Summary
Insulators are DNA-protein complexes that can mediate interactions in cis or trans between different regions of the genome. Although originally defined on the basis of their ability to block enhancer–promoter communication or to serve as barriers against the spreading of heterochromatin in reporter systems, recent information suggests that their function is more nuanced and depends on the nature of the sequences brought together by contacts between specific insulator sites. Here we provide an overview of new evidence that has uncovered a wide range of functions for these sequences in addition to their two classical roles.
Introduction
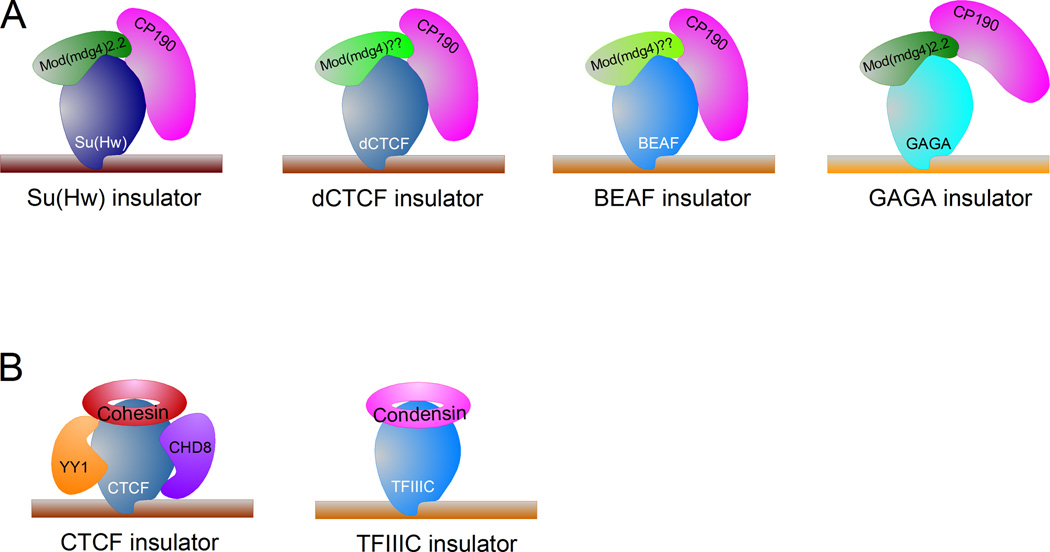
Insulators have now been found in most eukaryotes, from yeast to humans [1]. They were originally described in Drosophila and this organism remains noteworthy for the number of characterized insulators (Figure 1A). Drosophila insulators share two proteins, Mod(mdg4) and CP190, which interact with a variety of DNA binding proteins whose function appears to be limited to the recruitment of the shared components to different genomic locations, where they may play distinct roles [2]. Insulators in S. cerevisiae and S. pombe are mostly limited to RNA polymerase III promoter sequences containing binding sites for TFIIIC [3–5]. In vertebrates, the most widely studied insulator is CTCF, which requires cohesin for functionality and also associates with other co-factors, although their general requirement for CTCF function is not clear (Figure 1B) [6–8]. SINE elements and their associated Pol III promoters have been also shown to act as insulators in mammals [9], but the relevance of this initial observation had not been pursued in detail until recently.
Figure 1.
Diagram showing the structure of different Drosophila and vertebrate insulators. A. Each Drosophila insulator subclass contains a different binding protein that may define the specific function of the corresponding subclass. All insulators share the common protein CP190, although the role of this protein in the function of the GAGA insulator has not been demonstrated experimentally. In addition, all subclasses may also have one Mod(mdg4) isoform. The gypsy/Su(Hw) insulator contains Mod(mdg4)2.2. The dCTCF and BEAF insulators lack this isoform but contain a different variant of Mod(mdg4). GAGA has been shown to interact with Mod(mdg4)2.2. B. Structure of the vertebrate CTCF and TFIIIC insulators. Indicated are factors associated with CTCF such as cohesin, CHD8 and YY1
Results accumulated in the last two years suggest that insulators participate in numerous nuclear processes. In addition to blocking enhancer-promoter interactions, insulators can also direct enhancers to the appropriate promoters. Insulators can not only block the spreading of heterochromatin but they can also demarcate the boundaries between a variety of epigenetic states. Furthermore, the effect of insulators on genome biology goes beyond their involvement in transcription processes as they are also involved in regulating V(D)J recombination. Here we review recent results underscoring the variety of functional outcomes that arise as a consequence of the unique ability of insulators to mediate intra- and inter-chromosomal interactions between different regions of the genome. Although the name “insulator” may be counterintuitive in the context of these new functions, we will continue to use this name given its widespread use in the literature.
Regulation of enhancer-promoter interactions
Transcriptional enhancers have the ability to act over long distances to activate gene expression. This property gives enhancers great functional flexibility, being able to activate promoters of non-adjacent genes over distances of up to 1 Mb. But the same properties that are at the core of enhancer function are also a potential problem. How do enhancers find their target promoters and how is their activity confined to a specific gene? The CTCF insulator has been shown to confine enhancer action to a specific promoter by inhibiting its interaction with a second promoter in what is the archetypal example of endogenous insulator function. In the imprinted H19/Igf2 locus, endodermal enhancers present downstream of H19 are inhibited from activating the Igf2 gene by a CTCF insulator located between the two genes [10,11]. A similar function has now been described for the Drosophila dCTCF insulator [12]. This protein requires CP190 for proper function. In normal Kc cells, a CTCF site present between an enhancer and promoter of the Eip75B-RB gene is inactive due to the absence of CP190. When cells are treated with the hormone ecdysone, the Eip75B-RB promoter is turned on. After 3 hr of hormone treatment, CP190 is recruited to the dCTCF site, the insulator is activated, and the Eip75B-RB gene is turned off. In addition to demonstrating the effect of insulators on enhancer-promoter interactions, this example illustrates the principle that insulator activity can be modulated by recruitment of other protein components. In these two cases, the effect of the insulator on enhancer-promoter interactions is mediated by the formation of loops between individual insulator sites. As a consequence, the enhancer and promoter are partitioned to separate loops, precluding activation of transcription by mechanisms still unclear.
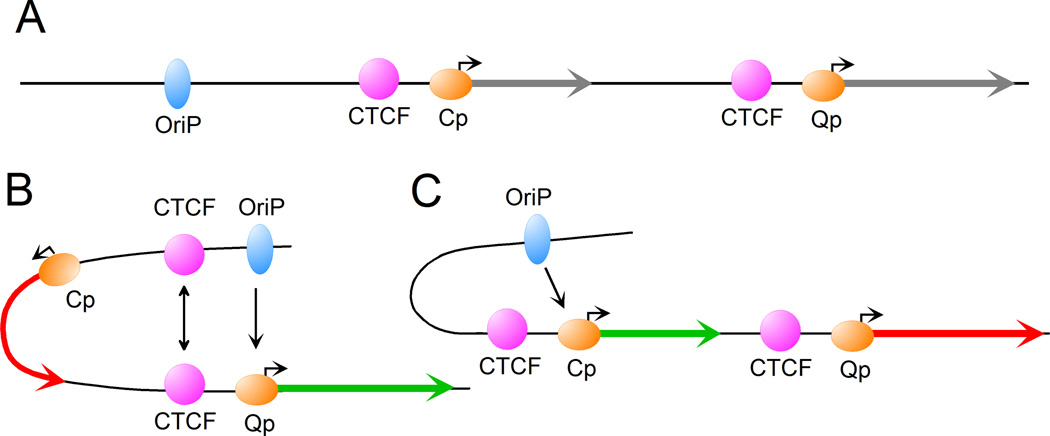
The ability of insulators to bring together sequences located far apart in the linear genome underlies their role in targeting enhancers to specific promoters. One interesting example is the use of CTCF for promoter selection and control of the latency cycle of the Epstein–Barr virus (Figure 2). In the genome of this virus, the enhancer OriP is shared by two downstream promoters, Cp and Qp. Cp determines the type III gene expression pattern for latency cycle III and Qp determines the type I expression program for latency cycle I [13–15]. CTCF binds both upstream of Cp and Qp. The default selection for OriP is the proximate promoter Cp. When Cp is active, deletion of either of the CTCF sites or depletion of CTCF does not affect the activity of Cp [13–15]. However, when CTCF-dependent interactions bring the distal OriP enhancer close to the Qp promoter, Cp is turned off. Mutations in either of the CTCF binding sites disrupt the interaction between OriP and Qp and lead to reactivation of Cp transcription. Thus, the insulator-mediated 3D structure of the chromatin contributes to promoter selection, probably by increasing the ability of Qp to compete out Cp. When Qp is not competent, Cp is selected by default [15].
Figure 2.
CTCF regulates enhancer-promoter interactions to control latency types of Epstein-Barr virus. A. linear organization of the EBV genome. B. Model showing CTCF-mediating looping that brings the OriP enhancer into contact with the Qp promoter in type I cells. C. In type III cells, the OriP enhancer interacts directly with the Cp promoter.
A similar logic underlies the expression of genes that regulate insulin synthesis and secretion, which are tightly controlled in the β-cells of the pancreas [16]. Functionally related genes in the genome of prokaryotes are organized into operons in order to co-regulate the synthesis of proteins that function in the same pathway. Eukaryotes may accomplish a similar end result by using insulators to bring together regulatory sequences of genes that participate in a common pathway but are located far away in the genome. The Insulin (INS) gene is located 300 kb away from the SYT8 gene, which is an important regulator of insulin secretion. CTCF sites are present next to INS and SYT8, and mediate long-range interactions between both genes. Glucose treatment increases the expression of INS and SYT8, and it enhances the interaction between the two genes. Depletion of CTCF attenuates the interaction as well as SYT8 expression [16]. Therefore, CTCF-mediated interactions can bring distal regulatory sequences located in the INS gene close to SYT8 to activate expression of this gene with the same tissue specificity as INS. The generality of this conclusion is supported by results from a genome-wide study of CTCF dependent intra- and inter-chromosomal interactions in mouse ES cells [17]. A total of 1,480 cis- and 336 trans-interacting loci were identified in this study. Using p300 as a mark for the location of enhancer sequences, the results show that CTCF-mediated loops bring these enhancers in close proximity to the promoters they regulate in mouse ES cells.
Establishment of chromatin domains
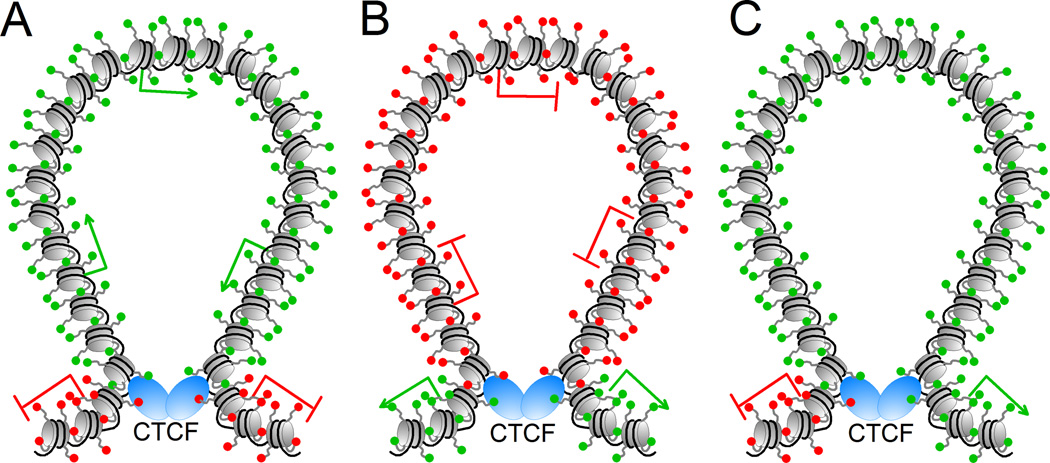
Loops formed by contacts between insulator sites can not only determine interactions between regulatory sequences but they can also establish distinct domains of chromatin structure defined by the presence of specific histone modifications. Genome-wide studies of CTCF distribution have uncovered a subset of CTCF sites localized at boundaries between active and repressive chromatin domains marked by acetylated histone H2A in Lys5 (H2AK5Ac) and trimethylated histone H3 in Lys27 (H3K27me3), respectively [18]. These regions are different between HeLa and CD4+ T cells, suggesting a possible role for CTCF-delimited domains in establishing lineage-specific gene regulation. This conclusion is supported by results from a the genome-wide study of CTCF dependent interactions mentioned above [17]. Alignment of CTCF-mediated loops with the location of histone modifications led to the finding of five different classes of loops, some of which demarcate chromatin domains with different epigenetic features (Figure 3). Category I loops are enriched in active histone modifications such as H3K4me1, H3K4me2 and H3K36me3, whereas the repressive marks H3K9me3, H3K20me3 and H3K27me3 are depleted inside but present outside of the loops. Category II loops show the opposite distribution of histone modifications, with H3K9me3, H3K20me3 and H3K27me3 inside of the loops, indicating the formation of repressive chromatin domains delimited by the presence of CTCF sites. Category III loops contain nucleosomes enriched in mono, di and trimethylated H3K4 at the boundaries of the loops, whereas the active transcription H3K36me3 and repressive H3K27me3 modifications are observed outside the loops on opposite sides [18].
Figure 3.
Structure of some of the domains created by interactions between CTCF insulators in mouse embryonic stem cells. Actively transcribed genes are represented by a green arrow and silenced genes by a red line; nucleosomes and the histone tails are represented in grey, with active histone modifications indicated as green spheres and repressive modifications as red spheres. DNA is represented in black and CTCF as blue ovals. A. CTCF forms a loop to separate a domain containing active histone modifications and transcribed genes from repressive marks and silenced genes. B. CTCF forms a loop to separate a domain containing repressive histone modifications and silenced genes from active marks and transcribed genes. C. CTCF forms a loop containing nucleosomes enriched in mono and dimethylated H3K4, and trimethylated H3K4 at the boundaries of the loops, whereas the active transcription modification H3K36me3 and repressive H3K27me3 mark are observed outside the loops on opposite sides.
The functional significance of the chromatin domains demarcated by CTCF is beginning to emerge from studies of specific loci. In human lung fibroblasts IMR90 cells the HOXA9-13 genes are silenced but other genes in the HOXA locus are transcriptionally active [19]. Consistent with this transcription pattern, the region containing HOXA9-13 is rich in H3K27me3 while the adjacent region containing the active HOXA1-7 genes is marked by H3K4me2, H3K4me3 and H3K36me3. The distribution of the repressive marks depends on the interaction between two CTCF insulators flanking HOXA9-13. Knockdown of CTCF results in the disruption of loop formation and spreading of silencing histone modifications, which causes downregulation of the adjacent HOXA6-7 genes [19].
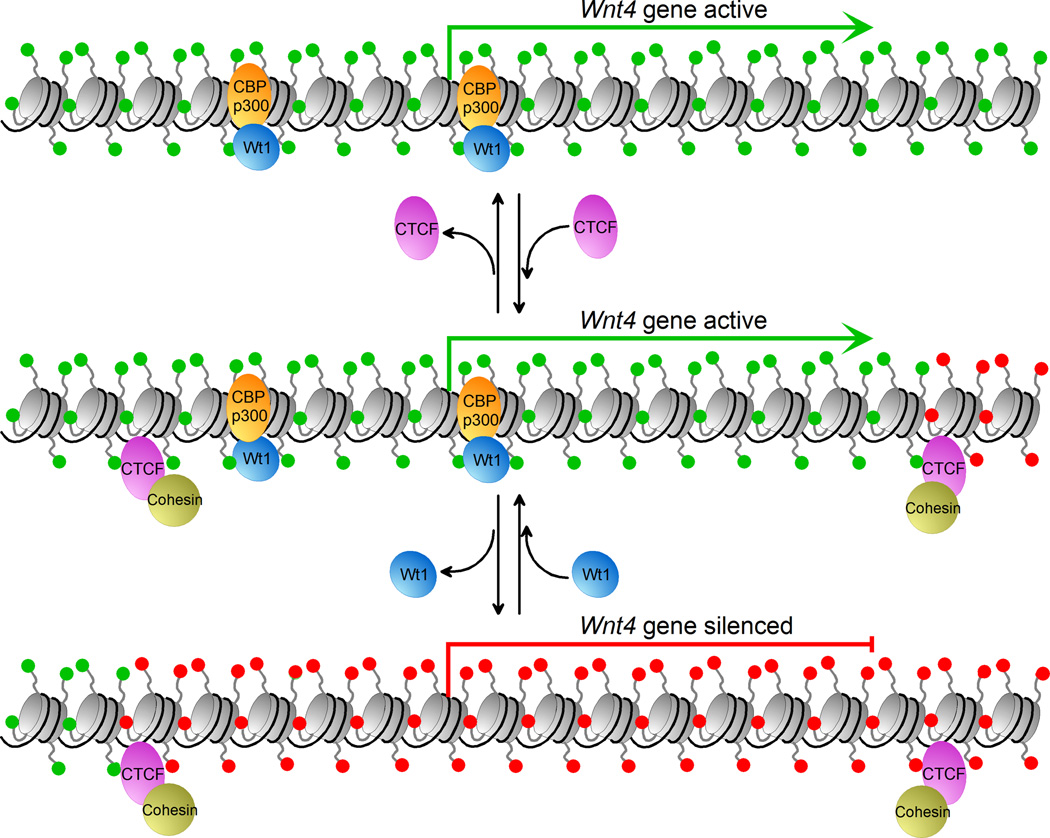
A second example of CTCF-delimited chromatin domains in the control of gene expression and the establishment of cell fates is that of the Wilm’s tumor protein (Wt1) in the regulation of epithelial-mesenchymal transitions during the development of the epicardium, and the kidney (Figure 4). These processes are controlled by Wt1 through the regulation of the expression of the Wnt4 gene [20]. Wt1 binds to the promoter of Wnt4 in both tissues with opposite outcomes. In kidney cells, Wt1 recruits the CBP/p300 coactivators and turns on transcription of Wnt4. In epicardial cells, Wt1 recruits the Basp1 co-repressor to silence Wnt4. These Wt1-induced changes in Wnt4 expression correlate with changes in chromatin structure. Kidney cells, in which the Wnt4 gene is active, contain H3K4me3, H3K9ac and H3K14ac in the Wnt4 locus. Epicardial cells, where Wnt4 is silenced, contain H3K27me3 instead. The Wt1-dependent changes of chromatin marks in the Wnt4 locus are confined to a region of the genome that is delimited by CTCF. The formation of a loop enclosing the Wnt4 locus may be the basis for the functional chromatin domain established by Wt1. In the absence of CTCF, the chromatin domain created by Wt1 spreads outside of its normal boundaries and alters the transcription of neighboring genes [20].
Figure 4.
CTCF forms boundaries at the Wnt4 Locus. The Wnt4 gene is flanked by two CTCF sites (middle row). The gene is active in mesenchymal kidney cells where it is bound by the transcription factor Wt1, which recruits the co-activators CBP/p300. The activate histone marks (green) are present in the domain defined by CTCF. Mutation of the Wt1 transcription factor in these cells causes repression of the Wnt4 gene and enrichment of repressive chromatin marks (red) within the CTCF domain (bottom row). The reverse effect is seen in epicardial cells (not shown). CTCF is critical for the establishment/maintenance of the chromatin domain by Wt1 at the Wnt4 locus. Loss of the CTCF protein causes the spreading of the active chromatin past the boundary (top row) and can result in the transcriptional activation of neighboring genes.
Regulation of recombination
The effects of insulators on genome function are not limited to transcription processes. At the Igh locus, insulators regulate V(D)J recombination through promoter selection to transcribe non-coding RNA from the proper promoter and by bringing together distant sequences to undergo specific patterns of recombination. DNA rearrangements in the Igh locus of pro-B cells are under temporal and spatial regulation during B cell development. The process begins with DH to JH rearrangement followed by rearrangement of a VH gene segment to DHJH. In mice, there are more than 100 VH genes spanning a 2.5 Mb region, whereas the JH genes occupy 2 kb. How do all the VH genes access the small JH cluster in the Igh locus? Insulators are critical for the execution of this complex pattern of DNA rearrangements. There are about 60 CTCF sites throughout the VH region and 2 clusters within other parts of the Igh locus in pro-B cells [21,22]. Of the two CTCF clusters not in VH, one is next to DH and the other is at the 3’ regulatory region of JH. These two sites interact strongly in pre-pro-B and pro-B cells, but only minimally in murine embryonic fibroblasts, promoting the selection of DH promoters over VH promoters before DH-JH combination takes place. These insulators also interact with an intronic enhancer (Eµ), which is required for the antisense transcription of DH. Antisense transcription through the DH locus precedes DH-JH rearrangement and has been proposed to make the DH region accessible for subsequent recombination [21–23]. Thus, CTCF-mediated interactions select the DH over the VH promoters for antisense transcription as well as to first bring together DH and JH instead of VH. Later, in pro-B cells, the locus compacts to bring VH genes close to the DH-JH region through interactions that also depend on CTCF [21,22]. As a consequence, insulator-mediated chromosome interactions regulate V(D)J recombination both spatially and temporally.
A twist on this effect of insulator-mediated chromatin architecture on DNA recombination is that described for the T-cell receptor α locus (Tcra) [24]. The formation of loops by CTCF requires the presence of cohesin, which can also mediate interactions between different sequences on its own to regulate the transcription of various genes. In mouse thymocytes, cohesin binding sites flank the TEA promoter and the Eα enhancer of Tcra. Deletion of the gene for the Rad21 component of cohesin in these cells at the time when the locus is rearranged leads to a defective chromatin architecture in the region. Cohesin is required enhancer-promoter interactions necessary for Tcra transcription and deposition of H3K4me3. This histone modification then recruits the recombination machinery, which causes Tcra rearrangements. Therefore, long-range interactions mediated by cohesin indirectly control rearrangements in the Tcra region by regulating the transcriptional state of the locus [24].
The TFIIIC insulator: more general than we thought?
Work carried out in S. cerevisiae and S. pombe indicates that RNA Polymerase III promoters can act as barriers and insulators. This property has been traced to the TFIIIC transcription factor [25]. In mammals, TFIIIC binds to human Alu sequences and murine SINE elements, both of which can act as insulators [9]. New results now suggest that Pol III promoters in human tDNA genes can act as both enhancer-blocking and barrier insulators [26]. It is not clear at this point whether this property can be exclusively assigned to the presence of TFIIIC. Insulator function of these sequences correlates with their ability to mediate interactions with other tDNAs in the genome. One possible interpretation of these results is that these interactions mediate the formation of Pol III transcription factories. TFIIIC sites are often found adjacent to CTCF sites in the genome, suggesting that the two proteins may function together to establish long-range interactions [27,28]. Interestingly, TFIIIC colocalizes with condensin in budding and fission yeast chromosomes, pointing to an interesting similarity to the interaction of CTCF with cohesin [29].
Concluding remarks
It is becoming increasingly clear that the role of insulators is to mediate inter- and intra-chromosomal interactions between different regions of the genome. In so doing, insulators can establish a specific three-dimensional organization of the genome within the nucleus of eukaryotic cells. An important question is whether this structure is a consequence or an effector of genome function. Loop structures created by some regulatory sequences, such as enhancers and promoters, are a consequence of their function. In the cases reviewed above, insulators mediate intra- and inter-chromosomal interactions in order to elicit a specific functional response. Thus, it is possible that the three-dimensional structure of the genome determines its function. As a consequence, insulators may be responsible for the establishment of patterns of three-dimensional architecture of the chromatin fiber that are cell-type specific and responsible for distinct blueprints of gene expression during development. Insulator-mediated 3D structure may carry epigenetic information that needs to be maintained during mitosis. Examination of these possibilities remains one of the main challenges for future research in the field.
ACKNOWLEGMENTS
Work in the authors’ laboratory is supported by U.S. Public Health Service Award GM35463 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Phillips JE, Corces VG. CTCF: Master Weaver of the Genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes & Development. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valenzuela L, Dhillon N, Kamakaka RT. Transcription Independent Insulation at TFIIIC-Dependent Insulators. Genetics. 2009;183:131–148. doi: 10.1534/genetics.109.106203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noma K-i, Cam HP, Maraia RJ, Grewal SIS. A Role for TFIIIC Transcription Factor Complex in Genome Organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 5.Iwasaki O, Tanaka A, Tanizawa H, Grewal SIS, Noma K-i. Centromeric Localization of Dispersed Pol III Genes in Fission Yeast. Molecular Biology of the Cell. 2010;21:254–265. doi: 10.1091/mbc.E09-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 7.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins Functionally Associate with CTCF on Mammalian Chromosome Arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proceedings of the National Academy of Sciences. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lunyak VV, Prefontaine GG, Núñez E, Cramer T, Ju B-G, Ohgi KA, Hutt K, Roy R, García-Díaz A, Zhu X, et al. Developmentally Regulated Activation of a SINE B2 Repeat as a Domain Boundary in Organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 10.Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters J-M, Murrell A. Cohesin Is Required for Higher-Order Chromatin Conformation at the Imprinted IGF2-H19 Locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proceedings of the National Academy of Sciences. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood AM, Bortle KV, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, Jones KC, Corces VG. Regulation of chromatin organization and inducibe gene expression by a Drosophila insulator. Molecular Cell. 2011;4:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tempera I, Wiedmer A, Dheekollu J, Lieberman PM. CTCF Prevents the Epigenetic Drift of EBV Latency Promoter Qp. PLoS Pathog. 2010;6:e1001048. doi: 10.1371/journal.ppat.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chau CM, Zhang X-Y, McMahon SB, Lieberman PM. Regulation of Epstein-Barr Virus Latency Type by the Chromatin Boundary Factor CTCF. J. Virol. 2006;80:5723–5732. doi: 10.1128/JVI.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tempera I, Klichinsky M, Lieberman PM. EBV Latency Types Adopt Alternative Chromatin Conformations. PLoS Pathog. 2011;7:e1002180. doi: 10.1371/journal.ppat.1002180. * This paper shows that EBV latency types adopt different chromatin architectures that depend on CTCF and mediate targeting of alternative promoters by the OriP enhancer.
- 16. Xu Z, Wei G, Chepelev I, Zhao K, Felsenfeld G. Mapping of INS promoter interactions reveals its role in long-range regulation of SYT8 transcription. Nat Struct Mol Biol. 2011;18:372–378. doi: 10.1038/nsmb.1993. This manuscript present results suggesting that two CTCF insulators interact to bring regulatory sequences of the INS gene close to the SYT8 promoter to co-regulate the expression of both genes
- 17. Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CWH, Ye C, Ping JLH, Mulawadi F, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. Results in this paper show for the first time the network of interactions among CTCF sites in the genome of mouse ES cells, and presents evidence for a function of this protein in demarcating different chromatin domains
- 18.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim YJ, Cecchini KR, Kim TH. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proceedings of the National Academy of Sciences. 2011;108:7391–7396. doi: 10.1073/pnas.1018279108. Results show an evolutionarily conserved role for CTCF in the establishment of a specific pattern of chromatin organization at the HoxA locus
- 20. Essafi A, Webb A, Berry RL, Slight J, Burn SF, Spraggon L, Velecela V, Martinez-Estrada OM, Wiltshire JH, Roberts SGE, et al. A Wt1-controlled chromatin switching mechanism underpins tissue-specific Wnt4 activation and repression. Developmental Cell. 2011;21 doi: 10.1016/j.devcel.2011.07.014. Results in this paper show that CTCF can define a chromatin domain on which the Wt1 transcription factor can act to activate or silence gene expression
- 21. Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, Vettermann C, Lin YC, Ju Z, Schulz D, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proceedings of the National Academy of Sciences. 2011;108:9566–9571. doi: 10.1073/pnas.1019391108. This and references 22 and 23 dissect the role of CTCF in organizing the three-dimensional architecture of the Igh locus necessary for proper recombination and transcription
- 22.Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Oltz EM, Sen R. Two Forms of Loops Generate the Chromatin Conformation of the Immunoglobulin Heavy-Chain Gene Locus. Cell. 2011;147:332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng H-L, Hansen E, Despo O, Bossen C, Vettermann C, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simms TA, Dugas SL, Gremillion JC, Ibos ME, Dandurand MN, Toliver TT, Edwards DJ, Donze D. TFIIIC Binding Sites Function as both Heterochromatin Barriers and Chromatin Insulators in Saccharomyces cerevisiae. Eukaryotic Cell. 2008;7:2078–2086. doi: 10.1128/EC.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Raab JR, Chiu J, Zhu J, Katzman S, Kurukuti S, Wade PA, Haussler D, Kamakaka RT. Human tRNA genes function as chromatin insulators. EMBO J. doi: 10.1038/emboj.2011.406. in press. Results in this paper demonstrate an insulator function for human tRNA genes, suggesting a general role for TFIIIC in genome organization
- 27.Carrière L, Graziani S, Alibert O, Ghavi-Helm Y, Boussouar F, Humbertclaude H, Jounier S, Aude J-C, Keime C, Murvai J, et al. Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic Acids Research. 2011 doi: 10.1093/nar/gkr737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–640. doi: 10.1038/nsmb.1794. Results show the presence of TFIIIC at non-canonical sites throughout the human genome in association with CTCF sites
- 29.D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes & Development. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]