methods
2.1 Notation
Let Y denote the binary disease status of case control individuals and X denotes the SNP data to be tested for association with Y. Our approach is to model the distribution of X given Y. Since not all SNPs in X are associated with Y, the goal is to select a subset of SNPs in X that are most likely associated with Y either marginally or jointly with other SNPs. It is extremely complicated to consider all possible interaction models among all SNPs. We therefore restrict our method to only detect saturated interactions of SNPs within a selected subset of SNPs (called cliques) and the pairwise interaction between cliques. We first define our notations:
Y = (y1,···, yN)T: the binary disease status of N individuals;
X = (x1, ···, xL): L SNPs, where each xi is a vector of genotypes in N individuals;
I = (I1, ···, IL): vector of L indicators, with Ii = 1 indicating SNP xi is associated with Y, and 0 otherwise;
X1: set of SNPs with indicator Ii = 1, i.e., disease associated SNPs;
C = (c1, ···, cK): partition of SNPs in X1 into K non-overlapping cliques;
Δ = {δij}1≤i < j≤K: K choose 2 indicators, with δij = 1 indicating interaction between cliques ci and cj, and 0 otherwise.
G = (C, Δ): an undirected graph built on SNPs in X1, called disease graph.
2.2 Probability Functions Built on a Graph
We first describe the probability functions used in our graphical model. The functions are designed for case control studies with dichotomous disease outcome and categorical genotypes. Given a collection Xc of k SNPs, there are 3k possible genotype combinations (genotype is the observed value at a SNP in an individual, and there are 3 possible genotype values). Let ni, mi denote the number of genotype combination i observed in cases and controls, respectively, and let pi, qi denote their frequencies, respectively. We write
Since pi, qi are unknown, we assume a Dirichlet prior for pi, qi with hyper-parameter , and we integrate out pi, qi to obtain
| (1) |
where denote the case and control sample sizes, respectively, and .
Formula (1) models the data observed in cases and controls separately, which is appropriate if SNPs in Xc are associated with the disease Y. On the other hand, if SNPs in Xc are not associated with the disease, we should have pi = qi for all i = 1, ···, 3k, and hence
| (2) |
A joint probability function of all SNPs in X1 (the set of disease associated SNPs) can be modeled by an undirected acyclic graph G = (C, Δ) conditioning on Y as
| (3) |
where each term in (3) is defined in (1). A justification of formula (3) can be found in Online Supplementary Material.
Correspondingly, if we assume that the set of SNPs in X1 are not associated with Y, we can model X1 given a (different) graph G′ as
| (4) |
2.3 Choice of Prior Distributions
We specify the prior distribution of SNP memberships I by a product of independent Bernoulli probabilities, i.e., Pr(Ii = 1) = 1 − Pr(Ii = 0) = p and , where |I| = Σi=1,···,L Ii. By default, we choose , and a larger p will help detecting weaker associations. It is also possible to specify Pr(Ii = 1) according to some biological knowledge, e.g., functional potential of each SNP. Given I (and hence X1), we specify the prior distribution of a clique partition C of SNPs in X1 by a Pitman-Yor process (Pitman and Yor, 1997). Assume X1 = (x1, ···, xp). Let Ci = (c1, ···, cKi) denote a partition of x1, ···, xi into Ki non-verlapping cliques. Let nk denote the number of SNPs in the kth clique. The probability of assigning a new SNP xi+1 into one of the existing cliques is
and the probability of assigning xi+1 into a new clique is
Here, α > 0 and β ∈ [0, 1) are two hyper-parameters in the Pitman-Yor process, with β = 0 yielding the standard Dirichlet process. By default, we let α = 10 and β = 0.5, where larger α and β will favor more cliques. Using Pitman-Yor process in clique partition allows us to determine the number of cliques probabilistically. Given I and C, we specify the prior distribution of interaction indicators Δ between cliques by a product of independent Bernoulli probabilities, i.e., Pr(δij = 1) = 1 − Pr(δ ij) = pD and Pr(Δ|I, C) = Πi<j Pr(δij). By default, we let pD = 0.1, where a larger pD will help detecting weaker interactions.
2.4 The Joint Probability Model
We write the joint probability function of all SNPs X, disease status Y, and parameters (I, G) as
| (5) |
where Pr(Y) denotes the distribution of disease status, Pr(G|I) denotes the prior distribution of G given I, and Pr(I) denotes the prior distribution of SNP indicators. Note that our model is a retrospective, i.e., we define the distribution of SNPs conditioning on the disease information.
We next define Pr(X|Y, G, I) in the form
| (6) |
Here, PA(·) denotes a probability function of X1 under the disease association hypothesis, and its form is given by (3). P0(·) denotes a baseline probability function (to be defined later) of X1 under the null hypothesis of no association. In (6), the first line is our definition; the second and the third line are due to the independence assumption between unassociated SNPs and disease information (Y, G), under the null hypothesis. We define the null model P0(X) to be invariant with respect to G and I, and thus we can treat P0(X) as a constant in our algorithm, yielding the last line of (6).
Our parameters of interest are the disease graph G and the SNP membership I, where X1 is uniquely determined by I. Plugging (6) back into (5), we obtain
| (7) |
It is shown in (7) that we avoid explicitly modeling the complicated dependence of all SNPs, except for a few disease associated SNPs. The total number of genome-wide SNPs could be in millions, modeling the dependence of which is computationally formidable. The number of true and detectable disease SNPs, however, is often small. As a result, our method saves a substantial amount of computation by avoiding explicit modeling of SNP dependence.
The choice of the baseline function P0(·) in (6) under the null hypothesis is critical. A simple Markov chain or a LD-block model cannot fully account for the complicated dependence among SNPs. Simple models only work when the disease models under the alternative hypothesis are equally restrictive. If a null model is overly simplistic compared to the disease graph model, numerous false positive interactions will be produced merely due to its over-simplicity. We define
| (8) |
where P0(X1|G′) is defined in (4). Here, we use graphs G′ (different from G) to define the baseline model, which can capture the unknown LD among SNPs. We do not infer G′ in practice, and thus we sum over all possible G′.
2.5 Markov Chain Monte Carlo Sampling
We iteratively sample the SNP membership variable I and the disease graph G from model (7). To update the indicator Ii of SNP i, let X1 and G denote the current set of disease SNPs and the disease graph, respectively, excluding SNP i. We add SNP i into the disease graph to obtain a new set of disease SNPs xi + X1, and a new disease graph G+i, conditioning on X1 and G, by calculating
| (9) |
We then sample Ii = 1 with probability γ/(1+γ), and Ii = 0 otherwise. Intuitively, if SNP i is more likely to be generated from the disease association model PA(·) than from the baseline model P0(·), we will have γ > 1 and thus the SNP is likely to be added into X1. On the other hand, if SNP i is sufficiently explained by the disease SNPs in X1 under the baseline model, i.e., it is explained by LD with SNPs that are already selected in X1, then PA(·) will be smaller than P0(·). As a result, γ < 1 and thus the SNP tends not to be selected. That is, LD effects are automatically accounted for by P0(·), and this occurs if and only if we update a SNP’s group membership, without any upfront computation.
Formula (9) involves computing
where denotes a graph of all SNPs in X1 and SNP i, and G′ denotes a subgraph of excluding SNP i. In practice, we generate many random samples of G′, denoted by , from the distribution Pr(G′|X1), and we approximate P0(xi|X1) by
| (10) |
To further improve the computation speed, we utlize an approximate sampling procedure. In each iteration, we simply let in (10). That is, rather than sampling G′, we directly use the current disease graph G, excluding SNP i, as the dependence graph G′ in (10). This step can dramatically improve the computation speed, although the sampling results will be biased. In all simulation studies we have checked, the bias produced by this approximate sampling step is very small, yet it substantially improves the computation speed by magnitudes. We therefore use this approximate sampling procedure as a tool to first screen for candidate SNPs in genome-wide scale. We then re-sample from the full model to estimate the true posterior distribution of the candidate SNPs.
To further update the disease graph including the clique partition C and the interaction indicator Δ, we sample G+i, conditioning on the set of SNPs xi + X1 and the subgraph G excluding xi, from the distribution
| (11) |
In practice, it is likely that the above sampling algorithm is trapped in a local mode. This is a common problem of MCMC algorithms. One possible solution is to implement advanced sampling schemes to achieve better mixing of the Markov chains. Alternatively, we suggest running the program several times independently and then check if the estimated posterior probabilities are in agreement across multiple runs. In a few cases we checked in our simulation study and in the real data analysis, we found that our algorithm converged within a hundred iterations.
3 Results
3.1 Simulation Study
We performed simulation study to evaluate the performance of BEAM3. We used HAPGEN (WTCCC, 2007) to simulate a large pool of individuals from the HapMap Phase II sample of European ancestry (CEU, parental individuals only) (The International Hapmap Consortium 2005). There are 2.7 million common SNPs in the HapMap Phase II CEU data, and thus the SNP density is about one SNP per kb. From the simulated pool of individuals, we randomly sampled cases and controls according to the following logistic regression model:
where p denotes the probability of disease, and the constant c relates to disease prevalence. c is not used in our simulation, because we used a retrospective simulation procedure with fixed numbers of cases and controls. In particular, given a number of case and control individuals, we first calculate a joint genotype frequency of all disease SNPs in cases and controls, respectively, based on the above logistic regression model. We then sample from the large pool of individuals according to the disease genotype frequency. More details of our simulation procedure can be found in Zhang and Liu 2007.
Our disease model contains 10 disease SNPs, including three marginally associated SNPs X1, X2, X3, two 2-way interactions (X4, X5), (X6, X7), and one 3-way interaction (X8, X9, X10). Each SNP takes value in {0, 1, 2}, denoting the minor allele counts at the SNP in each individual. Note that our choice of using a logistic regression model and coding SNPs by the minor allele counts does not favor our method.
Each simulated dataset contained 10,000 SNPs in 1000 cases and 1000 controls. All SNPs are distributed in 5 randomly selected regions (cover 10Mb) in the genome. The 10 disease SNPs were randomly selected from the 5 regions, with a preference giving to SNPs with a desired minor allele frequency (MAF). The distribution of the 10 disease SNPs is summarized in Table 1. We generated 50 datasets for each disease MAF=0.05, 0.1, 0.2, respectively. We also calculated the effect sizes of each disease SNP in the simulated data. Of the main effect SNPs (X1, X2, X3), their median effect size is 0.626, 0.590, 0.623 for MAF= 0.05, 0.1, 0.2, respectively. Of the 2-way interaction SNPs (X4 ~ X7), their median (main) effect size is 0.138, 0.300, 0.692, respectively. Of the 3-way interaction SNPs (X8, X9, X10), their median (main) effect size is 0.026, 0.149, 0.536, respectively.
Table 1.
Disease SNP distribution in 5 regions.
| Region | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Number of SNPs | 1000 | 2000 | 2000 | 2000 | 3000 |
| Disease SNPs | X1 | X2, X3 | X4, X5 | X6, X7 | X8, X9, X10 |
We compared BEAM3 with five existing algorithms: ChiSq, Mendel (Wu et al 2009), RandomJungle (Schwartz et al 2008), BEAM1 (Zhang and Liu 2007), and BEAM2 (Zhang et al 2011). ChiSq is a standard 2 × 3 contingency table test on the genotype counts, which we use to benchmark the performance of single-SNP tests. Mendel (Wu et al 2009) is a software package that implements the LASSO penalized regression method (Tibshirani, 1996). Mendel allows detecting both main effects and interaction effects of SNPs. We ran Mendel in two ways: 1) detecting single SNP association only, and 2) detecting both single SNP and pairwise interaction. We hereafter name the two approaches as Mendel-Single and Mendel-Pairwise, respectively. Ran-domJungle (Schwartz et al 2008) is a machine learning algorithm that constructs trees to classify individuals as affected versus normal, and evaluates the importance of SNPs with respects to classification accuracy. BEAM1 (Zhang and Liu 2007) is our first Bayesian partition model for interaction mapping, which utilizes a 1st-order Markov chain to account for LD between adjacent SNPs. BEAM2 (Zhang et al 2011) improves upon BEAM1 by utilizing a LD-block model to capture the block-like dependence among SNPs. Here we demonstrate that, for dense SNPs, both BEAM1 and BEAM2 are inadequate in modeling strong LD. In addition, BEAM1 and BEAM2 only allows detecting one interaction at a time, which loses power when multiple interactions exist.
3.2 Power Comparison
We first compared the power of all methods using a plot similar to the receiver operating characteristic (ROC) curve. Without referring to statistical significance, we define the power of each program as the fraction of disease SNPs captured by the top ranked SNPs. A disease SNP is “captured” if there is at least one top ranked SNP within 5kb to either side of the disease SNP. We checked the results using different window sizes, and the conclusion remained the same (an example using 50kb window can be found in Online Supplementary Material). SNP ranking was obtained from the outputs of each program. In particular, we ranked SNPs by the posterior probabilities output by BEAM3, BEAM1 and BEAM2, the p-values by Mendle-Single and Mendel-Pairwise, the importance score by RandomJungle, and the chi-square statistics by ChiSq, respectively.
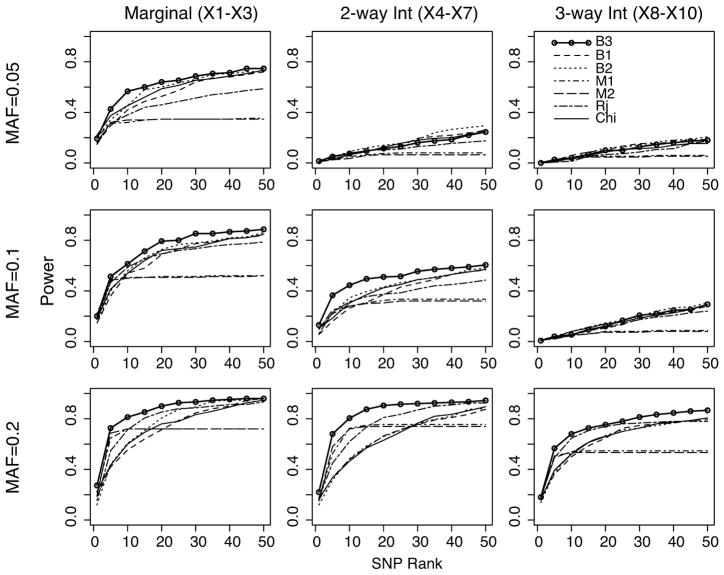
As shown in Figure 1, BEAM3 performed the best among all methods for detecting both marginal associations (first column in Figure 1) and 2-way and 3-way interactions (2nd and 3rd columns in Figure 1). Among the other programs, RandomJungle yielded good power for detecting 3-way interactions at MAF=0.2. Mendel (-Single and -Pairwise) yielded good power in the top 10 SNPs selected by LASSO penalized regression, yet its power remained almost flat when more top SNPs are included. This suggests that Mendel only detected a portion of the 10 disease SNPs (by default, we let Mendel select 50 best SNPs or SNP pairs). BEAM1 and ChiSq (and BEAM2 at MAF=0.2) yielded relatively low power than other methods. This is expected, because BEAM1 and ChiSq did not sufficiently account for LD, such that their top ranked SNPs are mostly clumped around just a few strongest associations. Overall, BEAM3 outperformed the other methods in almost all cases, except for the interactions at MAF=0.05, which are too weak to be detectable.
Figure 1.
Power comparison of BEAM3(B3, solid line with circle), BEAM1 (B1, dashed), BEAM2 (B2, dotted), Mendel-Single (M1, dotdash), Mendel-Pairwise (M2, longdash), RandomJungle (Rj, twodash), and ChiSq (Chi, solid black). Power (y-axis) is defined as the proportion of disease SNPs captured by the top ranked SNPs (x-axis) by each program. A disease SNP is captured if within its 5kb neighborhood there is at least one top ranked SNPs. All ranks are within-region ranks (i.e., within regions 1, 2 for marginally associated SNPs, within regions 3,4 for 2-way interactions, and within region 5 for 3-way interactions).
3.3 Number of Primary Disease SNPs
Using the output of BEAM3, we can estimate the number of primary disease SNPs (as oppose to secondary association created by LD) by summing over all SNPs the estimated posterior probabilities of marginal and interaction associations. Although BEAM1 and BEAM2 also output posterior probabilities, they tend to over-estimate the number of disease SNPs due to LD. We show in Table 2 the estimated numbers of disease SNPs by BEAM3, BEAM1 and BEAM2, from the simulated datasets at MAF=0.2. In these datasets, the truth is 3 marginal SNPs and 7 interacting SNPs. The other programs, Mendel, RandomJungle and ChiSq, do not estimate the number of disease SNPs. We instead computed the numbers of single SNPs and pairwise interactions selected by Mendel using Bayesian information criterion (BIC) (Diciccio et al 1997), and the number of significant SNPs by RandomJungle and ChiSq, respectively. The significance cutoff was estimated by permutation at the 0.05 level. We choose MAF=0.2 because the association signals in this scenario are strong and detectable, such that redundancy in SNP detection can be directly observed as over-estimation of the number of disease SNPs. The results for MAF=0.05 and 0.1 can be found in Online Supplementary Material, for which the data however have weak signals and thus the results are confounded by missing true SNPs.
Table 2.
Estimated numbers of disease SNPs by BEAM3 (B3), BEAM1 (B1), and BEAM2 (B2), and the numbers of SNPs selected by Mendel-Single (M1), Mendel-Pairiwise (M2), RandomJungle (Rj), and ChiSq (Chi). Disease MAF=0.2.
| dSNPs | True Size | B3 | B1 | B2 | M1 | M2 | Rj | Chi |
|---|---|---|---|---|---|---|---|---|
| X1 ~ X3 (Region 1, 2) | Single: 3 |
2.6a (0.5)b |
18.9 (15.3) |
20.5 (11.4) |
4.6 (2.4) |
2.8 (2.2) |
6.7 (6.2) |
63.8 (47.5) |
| Interact: 0 |
0.2 (0.2) |
0.2 (0.7) |
0.2 (0.6) |
n/a n/a |
9.8 (3.2) |
n/a n/a |
n/a n/a |
|
|
| ||||||||
| X4 ~ X10 (Region 3,4,5) | Single: 0 |
0.4 (0.5) |
79.4 (59.0) |
64.4 (35.6) |
11.3 (4.5) |
8.0 (3.3) |
38.0 (22.9) |
203.1 (129.8) |
| Interact: 7 |
7.6 (2.5) |
1.2 (1.0) |
2.8 (1.0) |
n/a n/a |
21.4 (2.7) |
n/a n/a |
n/a n/a |
|
most accurate estimation;
standard deviation.
As shown in Table 2, BEAM3 consistently obtained the most accurate estimates of the numbers of disease SNPs for both marginal and interaction association. In comparison, BEAM1 and BEAM2 substantially overestimated the number of marginal associations due to LD effects. They also underestimated the number of interacting SNPs, because they both used a saturated interaction model, and they only detect one interaction at a time. Among Mendel, RandomJungle and ChiSq, Mendel obtained the best shrinkage (fewest number) of SNP selection, after BIC. In contrast, ChiSq had no shrinkage at all, i.e., all marginally significant SNPs are reported including primary and secondary associations, 20~30 times larger than the true numbers. The number of SNPs selected by Mendel, however, still differ significantly from the truth. For instance, Mendel-Pairwise (which allowed both interaction and main effects) reported an average of 9.8 SNPs involved in pairwise interactions in regions (1, 2), where the regions only contained marginal associations. Mendel-Pairwise also reported an average of 8.0 SNPs involved in marginal associations in regions (3, 4, 5), where the regions only contained interaction associations. This result suggested that Mendel only selected a suboptimal set of SNPs. Overall, only BEAM3 accurately estimated the numbers of disease SNPs for both marginal and interactive association.
3.4 Structure of Interactions
BEAM3 outputs a (consensus) disease graph pertaining the interaction structures between the selected SNPs. The graph can be regarded as a reconstruction of the underlying disease model. Among the other methods we tested, only Mendel reconstructs a disease model via logistic regression. For easy comparison between BEAM3 and Mendel, we only considered marginal and pair-wise interactions, where high-order interactions output by BEAM3 were decomposed into pairwise interactions. Based on the outputs of BEAM3 and Mendel, respectively, we sequentially put SNPs and SNP pairs into a logistic regression model, one at a time, and we identified the best model by BIC. The SNP data were coded by minor allele counts. For BEAM3, SNPs and SNP pairs were entered into a logistic regression model according to their ranked posterior probabilities. For Mendel, SNPs and SNP pairs were entered into a logistic regression model according to their order given by Mendel’s output.
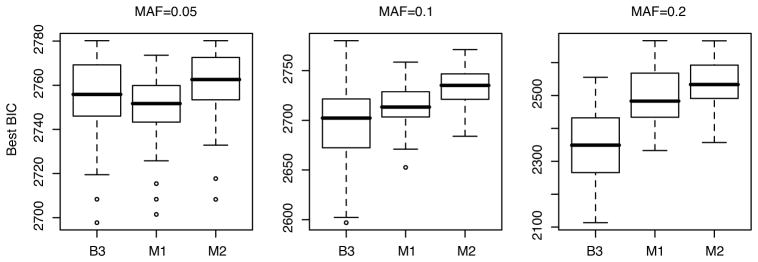
As shown in Figure 2, BEAM3 obtained the best model fitting (smallest BIC) at MAF=0.1 and 0.2, but performed slightly worse than Mendel-Single at MAF=0.05. By checking BEAM3’s output, we found that BEAM3 reported none or very few SNPs (at an arbitrary cutoff of 0.05 posterior probability) per dataset at MAF=0.05, because the association signals in these datasets were insignificant. Mendel, on the other hand, always outputs 50 SNPs and SNP pairs. Interestingly, Mendel-Pairwise consistently performed the worst in all cases, indicating that the interactions selected by Mendel-Pairwise were incorrect.
Figure 2.
Comparison of the best (smallest) BIC obtained by BEAM3(B3), Mendel-Single (M1) and Mendel-Pairwise (M2).
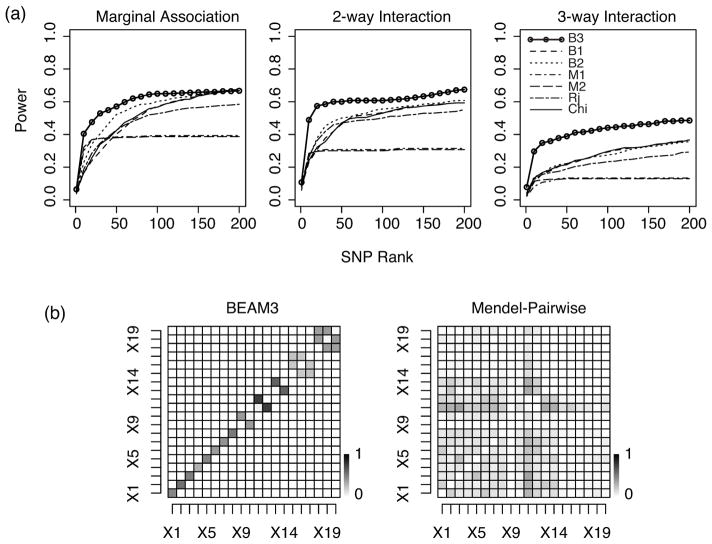
To check if BEAM3 and Mendel identified the correct interaction structures, we calculated the fraction of times an interaction between a pair of disease SNPs was detected by the two programs, respectively. An interaction between two disease SNPs is detected if there is an interacting SNP pair within 50kb to each of the two disease SNPs, respectively. As shown in Figure 3, BEAM3 accurately identified the true marginal association and interaction structures. In comparison, Mendel-Pairwise failed to capture the correct interactions. Mendel tended to select SNP pairs with stronger main effects (X1 − X3 had stronger main effects than X4 − X6, which in turn had stronger main effects than X7 − X10). We suspect that this result is partially due to the strong LD among SNPs. Also, Mendel uses a common penalty score to penalize the absolute value of both main and interaction effects. A different penalty parameter for the interaction effects may be desired in this problem. Additional results similar to Figure 3, but using a 5kb window size, can be found in Online Supplementary Material.
Figure 3.
Inferred interaction structures by (a) BEAM3 and (b) Mendel-Pairwise. The heatmap shows the proportion of times in 50 simulated datasets that an interaction is detected between two disease SNPs by each program. The x-axis and the y-axis of each heatmap denote the ten disease SNPs, where X1, X2, X3 do not interact, (X4, X5) and (X6, X7) are two independent pairwise interactions, and (X8, X9, X10) is a 3-way interaction. The diagonal cells show the fraction of times the disease SNPs are detected as marginally associated or as local interactions.
3.5 Large-scale Simulation
To evaluate the performance of BEAM3 in large-scale studies, we simulated 50 datasets containing 100,000 SNPs in 1,000 cases and 1,000 controls. In each dataset, we simulated 20 disease SNPs (MAF=0.1) according to the following logistic regression model:
Each dataset contained 10 SNPs with main effects (X1 ~ X9andX11), 6 SNPs involved in three pairwise interactions (X9 ~ X14), and 6 SNPs involved in two 3-way interactions (X15 ~ X20)). All disease SNPs are distributed in 7 randomly selected regions (of similar sizes and cover 100Mb) in the genome, with X1 ~ X3 in region 1, X4 ~ X7 in region 2, X8 ~ X10 in region 3, X11 ~ X13 in region 4, X14, X15 in region 5, X16, X17 in region 6, and X18 ~ X20 in region 7. As a result, the pairwise interaction between (X13,X14) and the 3-way interaction among (X15, X16, X17) are across regions.
We again calculated the rank power of all methods and the heatmap of the detected interaction structures by BEAM3 and Mendel-Pairwise on these large datasets. The calculation criteria were the same as those used in Figure 1 and Figure 3, respectively. As shown in Figure 4a, BEAM3 significantly outperformed all methods for detecting marginal associations, 2-way interactions, and 3-way interactions. This result is consistent with the result from the 10,000 SNP data, but the power gain by BEAM3 is more evident. As further observed in Figure 4b, BEAM3 “correctly” identified the true disease interaction structures, where Mendel-Pairwise failed again. We put a quotation mark on “correctly” because SNP X9 and X11 have both main and interaction effects, but BEAM3 detected them through interactions. In our model, we only distinguish between “joint association” and “marginal association” of SNPs, where ”main effect” and ”interaction effect” are only defined in regression models. An interaction by BEAM3 therefore does not necessarily mean pure interaction. Additional results using a different window size can be found in Online Supplementary Material.
Figure 4.
(a) Rank power comparison and (b) heatmap of the identified interaction structures by various methods on the 100,000 SNP datasets.
Mendel-Pairwise is a two-stage approach. It first selects a collection of candidate SNPs with main effects, the number of which is set at 1000 in this study. Mendel-Pairwise then adds all pairwise interaction terms of the candidate SNPs into the regression model to further identify interactions. Our result therefore suggests that BEAM3 can greatly outperform 2-stage approaches such as Mendel. In our simulation, SNP X11 has the strongest association with the disease, because its main effect and interaction effect are in the same direction, and interestingly, most pairwise interactions selected by Mendel-Pairwise involved X11.
BEAM3 is computationally efficient for analyzing all SNPs simultaneously. For most of the simulated datasets of 100,000 SNPs, BEAM3 were able to finish within 20 hours on a single CPU. The computation time of BEAM3 is mainly affected by the number of true disease SNPs, the total number of SNPs, and the sample size. BEAM3 dynamically and implicitly accounts for the unknown LD among SNPs, which effectively removes a large number of redundant SNPs from the disease graph, and hence achieves computational efficiency.
3.6 Application to Inflammatory Bowel Disease
We applied BEAM3 to an inflammatory bowel disease (IBD) dataset from the WTCCC project (2007). The dataset consists of 2,005 IBD affected individuals, 1,504 normal individuals from the 1958 British Birth cohort (58C), and 1,500 normal individuals from the UK Blood Service (UKBS). We removed individuals that were excluded by the original WTCCC analysis due to missing data, excessive heterozygosity, discrepancy between WTCCC and external identifying information, potential sample stratification, duplication and relatedness. We also removed SNPs with bad clustering results (WTCCC 2007), > 2% missing data (maximum genotype probability < 90%) in either cases or controls, unbalanced proportion of missing between cases and controls (at 10−3 level), discrepancy between the two control groups (at 10−6 level), and excessive Hardy-Weinberg disequilibrium in controls (at 10−6 level). The final dataset contained 1,748 cases, 2,962 controls (6% individuals removed), and 403,561 SNPs (20% SNPs removed).
To reduce the computation load, we first ran BEAM3 on all combinations of chromosome pairs separately. This may miss high-order interactions across more than two chromosomes, but can greatly reduce computation. This step took about 12 hours to complete. We next combined all results together and selected SNPs with average posterior probability > 0.05. This cufoff choice is arbitrary. We then included neighboring SNPs within 50kb to the selected SNPs. The filtered dataset contained 3,809 SNPs, a 100 times reduction from the original dataset. We reran BEAM3 on the filtered dataset to obtain the final posterior distribution of SNP association and interaction.
Figure 5 shows the top 32 loci identified by BEAM3, ranked by their posterior probabilities (≥ 0.1). Among the 32 loci, 20 (63%) were located within the previously reported loci (according to the reported location of these loci), including 15 (47%) confirmed IBD loci (Franke et al 2010) and 18 (56%) reported by WTCCC (2007). Most known IBD loci have large posterior probabilities. Interestingly, our method detected two confirmed IBD loci (rs744166, rs4263839) that were missed by the WTCCC (2007) analysis in the same data. SNP rs744166 is captured by an interaction association, and SNP rs4263839 is captured because another SNP (rs6478108) in its neighborhood showed additional association after accounting for the LD effects. We also observed that some moderate associations (marked by ∧ in Figure 5) reported by WTCCC had higher ranks in our results than the strongest associations (marked by +) reported by WTCCC. This is again because there were additional associations in the neighborhoods that leveraged the overall importance of the loci. Note that this would not be possible without accounting for LD. We further checked the weaker loci ranked by BEAM3 with posterior probability between [0.05, 0.1) (data not shown). We found 20 additional loci, of which one (rs7554511) was located at a known IBD locus and three (rs17309827, rs11119132, and a non-dbSNP at chr2:101.7Mb) were reported by a recent IBD meta analysis (Franke et al 2010).
Figure 5.
Top ranked IBD association output by BEAM3. Ranking is based on the sum of marginal (black) and interactive (grey) association posterior probabilities (y-axis) over a 100kb sliding window across the genome. Each bar along the x-axis represents a window, and the best candidate SNP within the window is labeled with dbSNP ID and chromosomal location (Mb). Known IBD loci are marked by ‘★’. Loci marked by ‘+’ and ‘∧’ are the strongest and the moderate associations, respectively, reported by WTCCC (2007).
In addition to marginal associations, BEAM3 detected some potential multi-SNP joint associations with IBD. There are two local joint associations located within known IBD loci. The first is between rs10489629 and rs10489628 (sum of marginal association probability 1.0 and interaction association probability 0.43). Both SNPs are located in the well-known IBD gene IL23R at 1p31 (Franke et al 2010). WTCCC (2007) only reported a single SNP association at this locus, but our result indicates that there is an interaction association (or perhaps joint-tagging) in IL23R on top of the known marginal association. The second local joint association is between rs2076756 (or rs7186163) and rs6500315 (sum of marginal association probability around each SNP equal to 0.62 and 0.03, respectively, and interaction probability 0.45) at 16q12. This region includes genes NDK1, SNX20 and NOD2, where NOD2 has been previously reported as a major IBD gene.
BEAM3 further detected an inter-chromosome interaction between rs744166 at 17q21 and rs4799144 at 18q23, with sum of marginal association probability around each SNP equal to 0.01 and 0.0, respectively, and sum of interaction probability equal to 0.25. We performed five expensive genome-wide permutation analysis, and observed that the maximum interaction probabilities from the permuted dataset were all < 0.1. It thus suggests that this IBD interaction is at least weakly significant in the genome-scale. SNP rs744166 lies in gene STAT3, which is a previously confirmed IBD locus (Franke et al 2010). WTCCC did not report this association, as its marginal association is insignificant. SNP rs4799144 lies in gene PARD6G, which is a novel gene. The protein coded by PARD6G contains a pseudo-CRIB domain that, together with PDZ domain (coded by RAPGEF6 at 5q31, a known IBD gene), are required to interact with Rho small GTPases. Rho GTPases are known to mediate a plethora of cellular effects, such as proliferation, apoptosis/survival (Benitah et al 2004). Rho GTPases are also considered essential for many biological processes, including immune and inflammatory responses (Benitah et al 2004). Interestingly, Rac1, a member of the Rho family, has been found to activate STAT3 (Turk-son et al 1999), suggesting that the interaction between rs744166 and rs4799144 are potentially interesting.
We obtained an independent dataset from NIDDK IBDGC study (Duerr et al 2006) to further validate this interaction. The SNPs genotyped in the two studies were different. We therefore checked the proxy SNPs in NIDDK IBDGC data within 100kb to rs744166 and rs4799144, respectively. We calculated LD of the proxy SNPs using HapMap Phase II CEU samples, and we removed SNPs in low LD (|r| < 0.2) with the two targeting SNPs, respectively. There were 11 proxy SNPs near rs744166 and 6 proxy SNPs near rs4799144, yielding 66 SNP pairs to be tested for interaction. The most significant marginal association of the 17 proxy SNPs is rs1905339 (67kb downstream of rs744166), with nominal p-value 0.002 and adjusted p-value 0.016 by 1000 permutations. For the pure interaction effects of the 66 SNP pairs, the most significant result is between rs9897702 (100kb downstream of rs4799144, |r| = 0.30, |D′| = 0.63) and rs6506816 (17kb downstream of rs4799144, |r| = 0.48, |D′| = 1), with nominal p-value 0.0012 and permutation adjusted p-value 0.025. We further used logistic regression to estimate the effects of each genotype combination between rs744166 and rs4799144 in WTCCC, and between rs9897702 and rs6506816 in NIDDK IBDGC, respectively. As shown in Table 3a, the direction and the magnitude of the effects are similar between WTCCC and NIDDK IBDGC, suggesting that this interaction is replicated.
Table 3.
Estimated effect and standard deviation of two 2-way interactions in WTCCC and the corresponding estimates of their proxy SNPs in NIDDK. Results are obtained by logistic regression with genotype combination AA/AA used as baseline. Significant effects (p-value < 0.05) are shown in bold.
| (a) Interaction between (rs744166, rs4799144) in WTCCC, compared with interaction between (rs9897702, rs6506816) in NIDDK. LD between (rs744166, rs9897702) is |r| = 0.30, |D′ | = 0.63; LD between (rs4799144, rs6506816) is |r| = 0.48, |D′ | = 1, in HapMap CEU sample. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| rs744166 | rs9897702 | ||||||||
| AA | Aa | aa | AA | Aa | aa | ||||
|
|
|
||||||||
| rs4799144 | AA | −0.39 (0.05) |
−0.23 (0.07) |
−0.27 (0.09) |
rs6506816 | AA | −0.26 (0.07) |
0.16 (0.12) |
0.49 (0.22) |
| Aa | 0.33 (0.17) |
0.22 (0.15) |
−1.77 (0.48) |
Aa | 0.19 (0.15) |
0.12 (0.18) |
−1.07 (0.51) |
||
| aa | 1.49 (1.16) |
−11.2 (197.0) |
NA NA |
aa |
1.43 (0.52) |
−0.55 (0.61) |
0.67 (0.92) |
||
| (b) Interaction between (rs16893872, rs2579176) in WTCCC, compared with interaction between (rs7729215, rs1248678) in NIDDK. LD between (rs16893872, rs7729215) is |r| = 0.32, |D′| = 1; LD between (rs2579176, rs1248678) is |r| = 0.43, |D′| = 1, in HapMap CEU sample. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| rs16893872 | rs7729215 | ||||||||
| AA | Aa | aa | AA | Aa | aa | ||||
|
|
|
||||||||
| rs2579176 | AA | −0.42 (0.05) |
−0.27 (0.47) |
NA NA |
rs1248678 | AA | −0.13 (0.07) |
−0.31 (0.18) |
−1.25 (0.79) |
| Aa | −0.21 (0.07) |
1.52 (0.37) |
12.0 (197.0) |
Aa | −0.08 (0.11) |
0.71 (0.23) |
−0.97 (1.16) |
||
| aa | −0.37 (0.10) |
0.42 (0.71) |
NA NA |
aa | −0.05 (0.21) |
−0.16 (0.77) |
12.7 (324.7) |
||
Another potential distant interaction detected by BEAM3 is between rs16893872 and rs2579176 (marginal association probability 0.0 at both SNPs and interaction probability 0.08). Although this interaction is relatively weak (not shown in Figure 5), SNP rs2579176 is located 21kb upstream of gene DLG5 at 10q22, and DLG5 has been found to be associated with perianal Crohn disease (Ridder et al 2007). We again used NIDDK IBDGC data to validate this interaction. Both SNPs were not genotyped in NIDDK IBDGC. We thus obtained the proxy SNPs within 100kb to rs2579176 and rs16893872, respectively, and we removed SNPs in low LD (|r| < 0.2) with the targeting SNPs in HapMap Phase II CEU samples. There were one proxy SNP (rs7729215) near rs16893872 and four proxy SNPs (rs1866437, rs2579159, rs1248678, rs1248688) near rs2579176, yielding 4 SNP pairs to be tested for interaction. There were no significant marginal associations of the 5 proxy SNPs, but the most significant pure interaction effect was between rs7729215 (45kb downstream of rs16893872, |r| = 0.32, |D′| = 1) and rs1248678 (78kb downstream of rs2579176, within DLG5, |r| = 0.43, |D′| = 1), with nominal p-value 0.0027 and permutation adjusted p-value 0.005. As shown in Table 3b, the direction and the magnitude of the effects of genotype combinations between rs16893872 and rs2579176 in WTCCC are in agreement with that of the proxy SNPs rs7729215 and rs1248678 in NIDDK IBDGC, again suggesting that this interaction is replicated.
4 Discussion
In this paper, we introduced a novel Bayesian graphical model for multi-locus disease association mapping in genome-wide case control studies. The method simultaneously detects single-SNP association, multi-local-SNP association that may jointly tag an unobserved disease mutation, and long distance SNP-SNP interaction in high orders. Distinct from most existing methods, BEAM3 uses graphs to implicitly account for SNP LD. Our simulation study using HapMap Phase II SNPs (The International HapMap Consortium, 2005) clearly demonstrated that our graphical model is more effective in capturing the complex LD patterns of dense SNPs than Markov chains and regression methods. This is particularly useful for GWAS with an increasing number of SNPs genotyped in the future. Our method fits a joint probability function to all SNPs simultaneously and detects the primary disease association, such that our results are cleaner than those produced by hypothesis testing procedures and other joint modeling approaches. Our novel modeling of LD can also reduce the computational burden, because secondary associations will not enter the disease graph when better candidates are already included. This is critical for interaction mapping, otherwise a large number of redundant and false “interactions” will be detected, which then requires further ad hoc filtration. Our method explores a variety of combinations of SNPs and interaction structures via regularized probability functions, by which we mean that all model parameters are treated as random in a Bayesian framework, and we analytically integrate out all nuisance parameters, such that the complexity of various interaction structures are automatically standardized by the normalizing constants of their corresponding distributions. BEAM3 therefore automatically strikes a balance between the power of disease mapping and the model complexity. As demonstrated by simulation, BEAM3 outperformed all other tested methods and identified the most accurate disease models.
We successfully applied BEAM3 to WTCCC IBD data (WTCCC 2007), which consists of 4,720 individuals and 401,473 SNPs after quality control. Our method is computationally affordable to analyze this huge dataset. Not only we detected all loci that were previously reported in this data, but also we recovered two missed IBD loci. We further detected a few novel IBD loci and interaction associations. In particular, the interaction between genes STAT3 (a known IBD gene but missed by WTCCC) and PARD6G (a novel locus) has biological connections and is further validated by the NIDDK IBDGC dataset (Duerr et al 2006). Another potential interaction is between DLG5 and an intergenic region at 5p14. Again, DLG5 is a known IBD gene missed by WTCCC (2007), and we validated this interaction in the NIDDK IBDGC dataset.
The WTCCC IBD dataset has been analyzed by others. Liu et al (2011) reported two interactions in the WTCCC IBD data. The first interaction is between rs7522462 and rs11945978, which has a combined p-value (main + interaction effect) 0.039 after multiple comparison adjustment. This is done using expanded controls, i.e., including patients from other diseases as controls. Using our method, however, we found the two SNPs are more likely to affect the disease risk independently. Despite that we only used the normal people as controls, the discrepancy between the two studies may be attributable to the different models. Liu et al. (2011) treated the SNP effects as unknown but constant parameters, while we treated genotype frequencies as random variables following a Dirichlet distribution. Furthermore, our model assumes that the frequencies of genotype combinations are all different between cases and controls under the alternative hypothesis. The two SNPs, however, only showed frequency difference in 2 out of 9 genotype combinations, where 7 combinations confirmed almost perfectly to independence. As a result, our marginal association model has likelihood than our interaction model, because the former has less complexity and variability. A larger sample size would resolve this discrepancy. The second interaction reported by Liu et al (2011) is between rs153423 and rs748855, with adjusted p-value 0.146 using expanded controls. When testing the two SNPs alone, the interaction is indeed identifiable by our method. When testing together with their neighboring SNPs, very interestingly, the interaction disappeared. SNP rs748855 lies in NOD2, a well-known IBD gene. There are several other SNPs in NOD2 with much stronger associations with IBD. The approach taken by Liu et al (2011) ignores the dependence between SNPs. The interaction is thus likely created by LD effects. This interaction in fact was not replicated by the proxy SNPs (Liu et al 2011) in NIDDK IBDGC.
Our method can be improved in several aspects. Most association signals in GWAS are weak, particularly for multi-SNP associations. Our current model is still restrictive in that it assumes all allele combination frequencies in a SNP set are all different between cases and controls. This assumption could be relaxed by allowing subset frequency differences using a mixture model, which will then improve the power of detecting subtle interactions. It is also desirable to include expert knowledge of disease loci and potential interactions between genes involved the same biological processes as a prior in our model. This is straightforward to implement in a Bayesian framework. In addition, the current model does not include covariates, such as environment factors and population structures. One possible way to incorporate covariates is to replace all the probability functions in our model by conditional probability functions giving the covariates. Finally, the computation speed of our method could be further improved. One solution is to utilize parallel computing infrastructures to split the task into smaller pieces and run each piece in parallel. Although we applied a similar idea in analyzing the WTCCC IBD data, a real parallel implementation is needed for analyzing larger datasets with hundreds of thousands individuals and many millions of SNPs.
Supplementary Material
Acknowledgments
YZ is supported by NIH grant R01-HG004718. The data of the WTCCC IBD study is obtained from the Wellcome Trust Case-Control Consortium. The data of the NIDDK IBDGC study (phs000130.v1.p1) is obtained from dbGaP. All the chromosomal positions are in NCBI build 35 coordinates.
Footnotes
Competing interests: none
References
- 1.Benitah SA, Valern PF, van Aelst L, Marshall CJ, Lacal JC. Rho GTPases in human cancer: an unresolved link to upstream and downstream transcriptional regulation. Biochim Biophys Acta. 2004;1705(2):121–32. doi: 10.1016/j.bbcan.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 3.Cook NR, Zee RY, Ridker PM. Tree and spline based association analysis of gene-gene interaction models for ischemic stroke. Stat Med. 2004;23:1439–1453. doi: 10.1002/sim.1749. [DOI] [PubMed] [Google Scholar]
- 4.Cordell HJ. Detecting gene-gene interactions that underline human diseases. Nat Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Culverhouse R, Klein T, Shannon W. Detecting epistatic interactions contributing to quantitative traits. Genet Epidemiol. 2004;27:141–152. doi: 10.1002/gepi.20006. [DOI] [PubMed] [Google Scholar]
- 6.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 7.DiCiccio TJ, Kass RE, Raftery A, Wasserman L. Computing Bayes factors by combining simulation and asymptotic approximations. Journal of American Statistical Association. 1997;92:902–915. [Google Scholar]
- 8.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Mont-gomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wijmenga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Bning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, De Vos M, D’Inca R, Dubinsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van Gossum A, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Pans J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D’Amato M, Weersma RK, Kugathasan S, Griffths AM, Mansfield JC, Vermeire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42(12):1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kooperberg C, Ruczinski I. Identifying interaction SNPs using Monte Carlo logic regression. Genet Epidemiol. 2005;28:157–170. doi: 10.1002/gepi.20042. [DOI] [PubMed] [Google Scholar]
- 12.Kuno S, Taniguchi A, Saito A, Tsuchida-Otsuka S, Kamatani N. Comparison between various strategies for the disease-gene mapping using linkage disequilibrium analyses: studies on adenine phosphoribosyltransferase deficiency used as an example. J Hum Genet. 2004;49:463–73. doi: 10.1007/s10038-004-0175-y. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Xu H, Chen S, Chen X, Zhang Z, Zhu Z, Qin X, Hu L, Zhu J, Zhao GP, Kong X. Genome-wide interaction-based association analysis identified multiple new susceptibility Loci for common diseases. PLoS Genet. 2011;7(3):e1001338. doi: 10.1371/journal.pgen.1001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- 15.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 16.Moore JH, Williams SM. New strategies for identifying gene-gene interactions in hypertension. Ann Med. 2002;34:88–95. doi: 10.1080/07853890252953473. [DOI] [PubMed] [Google Scholar]
- 17.Nelson MR, Kardia SL, Ferrell RE, Sing CF. A combinatorial partitioning method to identify multilocus genotypic partitions that predict quantitative trait variation. Genome Res. 2001;11:458–470. doi: 10.1101/gr.172901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pitman J, Yor M. The two-parameter Poisson-Dirichlet distribution derived from a stable subordinator. Ann Prob. 1997;25:855–900. [Google Scholar]
- 19.de Ridder L, Weersma RK, Dijkstra G, van der Steege G, Benninga MA, Nolte IM, Taminiau JA, Hommes DW, Stokkers PC. Genetic susceptibility has a more important role in pediatric-onset Crohn’s disease than in adult-onset Crohn’s disease. Inflamm Bowel Dis. 2007;13(9):1083–92. doi: 10.1002/ibd.20171. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz DF, Ziegler A, Knig IR. Beyond the results of genome-wide association studies. Genet Epidemiol. 2008;32:671. [Google Scholar]
- 21.Tibshirani R. Regression shrinkage and selection via the lasso. J R Statist Soc B. 1996;58:267–288. [Google Scholar]
- 22.Turkson J, Bowman T, Adnane J, Zhang Y, Djeu JY, Sekharam M, Frank DA, Holzman LB, Wu J, Sebti S, Jove R. Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in Stat3 transcriptional activity induced by the Src oncoprotein. Mol Cell Biol. 1999;19(11):7519–28. doi: 10.1128/mcb.19.11.7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan X, Yang C, Yang Q, Xue H, Fan X, Tang NL, Yu W. BOOST: A fast approach to detecting gene-gene interactions in genome-wide case-control studies. Am J Hum Genet. 2010;87(3):325–40. doi: 10.1016/j.ajhg.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu TT, Chen YF, Hastie T, Sobel E, Lange K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics. 2009;25(6):714–21. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K, Calabrese P, Nordborg M, Sun F. Haplotype structure and its applications to association studies: power and study designs. Am J Hum Genet. 2002;71:1386–1394. doi: 10.1086/344780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Liu JS. Bayesian Inference of Epistatic Interactions in Case-Control Studies. Nat Genet. 2007;39:1167–1173. doi: 10.1038/ng2110. [DOI] [PubMed] [Google Scholar]
- 28.Zheng T, Wang H, Lo SH. Backward genotype-trait association (BGTA) - based dissection of complex traits in case-control design. Hum Hered. 2006;62:196–212. doi: 10.1159/000096995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Zhang J, Liu JS. Block-based Bayesian Epistasis Association Mapping with Application to WTCCC Type 1 Diabetes Data. Ann Appl Stat. 2011 doi: 10.1214/11-AOAS469. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.