Abstract
Motivation: Analyses and algorithmic predictions based on high-throughput data are essential for the success of systems biology in academic and industrial settings. Organizations, such as companies and academic consortia, conduct large multi-year scientific studies that entail the collection and analysis of thousands of individual experiments, often over many physical sites and with internal and outsourced components. To extract maximum value, the interested parties need to verify the accuracy and reproducibility of data and methods before the initiation of such large multi-year studies. However, systematic and well-established verification procedures do not exist for automated collection and analysis workflows in systems biology which could lead to inaccurate conclusions.
Results: We present here, a review of the current state of systems biology verification and a detailed methodology to address its shortcomings. This methodology named ‘Industrial Methodology for Process Verification in Research’ or IMPROVER, consists on evaluating a research program by dividing a workflow into smaller building blocks that are individually verified. The verification of each building block can be done internally by members of the research program or externally by ‘crowd-sourcing’ to an interested community. www.sbvimprover.com
Implementation: This methodology could become the preferred choice to verify systems biology research workflows that are becoming increasingly complex and sophisticated in industrial and academic settings.
Contact: gustavo@us.ibm.com
1 BACKGROUND AND PHILOSOPHY OF SYSTEMS BIOLOGY VERIFICATION
1.1 What is verification?
In the past two decades molecular biology has experienced an increase in the amount and diversity of data that are produced to answer key scientific questions. Systems biology has emerged as a new paradigm for the integration of experimental and computational efforts. This uses algorithmic analyses to interpret the data and mathematical models are built to predict yet unmeasured states of the biological system. However, algorithms and models are not unique and the determination of the right algorithm and model leading to the true interpretation of the natural phenomena under study becomes a fundamental question that falls within the realm of the philosophy of science.
Popper postulated (Popper, 1959) that a hypothesis, proposition, theory or in the case of systems biology a model, is ‘scientific’ only if it is falsifiable. In Popper's thesis, a theory can be proven wrong by producing evidence that is inconsistent with the theory. In contrast, a theory cannot be proven correct by evidence because other evidence, yet to be discovered, may exist that will falsify the theory. Conversely, according to the verificationist school, a scientific statement is significant only if it is a statement of logic (such as a mathematical statement deduced from axioms) or if the statement can be verified by experience (Ayer, 1936). Statements that do not meet these criteria of being either analytic or empirically verifiable are judged to be non-sensical.
The McGraw-Hill Concise Dictionary of Modern Medicine© (2002) defines verification as: ‘The process of evaluating a system, component or other product at the end of its development cycle to determine whether it meets projected performance goals’ (http://medical-dictionary.thefreedictionary.com/verification). For systems biology, a fundamental question to address is how to verify the correctness of a model that integrates vast amounts of data into a representation of reality. These data are not only high-dimensional but noisy given the biological variability, sample preparation inconsistencies and measurement noise inherent to the sensor instrumentation. While the concept of verification may be applied to different contexts with slightly different meanings, here we always use verification as checking for the truth or correctness of either data (i.e. whether the data represents what we wish to measure) or the correctness of a theory's predictions.
1.2 Crisis in peer-review/slow and low throughput
The quality of a scientific prediction or the accuracy of a scientific model is the subject of rigorous scrutiny, usually by the researchers themselves or by colleagues in the peer-review process that is at the heart of scientific publishing (Spier, 2002). As stated by the editors of the journal Science (Alberts et al., 2008),
peer review is under increasing stress […] The growth of scientific publishing is placing a burden on the entire scientific enterprise. Papers today are more interdisciplinary, use more techniques, and have more authors. Many have large volumes of data and supplementary material.
The coming of age of systems biology and its computational methods such as data-interpreting algorithms are challenging the peer-review process as large numbers of simultaneous predictions are generated, but only a small minority is tested. In the best cases, a very small sampling of predictions are verified using sound experimental assays and methods and then are presented as representative confirmation of the soundness of the entire set of predictions. Typically, this verification method lacks sufficient rigor, objectivity and a clear characterization of the relative strengths and weaknesses of the algorithms (Dougherty, 2010; Jelizarow et al., 2010; Mehta et al., 2004).
The same lack of rigor in verification of model predictions can be found in many areas of science where complex systems are measured, analyzed and modeled. For example, in systems biology, high-throughput data are collected and analyzed together with insufficient verification. Specifically, false positive and, equally important, false negative rates, are rarely considered a requisite for verification of the analysis for publication. Consider that the first experimentally-generated, genome-wide interactomes in yeast (Gavin et al., 2006; Ito et al., 2001; Uetz and Hughes, 2000) showed minimal overlap, generating some concerns within the scientific community that the data and methodologies were unreliable. Later work showed that high quality interactome maps could be generated by including controls and quality standards in data collection, careful verification of all interacting pairs and validation tests using independent, orthogonal assays (Dreze et al., 2010). Similarly, Genome-Wide Association Studies (GWAS) generate a high rate of false positives as correlations are found for single nucleotide polymorphisms with no direct effect on the phenotype. The community responded by defining a quality-control process and software package for analysis (Purcell et al., 2007). Similar problems are found in other fields including protein structure prediction (Moult et al., 1995), prediction of docking between proteins (Wodak and Mendez, 2004), text mining from scientific literature (Hirschman et al., 2005) and biological network inference (Stolovitzky et al., 2007). In these cases the response has been to set up community-based efforts, as discussed below.
1.3 Proposed community approaches for science verification
The difficulties in verifying complex science with traditional methods is driving changes in the methods of evaluation. Advances in web technology (called web 2.0) have allowed communities to stay tightly in touch to develop their interests, even when they are geographically dispersed. The journal Nature developed in 2006 an experiment allowing an online public review of manuscripts that in parallel were undergoing peer-review (http://www.nature.com/nature/peerreview/). Faculty of 1000 is an annotation service that allows researchers to locate outstanding or influential papers from the whole body available that can completely overwhelm the individual. Faculty of 1000 has domain experts cull, rate and summarize both the importance of the paper's findings and context within the field and hence is a good example of new practices in research evaluation that go far beyond simple indexing and content annotation (as in PubMed, for example). The journal PLoS ONE and now even mainstream sites like Twitter have become places where manuscripts are publicly criticized (Mandavilli, 2011). We think that these changes in research evaluation, while valuable, will not have sufficient rigor and consistency for the needs of research workflows verification.
2 COMMUNITY APPROACHES FOR SCIENCE VERIFICATION
2.1 Community consensus as criteria of science done right
A natural evolution of allowing community feedback has been the development of crowd-sourcing, a modality of distributed problem-solving. Challenges are broadcasted to potential interested stakeholders (solvers) in the form of an open call for participation. Participants submit solutions for the challenges, and the best solutions are typically chosen by the crowd-sourcer (the entity that broadcasted the challenge). The top performing participants are sometimes rewarded either with monetary awards, prizes, certificates or with recognition. We think that such directed community approaches could complement and enhance the peer-review process. Most importantly, we think that these could serve as a tool to verify the scientific results and fulfill the ultimate goal of scientific research that is to advance our understanding of the natural world (Meyer et al., 2011).
Community-based approaches to verify scientific research can be considered a more focused attempt to tap the consensus building that historically occurs in scientific progress. Kuhn understood progress in science as an eminently social process, in which the scientific worldview is dominated by the paradigm embraced by the scientific community at any given time (Kuhn, 1962). When the number of anomalies accumulated under the current paradigm generates distrust, the community may adopt a new paradigm that now guides how research is conducted. In this view, the scientific community, and not just nature itself, needs to be taken into account when considering what is accepted as ‘verified science’. For our purposes, we abbreviate the typical definition of verification given in the first paragraph to: ‘science done right’, where the ‘right’ refers to the accepted best practices of the scientific community or similar criteria. Accepted best practices means that there is a consensus in the community as to the proper collection and analysis of a data modality. Obviously, a modality must already be accessible to a wide community for the consensus to form. For newly developed modalities, crowd-sourcing provides a means to a rapid consensus as to the best collection and analysis methodologies.
2.2 Summary of community approaches for verification in other fields
Recent practices involving a new form of research quality control have become well-established during the last decade and a half. These efforts have merged the need of scientific verification of methods used in research, with the widespread practice of crowd-sourcing, to create a sort of collaboration-by-competition communities. The practice of this idea has been sufficiently well-established to become the business model of for-profit companies. In this section, we summarize three relevant community-based verification approaches with overlapping objectives but different focus areas. Some relevant details of these efforts are listed in Table 1
Knowledge Discovery and Data Mining Cup (KDD Cup) is an annual competition organized by the Association for Computing Machinery (ACM) Special Interest Group on Knowledge Discovery and Data Mining, the leading professional organization of data miners (Fayyad, 1996). KDD goals are to achieve a better understanding and analysis of data in many knowledge domains, such as medical informatics, consumer recommendations, diagnostics from imaging data and Internet user search query categorization.
InnoCentive, a spin-off of Eli Lilly, was founded in 2001 to match problems in need of solutions with problem solvers. The main entry point of InnoCentive is a web portal where solutions to scientific and business problems are solicited on behalf of organizations seeking innovations. An example of a recent challenge is ‘Solutions to Respond to Oil Spill in the Gulf of Mexico’. InnoCentive works with seekers to design the challenge, score/judge solutions and manage the intellectual property transfer. There is usually a cash award to the winning solver.
Netflix Prize was a competition to produce a better algorithm to substantially improve the accuracy of predictions about how much a customer is going to enjoy a movie based on their past movie preferences. The results were measured against the predictions proposed by Cinematch, the algorithm then used by Netflix for customer preference prediction. In 2009, the $1M Grand Prize was awarded, and the description of the best performing algorithm (if not the source code) was made publicly available.
Table 1.
Additional information for the eight community-based efforts described in the paper. The last row describes other efforts not discussed in the main text
| Name | Domain and Regularity | Website |
|---|---|---|
| KDD Cup | Knowledge discovery and machine learning in various domains. | http://www.sigkdd.org |
| Knowledge Discovery and Data Mining. Every year since launch in 1997. | ||
| InnoCentive | The name mixes Innovation and Incentive. | http://www.innocentive.com/ |
| Crowd-sourcing for problems of commercial interest. | ||
| Founded in 2001. New challenges are released on a rolling schedule. | ||
| Netflix Prize | The name comes from the sponsoring company, Netflix. | http://www.netflixprize.com//index |
| Prediction of user ratings for films, based on previous ratings. | ||
| Only challenge so far, released in 2006, lasted 3 years to complete. | ||
| CASP | Critical Assessment of Techniques for Protein Structure Prediction. | http://predictioncenter.org/ |
| Protein 3D structure prediction assessment. | ||
| Every 2 years since 1994. | ||
| CAPRI | Critical Assessment of PRedicted Interactions. Assessment of predictions of | http://www.ebi.ac.uk/msd-srv/capri |
| protein–protein docking or protein–DNA interaction from 3D structure. | ||
| Goes by Round 22 since 2001. Starts whenever an experimentalist offers an adequate target. | ||
| Predicted structures are submitted 6–8 weeks later. | ||
| DREAM | Dialogue for Reverse Engineering Assessments and Methods. | http://www.the-dream-project.org/ |
| Assessment of quantitative modeling in systems biology. | ||
| Every year since 2006. | ||
| BioCreAtIve | Assessment of Information Extraction Systems in Biology. Evaluating text mining | http://www.biocreative.org |
| and information extraction systems applied to the biological literature. | http://biocreative.sourceforge.net | |
| Every 2 years beginning in 2004. | ||
| FlowCAP | Flow Cytometry Critical Assessment of Population Id Methods. | http://flowcap.flowsite.org/ |
| Evaluation of automated analysis of flow cytometry data. | http://groups.google.com/group/flowcap | |
| Only one iteration on 2010, second one on planning phase. |
Others efforts TunedIT: http://tunedit.org/, RGASP-RNAseq Genome Annotation Assessment Project: www.sanger.ac.uk/PostGenomics/encode/RGASP.html Pittsburgh brain competition: http://pbc.lrdc.pitt.edu/ CAMDA Critical Assessment of Microarray Data Analysis: http://camda.bioinfo.cipf.es/camda2011/ Genome Access Workshop evaluation of statistical genetics approaches: http://www.gaworkshop.
2.3 Summary of community approaches for verification in the bio-sciences
In this section, we summarize five different verification approaches in the bio-sciences, with overlapping objectives but different scientific focus. A summary of these efforts is listed in Table 1.
CASP (Critical Assessment of protein Structure Prediction) is used to objectively test structure prediction methods against experimentally found structures in a worldwide-community context (Moult et al., 1995; Moult, 1996; Shortle, 1995). Even though the primary goal of CASP is to advance the methods of predicting protein 3D structure from its amino acid sequence, the pioneering efforts started by CASP have inspired other similar collaboration-by-competition challenges, such as those listed below.
CAPRI (Critical Assessment of PRediction of Interactions) is a community-wide experiment designed on the model of CASP (Wodak and Mendez, 2004). Both CASP and CAPRI are blind prediction experiments that rely on the willingness of structural biologists to provide unpublished experimental structures as targets. CAPRI is a blind test of the ability of protein–protein docking algorithms to predict the mode of association of two proteins based on their 3D structure.
DREAM (the Dialogue for Reverse Engineering Assessment and Methods) is a community-based effort whose goal is to help improve the state of the art in the experimental design, application and assessment of systems biology models. DREAM organizers do this through annual reverse-engineering and modeling challenges and conferences (Prill et al., 2010; Stolovitzky et al., 2007; Stolovitzky et al., 2009). The challenges, based on either new or pre-existing but obfuscated datasets, test participants in biological network inference and model predictions. Overall, a handful of best-performer teams are identified in each challenge, while some teams make predictions equivalent to random. As observed in many DREAM challenges, the aggregation of the predictions of all the teams improves the predictive power beyond that of any single method (G.Stolovitzky, personal communication), providing a sort of community wisdom that truly gives meaning to the notion of collaboration by competition.
BioCreAtIve is the Critical Assessment of Information Extraction systems in Biology. Patterned on CASP, BioCreAtIve is a community-wide project for assessing the application of information retrieval, information extraction and text mining to the biomedical literature. An example of a BioCreAtIve task is the recognition of gene names in sentences. Tasks are released biannually, with associated workshops for dissemination of the methods applied to the tasks by the participating researchers. Results and level of participation in BioCreAtIve I and II are detailed in (Hirschman et al., 2005; Morgan, Lu et al., 2008), where the lessons learned and the remaining opportunities in this important area of systems biology are also discussed.
FlowCAP is a community-based effort to develop new methods for flow cytometry applications. The motivation for the project comes from the rapid expansion of flow cytometry applications that have outpaced the functionality of traditional analysis tools used to interpret flow cytometry data. Hence, scientists are faced with the daunting prospect of manually identifying interesting cell populations in 20 dimensional data from a collection of millions of cells. For this reason a reliable automated approach to flow cytometry analysis is becoming essential. FlowCAP is a community-based project to assess the interpreting flow cytometry data and automated ‘gating’ of single-cell multi-variate data compared with gold standards based on manual gating.
2.4 Lessons from community approaches for verification in the biosciences
The discussion in the previous section supports the notion that different communities have embraced crowd-sourcing and collaborative-competition as an aid toward science verification and problem solving. The value of these efforts is well-demonstrated by the level of acceptance by their respective communities. The main goals of approaches such as CASP or DREAM are, within their respective areas of focus, to determine the state of the art in predictive models, to identify progress over time, to reveal bottlenecks that stymie progress and to show where effort may best be focused.
For all these efforts, clear ‘gold standards’ and metrics are necessary to quantify and score the entries of the participants. Three kinds of gold standards are commonly used. In one case, evoking the classical machine learning paradigm, some of the data is released as a training set whereas the remainder of the data is withheld as a gold standard test set. The second case consists of using an established method, a technology or a database accepted by the community as a reference. The third case consists of combining numerous datasets, algorithms or techniques, to get a closer estimate of the ground truth. A complication is that gold standard datasets are typically hard to obtain, and in many cases, are presently unobtainable in biology. For example, in protein structure prediction or macromolecular interactions, unpublished experimental structures can be hard to obtain, depending on the willingness of structural biologists to share their pre-publication data. On the other hand, the complete connectivity of a signaling network in a cell may be unobtainable with today's technology. Therefore, gold standards for signaling networks are lacking. There are solutions to this, however, such as requesting participants to train their network models to be consistent with measured levels of phospho-proteins provided in a training set, while testing the resulting models on their ability to predict levels of phospho-proteins under previously unseen perturbations provided in the test set (Prill et al., 2011).
Establishing a performance metric for scoring a challenge is another far-from-trivial task, which is central to challenge design. There is no unique or perfect scoring metric. The three main steps involved in evaluation are: (i) identification of a suitable metric (such as the area under the ROC and root mean square between prediction and measurement); (ii) simulation of a null distribution for the chosen metric by evaluation of randomly sampled predictions; and (iii) assignment of a P-value for a prediction with respect to the null distribution for the metric.
The choice of a useful scoring metric involves complexities that may not be as straightforward as one's intuition might suggest. Consider the case of CASP in which participants' predictions are compared with measured 3D structures. Early experience with matching only α-carbon position rather than side chains led to artifacts and over-fitting that were later addressed by more complex metrics than in averaged structure similarities over multiple spatial scales (Ben-David et al., 2009).
The invariance of the metric under different transformations of the data is another issue to take into account when scoring. For example, when testing a model prediction that spans a large dynamic range (such is the case in phosphoproteomics and gene expression measurements), a root mean square of the differences between predicted and measured variables may depend on the scale of interest. For example, the sum of differences squared in linear scale could overemphasize the difference over the large scales, whereas the sum of differences squared after log transforming the data amplifies the differences at the smaller values of the predictions. The results of such different measures of proximity could yield different best performers. Thus, aggregation of metrics plays an important role to balance the different biases imposed when choosing a metric.
Even in the simple case of binary classification, metrics such as area under the ROC curve, may be misleading if the positive and negative sets are very unbalanced, and it may need to be complemented with the area under the precision recall curve (Davis and Goadrich, 2006; Stolovitzky et al., 2007). Other typical performance metrics involve the correlation between the predicted values and the gold standard values. Potential correlation methods include rank correlation, linear correlation, correlation of the log-values and mutual information.
Combined community predictions can yield meta-predictions that are robust and often more accurate than any of the individual predictions. In CASP, Meta-servers that poll the results of automatic servers are among the best performers. Similar observations have been made for some of the DREAM challenges (Marbach et al., 2010; Prill et al., 2010).
Lessons from DREAM suggest that in the absence of first principle understanding, algorithms should be simple to avoid over-fitting to a particular dataset. In general, there is no one-size-fits-all algorithm, as the DREAM results have shown that the best algorithm depends on the subtleties of the data or on the system studied. For example, gene network reconstruction algorithms that may work very well in prokaryotes do not translate to eukaryotes, and data based on gene deletions have to be treated differently than data based on gene overexpression in network inference tasks.
The community-wide acceptance of these crowd-sourcing methodologies can be thought of in the context of the discussions between verificationists and falsificationists on when a theory is correct or not. Instead of choosing between validation and refutation the option is finding a practical solution that is accepted by the community. Of course, this acceptance is not arbitrary as the scientific community is the guardian of rigor and good science. The community acceptance of the efforts described here gives credibility to the use of the same techniques and challenges to check theories, hypothesis and models. How we can use this credibility to implement a methodology to verify systems biology results will be discussed next.
3 PROCESS OF VERIFICATION IN INDUSTRIAL RESEARCH
3.1 IMPROVER methodology: research workflow and building blocks
Among the lessons that we extracted from the community approaches described in the previous section, the notion that challenges can be used for science verification is paramount. In this section, we embrace that concept and present a methodology for process verification that can be used in industrial research workflows and other settings. We call this methodology IMPROVER, for Industrial Methodology for Process Verification in Research. IMPROVER evaluates the robustness of a research workflow by dividing it into building blocks that are relatively small and verifiable (Meyer et al., 2011). A building block is the small functional unit of a research pipeline that has a defined input (data, samples or materials), resulting in a defined output (data analyses, samples, high-throughput data or materials). Functionally, a building block is a discrete research operation at the small end of the scale that is amenable to verification. Similar divide and conquer approaches are employed in other fields. Typically, however, building blocks are developed around rigidly defined criteria in which the output is a known function of the input. In contrast, IMPROVER building blocks need to accommodate a priori unknown input–output functions. The development of appropriate scoring metrics is a key element to the verification methodology that helps identify the strength or weakness of a building block when a precise knowledge of an input–output relationship is not possible. The verification can be done internally by members of a research group, or externally by crowd-sourcing to an interested community. IMPROVER is, therefore, a mix of internal/non-public as well as external/public assessment tests or challenges.
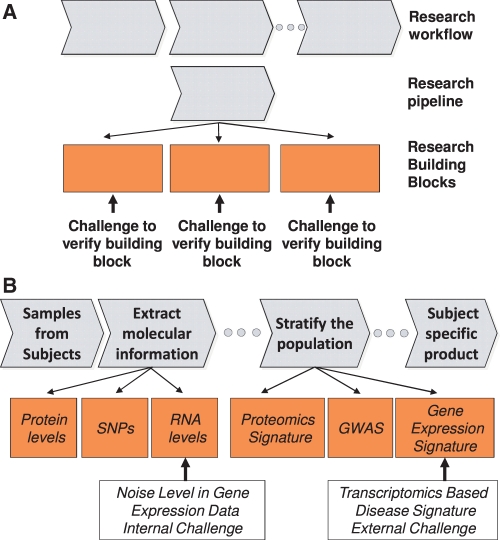
The concepts of research workflow and building blocks are clarified in Figure 1. The chain resulting from linking together the building blocks is a research ‘pipeline’ (Fig. 1A). The integration of several pipelines forms a research workflow. Note that there is no unique way of parceling a research pipeline into modules and building blocks. In general, however, any decomposition will ultimately have some interdependence on natural functional boundaries and the ability to isolate and verify the building block. In order to be verified, a research building block has to be recast into a challenge (similar to the challenges of the crowd-sourcing efforts discussed in the previous section), that may be assessed internally or broadcasted externally to stakeholders in the interested community. In both cases, the challenge construction has critical features such as producing the gold standard datasets that will be used as an anchor against which to compare the predictions of a challenge output, and the scoring methodology to assess the performance of the predictions.
Fig. 1.
Organization of a research workflow by decomposition into building blocks amenable to verification. (A) Research pipelines are indicated by the gray arrows, whereas the orange blocks are the more specific building blocks necessary to execute the pipeline. A concatenation of research pipelines forms a research workflow. Each of the building blocks in this diagram can be verified by the challenges indicated by the black arrows emerging from the orange blocks. (B) Example of a research pipeline including the challenges discussed in Section 3. For the internal challenge example, levels of RNA extracted from tissue or cells are measured with 2 different technologies, one of which is used as reference. For the external challenge example, gene expression data from patients and control subjects are used to test whether a disease signature can be extracted and verified.
Although IMPROVER has some commonalities with other crowd-sourcing methods, fundamental differences exist. Here we briefly highlight the differences between DREAM and IMPROVER. DREAM is a forum to organize the academic systems biology research community around challenges. These challenges are chosen by the DREAM organizers in collaboration with the community and are mostly structured to tackle independent problems in systems biology, with no specific link between challenges. DREAM challenges are widely advertised to the community, and its results are publicly announced. Conversely, IMPROVER challenges are designed following the interests of a research organization. These challenges, in turn, are designed to verify building blocks that work synergistically in a research workflow. Challenges performed to verify these building blocks can help the organization determine a way forward with respect to a previously laid plan: if the task that a building block was supposed to perform at a given level of accuracy is not verified, then the building block has to be modified. If a building block is verified, then its outcomes can be trusted with a higher degree of confidence. Examples of building block tasks and possible challenges to verify them are shown in Figure 1B. IMPROVER can pose its challenges internally, that is within the organization, or externally, to a wide community.
3.2 Internal challenges
An organization will use internal assessment challenges to verify in-house data generation, analysis and interpretation, either because of proprietary concerns or because the scope does not require a community effort. An IMPROVER challenge internal to an organization could help researchers identify building blocks that need either improvement or replacement with a new technology. As it will be described for external challenges, internal assessment challenges should be scored by an objective third party, who will not participate in the challenge but that could be from another group within the same company or institution. An internal challenge could be designed to evaluate the quality of data used for an external challenge. While data production can be ensured by Good Laboratory Practices (OECD 1998), the robustness of the technology used to collect the data may evolve in time, and therefore the quality of the data collection process itself may need to be verified (exemplified by the ‘Noise Level in Gene Expression Data’ challenge in Fig. 1B).
Consider that an organization must decide if the output data from the Gene Titan System for gene expression profiling from Affymetrix is of sufficient quality to consider its adoption. This technology allows researchers to process hundreds of samples in one experiment with minimal hands-on time, thus considerably increasing gene expression profiling throughput. An internal challenge is then constructed to compare the Gene Titan platform with the more established standard using Affymetrix single cartridge technology. A first verification challenge could consist of profiling a gold standard mRNA references sample, containing known quantities of spiked RNA. These reference samples, when hybridized on both technology arrays, would allow for the comparison of the sensitivities and error levels of both technologies. What is essential here is that the assessment be done by an objective third party who knows the composition of the reference sample that is unknown to the experimenter. In general, the IMPROVER internal challenge contribution to a research workflow will result in an understanding of the limitations of the methodology used in a pipeline. This understanding could be used to improve the results expected from a building block, thus increasing the robustness and value for the larger research pipeline.
3.3 External challenges/the first IMPROVER challenges
An external challenge can be designed to achieve multiple goals when aimed at verifying a building block within a pipeline. First, a public challenge invites novel approaches to a problem, not considered by the internal researchers. Second, a blended prediction aggregated from the entire community of predictions is often more accurate than any individual prediction (G.Stolovitzky, personal communication). Third, the public discourse centered on a challenge, including conference presentations and papers on the best-performing methods, can rapidly build a consensus in the community as to which approaches are the most fruitful for a given task. Fourth, if despite wide participation, no single team manages to achieve a good performance at solving the challenge, then the building block can be considered as non-verified, increasing the risk of failure of that building-block's pipeline.
Wide participation by the community is particularly important. While financial incentives are only one approach to increase participation, other incentives could be just as attractive, including the opportunity to verify the algorithm predictions against newly collected experiments, ‘bragging rights’ for the best algorithm, the ability to publish and to drive the field for purely academic interests.
We illustrate the concept of an IMPROVER external challenge using as an example the search for robust signatures to perform diagnosis of diseases based on commonly available transcriptomics data. There are examples of gene expressions signature in use today, such as Oncotype DX and MammaPrint, two FDA approved tests that provide prognostic value and can guide treatment in subsets of breast cancer patients (Paik et al., 2004; van de Vijver et al., 2002). While diagnostic signatures exist in limited cases, the wide availability of high-throughput transcriptomics data makes plausible the discovery of diagnostic signatures for a multitude of diseases. The community has recognized the need for robust genomic and gene expression signatures as important enablers for personalized medicine, as patients could directly benefit from treatments tailored to the individual (Subramanian and Simon, 2010).
While there has been a clear need for diagnostic signatures, efforts to discover such signatures in commonly available transcriptomics data have generally fallen short of expectation. There are many reports in the literature in which the lists of differentially expressed genes purported to distinguish between two biological conditions showed little overlap when the data were taken from different cohorts or when experiments were performed in different laboratories with different platforms (Ioannidis, 2005). Hence, the discovered signatures do not generalize and perform poorly when classifying datasets other than the ones used to develop the methods. Even with good control over data collection and patient selection, signature discovery can be inhibited by inherent variability in gene expression. One proposed method to discover robust classifiers in spite of inherent variability is to separate ‘driver genes’ from the ‘passenger genes’ (Lim et al., 2009). The driver genes (sometimes referred to as master regulators) are upstream controllers that are proposed to be better indicators of disease state than the downstream regulated genes that can show more inherent variability.
The first set of IMPROVER challenges, termed the Diagnostics Signature Challenge, addresses the problem of diagnostics from transcriptional data in a biomedical context. (This challenge is being organized at the time of this writing.) The need to find biomarkers that stratify a population into segments characterized by a given phenotype is felt not just in biomedicine but also in other contexts such as the pharmaceutical industry, where a similar IMPROVER challenge could be deployed. We consider four prevalent diseases: multiple sclerosis (MS), psoriasis, lung cancer and chronic obstructive pulmonary disease. The building block that this challenge is designed to verify is ‘Find Gene Expression Signature’ (Fig. 1B). In other words, what needs to be verified is the hypothesis that transcriptomics data contains enough information for the determination of these human disease states. In a context such as the pharmaceutical industry, a test of validity of the notion of transcriptomics-based signatures would be a pre-requisite to attain the research pipeline goal of finding a product (such as a drug) tailored for each individual (Fig. 1B).
We will now describe the operational steps for the Diagnostic Signature Challenge taking out of the four diseases, MS as an example. MS is an inflammatory disease, believed to be an autoimmune disease that affects the central nervous system. The trigger of the autoimmune process in MS is unknown, but it is believed that MS occurs as a result of some combination of genetic, environmental and infectious factors (Compston and Coles, 2008), and possibly other factors such as vascular problems (Minagar et al., 2006). The symptoms of the disease result from inflammation, swelling and lesions on the myelin and in 85% of patients start with a relapse-remitting stage of MS (RRMS). Finding a robust genetic signature would be of great importance, as diagnosis by a neurologist usually involves ruling out other nervous system disorders with invasive and expensive tests (NINDS Multiple Sclerosis Information Page, http://www.ninds.nih.gov/disorders/multiple_sclerosis/multiple_sclerosis.htm) and recently drugs can delay the progression of MS when RRMS, is diagnosed early on (Rudick et al., 2006).
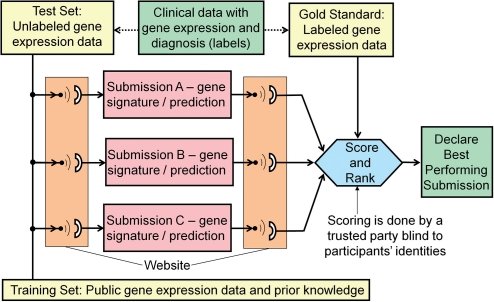
IMPROVER organizers will procure from the public literature, a training set of gene expression data from peripheral blood mononuclear cells (PBMCs) corresponding to MS and healthy patients (Fig. 2). In this challenge, the test set corresponds to an unpublished cohort of 129 samples whose labels will be hidden from the participants. This set of samples obtained from patients that were determined as healthy or RRMS by a physician will constitute the gold standard. A wealth of additional useful gene expression data is also available through databases such as the Gene Expression Omnibus or ArrayExpress. Participants can use the training set, open literature information and any other publicly available data. With this data at hand, participants will generate the transcriptomics-based molecular signature that can differentiate between healthy and RRMS patients. Participants will be asked to submit for each sample a confidence of the prediction to belong to the RRMS class. The confidence of the classification should have a value between 1 and 0, 1 being the most confident and 0 the least confident.
Fig. 2.
Schematic diagram of MS Disease signature challenge organization. A dataset with both gene expression and corresponding clinical diagnoses or prognosis forms the basis of the challenge. The test data contains the gene expression data generated only and is transmitted to the participants via a web portal. There are three participants shown, the actual challenges could involve many more. The participants generate predictions-based gene signatures that are submitted back via the website. A trusted party will blindly score and rank the prediction by comparing to the gold standard dataset that contains both the gene expression data and actual clinical outcomes.
After predictions from participants are collected via website submissions, the results will be scored using metrics such as the Area Under the Precision versus Recall (AUPR) curve. Precision is defined as the fraction of correct positive set predictions, and recall is the proportion of correct positive set predictions out of all patients in the positive set. Other metrics for binary classification assessment will also be evaluated. Teams will be ranked according to their overall performance based on those metrics. Figure 2 illustrates how the MS disease signature challenge will be organized in order to verify through the IMPROVER methodology whether a robust MS gene signature can be found. A diagnostic signature for those phenotypes can be accepted as existing, and the building block ‘Find a Transcriptomics-based signature for control versus RRMS’ verified, only if there is at least one participating team who classified in the correct class a statistically significant number of subjects. A subsequent verification of the molecular signature discovered by the best performer could be further tested by evaluating its performance in a similar, but biologically independent dataset. Finally, if no team managed to distinguish the RRMS patients from healthy donors from PBMC transcriptomics data, then we can assert that the building block failed verification, and an alternative way of classification should be explored.
If the building block was verified, an obvious by-product of the challenge is the identification of the best diagnostic signature and the corresponding discovery algorithm for each of the diseases. Other expected advantageous outcome of the IMPROVER challenge is that it enables a fair comparison of competing methods, as the IMPROVER format requires blind prediction by the participants and blind scoring of the submissions (Fig. 2). This approach will alleviate many of the problems that produce overestimation of results when the authors of an algorithm compare their own method with other existing methods (Norel et al., 2011). For example, over-fitting and information leakage between training and test datasets are two common pitfalls that can be avoided. A final advantage of the methodology is that it allows for an assessment of the performance of submissions across both participants and diseases. This will provide an unparalleled opportunity to assess whether the diagnostic signature discovery approaches can be applied across different diseases. Such a controlled assessment is harder to reach with traditional scientific approaches, as it requires a wide variety of participants using different methodologies on the same data and scored under the same metrics.
3.4 Gold standard and metrics of performance
A foremost concern in designing a challenge for IMPROVER is to obtain a gold standard dataset against which a set of predictions can be scored in order to verify a building block. While designing a challenge to verify a building block, the possibility exists that a gold standard cannot be defined or is considered suboptimal as an adequate database, unpublished good quality data or an accessible expert in the field is unavailable. In this case, the rationale behind the challenge has to be altered and the challenge must be redesigned before the building block can be verified. Redesigning a challenge can be laborious as it might imply obtaining data for a new gold standard and change assumptions that simplified the underlying biology and favored a good challenge formulation.
A building block can be considered as verified if the predictions made within the challenge are close enough to the known gold standard. For each challenge, a quantitative metric of performance must be defined. Like the gold standard, the performance metric is central and should be an integral part of the challenge formulation. This performance metric can also be used to assign a risk that the verification was a fluke (e.g. computing a P-value). It is also possible that a challenge results in lack of verification: none of the participating teams could find an acceptable solution to the problem.
There is generally no a priori reason why one metric should be better than the others. As a rule of thumb, aggregating the several metrics into one overall metric may have advantages and provide less arbitrary performance metric. In other cases, however, the nature of the problem guides the choice of metric. For example, the large dynamic range of gene expression data suggest a performance metric in which the values are represented in logarithmic scale.
4 CONCLUSION AND FUTURE DIRECTIONS
The great opportunities made possible by the emergence of high-throughput data in all realms of science and technology have also resulted in the problem of extracting knowledge from these massive datasets. The proliferation of algorithms to analyze this data creates the conundrum of choosing the best algorithms among the multiple existing ones. Crowd-sourcing efforts that take advantage of new trends in social networking have flourished. These initiatives, summarized in Section 2, match discipline-specific problems with problem solvers, who are motivated by different incentives to compete and show that their solution is the best. In this way, the best method available to solve a given problem can be found in an unbiased context.
Interestingly, these crowd-sourcing methodologies also have an epistemological value, shedding light to the question of when a theory is correct or not. Instead of tasking a researcher to self-assess (a process suspect of biases) the truth of a model or methodology, the alternative is finding how it fares in an unbiased and rigorous test. The community acceptance of the efforts described in the first part of this article gives some credibility to the use of similar approaches to verify the sometime elusive results attained in systems biology research.
Extrapolating the idea of using challenges for verification of scientific results, we propose the IMPROVER methodology to assess the performance of a research workflow in contexts such as industrial research. A main concept in IMPROVER is the formalization of a process to determine a go or no-go decision for the research pipeline in an industrial context (internal and external challenges), as well as better methods inspired by the community participation (external challenges). If the results are positive, that is, if the pipeline passes all the challenges and there is active community participation, then the credibility of the data, analysis and of the subsequent results would be enhanced in the eyes of the scientific community and regulatory agencies.
The challenge-based approach creates a metric for comparison between possible solutions to a challenge designed to verify a building block. Superior performance by one methodology could promote acceptance by the community of the best performer methodology as a reference standard. IMPROVER could offer a complement and enhancement to the peer-review process in which the results of a submitted paper are measured against benchmarks in a double-blind challenge, a process that can well be called challenge-assisted peer-review. The IMPROVER approach could be applied to a variety of fields where the outputs of a research project are fed into the input of other projects, such as is the case in industrial research and development, and where the verification of the individual projects or building blocks is elusive, as is the case in systems biology.
ACKNOWLEDGEMENTS
We thank Robert J. Prill and Alberto de la Fuente for useful discussions and Hugh Browne and Jennifer Galitz McTighe for a careful reading of the manuscript.
Funding: IBM and PMI authors performed this work under a joint research collaboration funded by PMI.
Conflict of Interest: none declared.
REFERENCES
- (2002) SEGEN JC. McGraw-Hill Concise Dictionary of Modern Medicine©. McGraw-Hill.
- Alberts B., et al. Reviewing peer review. Science. 2008;321:15. doi: 10.1126/science.1162115. [DOI] [PubMed] [Google Scholar]
- Ayer A.J. Language, Truth, and Logic. Oxford University Press; 1936. [Google Scholar]
- Ben-David M., et al. Assessment of CASP8 structure predictions for template free targets. Proteins. 2009;77(Suppl. 9):50–65. doi: 10.1002/prot.22591. [DOI] [PubMed] [Google Scholar]
- Compston A., Coles A. Multiple sclerosis. Lancet. 2008;372:1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- Davis J., Goadrich M. Proceedings of the 23rd International Conference on Machine learning. Pittsburgh, Pennsylvania: ACM; 2006. The relationship between Precision-Recall and ROC curves; pp. 233–240. [Google Scholar]
- Dougherty E.R. Validation of gene regulatory networks: scientific and inferential. Brief Bioinform. 2011;12:245–252. doi: 10.1093/bib/bbq078. [DOI] [PubMed] [Google Scholar]
- Dreze M., et al. High-quality binary interactome mapping. Methods Enzymol. 2010;470:281–315. doi: 10.1016/S0076-6879(10)70012-4. [DOI] [PubMed] [Google Scholar]
- Fayyad U., et al. The KDD process for extracting useful knowledge from volumes of data. Commun. ACM. 1996;39:27–34. [Google Scholar]
- Gavin A.C., et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Hirschman L., et al. Overview of BioCreAtIvE: critical assessment of information extraction for biology. BMC Bioinformatics. 2005;6(Suppl. 1):S1. doi: 10.1186/1471-2105-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis J.P.A. Microarrays and molecular research: noise discovery? Lancet. 2005;365:454–455. doi: 10.1016/S0140-6736(05)17878-7. [DOI] [PubMed] [Google Scholar]
- Ito T., et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. In. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelizarow M., et al. Over-optimism in bioinformatics: an illustration. Bioinformatics. 2010;26:1990–1998. doi: 10.1093/bioinformatics/btq323. [DOI] [PubMed] [Google Scholar]
- Kuhn T. The structure of scientific revolutions. University of Chicago Press; 1962. [Google Scholar]
- Lim W.K., et al. Master regulators used as breast cancer metastasis classifier. Pac. Symp. Biocomput. 2009:504–515. [PMC free article] [PubMed] [Google Scholar]
- Mandavilli A. Peer review: trial by Twitter. Nature. 2011;469:286–287. doi: 10.1038/469286a. [DOI] [PubMed] [Google Scholar]
- Marbach D., et al. Revealing strengths and weaknesses of methods for gene network inference. Proc. Natl. Acad. Sci. USA. 2010;107:6286–6291. doi: 10.1073/pnas.0913357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta T., et al. Towards sound epistemological foundations of statistical methods for high-dimensional biology. Nat. Genet. 2004;36:943–947. doi: 10.1038/ng1422. [DOI] [PubMed] [Google Scholar]
- Meyer P., et al. Verification of systems biology research in the age of collaborative competition. Nat. Biotech. 2011;29:811–815. doi: 10.1038/nbt.1968. [DOI] [PubMed] [Google Scholar]
- Minagar A., et al. Multiple sclerosis as a vascular disease. Neurol. Res. 2006;28:230–235. doi: 10.1179/016164106X98080. [DOI] [PubMed] [Google Scholar]
- Morgan A.A., et al. Overview of BioCreative II gene normalization. Genome Biol. 2008;9(Suppl 2):S3. doi: 10.1186/gb-2008-9-s2-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moult J., et al. A large-scale experiment to assess protein structure prediction methods. Prot. Struct. Func. Bioinform. 1995;23:ii–iv. doi: 10.1002/prot.340230303. [DOI] [PubMed] [Google Scholar]
- Moult J. The current state of the art in protein structure prediction. Current Opinion in Biotechnology. 1996;7:422–427. doi: 10.1016/s0958-1669(96)80118-2. [DOI] [PubMed] [Google Scholar]
- Norel R., et al. The self-assessment trap: can we all be better than average? Mol. Syst. Biol. 2011;7:537. doi: 10.1038/msb.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organisation for Economic Cooperation and Development. OECD Good Laboratory Practice - Principles and Guidance for Compliance Monitoring. OECD press; 1998. [Google Scholar]
- Paik S., et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. New Engl. J. Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- Popper K.R. The Logic of Scientific Discovery. Routledge Classics; 1959. [Google Scholar]
- Prill R.J., et al. Towards a rigorous assessment of systems biology models: the DREAM3 challenges. PLoS One. 2010;5:e9202. doi: 10.1371/journal.pone.0009202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prill R.J., et al. Crowdsourcing network inference: the DREAM predictive signaling network challenge. Sci. Signal. 2011;4:mr7. doi: 10.1126/scisignal.2002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudick R.A., et al. Natalizumab plus interferon beta-1a for relapsing multiple sclerosis. New Engl. J. Med. 2006;354:911–923. doi: 10.1056/NEJMoa044396. [DOI] [PubMed] [Google Scholar]
- Spier R. The history of the peer-review process. Trends Biotechnol. 2002;20:357–358. doi: 10.1016/s0167-7799(02)01985-6. [DOI] [PubMed] [Google Scholar]
- Stolovitzky G., et al. Dialogue on reverse-engineering assessment and methods. Ann. NY. Acad. Sci. 2007;1115:1–22. doi: 10.1196/annals.1407.021. [DOI] [PubMed] [Google Scholar]
- Stolovitzky G., et al. Lessons from the DREAM2 challenges. Ann.NY. Acad. Sci. 2009;1158:159–195. doi: 10.1111/j.1749-6632.2009.04497.x. [DOI] [PubMed] [Google Scholar]
- Subramanian J., Simon R. What should physicians look for in evaluating prognostic gene-expression signatures? Nat. Rev. Clin. Oncol. 2010;7:327–334. doi: 10.1038/nrclinonc.2010.60. [DOI] [PubMed] [Google Scholar]
- Uetz P., Hughes R.E. Systematic and large-scale two-hybrid screens. Curr. Opin. Microbiol. 2000;3:303–308. doi: 10.1016/s1369-5274(00)00094-1. [DOI] [PubMed] [Google Scholar]
- van de Vijver M.J., et al. A gene-expression signature as a predictor of survival in breast cancer. New Engl. J. Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Wodak S.J., Mendez R. Prediction of protein-protein interactions: the CAPRI experiment, its evaluation and implications. Curr. Opin. Struct. Biol. 2004;14:242–249. doi: 10.1016/j.sbi.2004.02.003. [DOI] [PubMed] [Google Scholar]