Abstract
Summary: Molecular networks are often studied in diverse cellular or experimental contexts, with highly context-specific details. Modelling introduces further choices as to levels of mathematical description. The resulting possibilities are difficult to explore rapidly, hampering the integration of modelling and experiment. We have developed Proteus, a web-based, context-specific tool for building compartmentalized, ordinary differential equation (ODE) models. It is inspired by the idea of a molecular ‘toolkit’ for Ca2+ signalling. Toolkits in Proteus are context-independent representations of biological systems as sets of components, which may correspond to mechanisms of differing levels of complexity. Users pick and choose components from a toolkit and, for each component, pick and choose from different mechanisms, each of which describes a different instantiation of the component's mechanism. Proteus combines these choices into a system of ODEs, which may then be downloaded in SBML (Systems Biology Markup Language), Matlab or Fortran format and independently analyzed. Toolkits, components and mechanisms are user-constructible, either de novo or by cannibalizing existing models, including all those in the Biomodels database. A wide variety of context-specific models may thereby be rapidly built, modified and explored.
Availability and implementation: Proteus, implemented in C#, and a prototype toolkit for modelling calcium signalling are freely and universally available at www.modularmodeling.com
Contact: gnad.florian@gene.com; jeremy@hms.harvard.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Mathematical models are increasingly used with experimental approaches to analyze the functional complexities of molecular networks. One of the challenges in doing this is model re-construction in the light of new experiments, new knowledge, changing biological context and changing levels of mathematical description. New approaches have emerged to shift the burden of model re-construction to software. The ‘models as programs’ approach has brought programming principles to bear, both at the low level of molecular domains, through rule-based methodologies, (Faeder et al., 2009; Hlavacek et al., 2006), and, at a higher level of modularity, through the functional paradigm, (Ginkel et al., 2003; Mallavarapu et al., 2009). Here, we take a complementary approach inspired by biological ideas. Compared to other tools, the main advantage of Proteus is modularity allowing the user to model complex systems by combining simpler components via a web-based ‘pick and choose’ approach. Moreover, Proteus allows the establishment of a user community that manages and shares toolkits and mechanisms.
Experimental work typically focusses on a specific biological context, such as the ‘NF-κB pathway’ or ‘calcium signalling’. Such contexts allow for much variation in detail but also for constraining assumptions about the molecular players and their underlying mechanisms. For instance, calcium signalling is thought to involve a ‘calcium signalling toolkit’, with a limited repertoire of functional parts—transporters, pumps, calcium-sensitive enzymes, buffering proteins, organelle stores and leak currents—from which mammalian cells mix and match-specific molecular mechanisms to orchestrate the calcium response appropriate to their biological context, (Berridge et al., 2003). We exploit the biological idea of a toolkit by analogy at the software level.
2 USING A TOOLKIT
A toolkit in Proteus is a collection of components that provide a context-independent representation of a biological context. Components are modular subsystems of a whole model and can encompass many levels of granularity, from individual proteins, to entire subnetworks, such as multi-protein complexes or kinase cascades. Context-dependency arises from choosing which components are relevant to the context and, for each component, which mechanism is appropriate. Mechanisms can reflect both biological distinctions, such as splice variants or regulatory changes between cell types, as well as differences in the granularity of mathematical assumptions, such as numbers of post-translational modification states.
Users may create models by opening an existing toolkit (in the ‘Open Toolkit’ menu), choosing components and, for each component, a corresponding mechanism, in a modular way (Fig. 1). In the case of Ca2+ signalling, components interact largely through Ca2+ itself, by regulating its transfer between different stores. This makes the modularity relatively easy to implement. In other cases, juxtaposing mechanisms for different components may result in more complex interactions, such as binding or enzymatic activity, that create new chemical entities not necessarily present in the individual mechanisms, (Mallavarapu et al., 2009). At present, the user has to address these context-dependencies on an ad hoc basis. Any model may be compiled into Systems Biology Markup Language (SBML) (Novére et al., 2006), which serves as a publicly-accessible representation. It may also be compiled into Matlab functions or, for those requiring more control over numerical integration, into Fortran code. Compiled versions may be saved locally on the user's computer and independently simulated and analyzed.
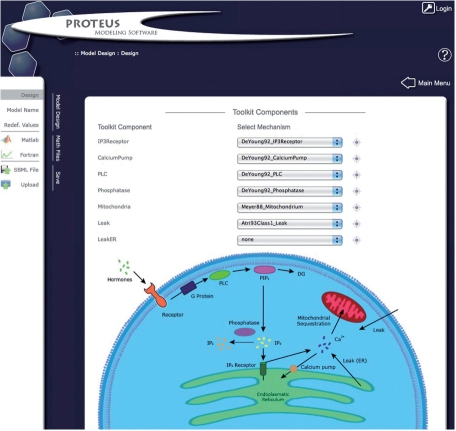
Fig. 1.
Calcium toolkit in Proteus. The screenshot shows the menu when the user is building a model from the calcium toolkit. A list of components is shown, for each of which the user can access a pulldown menu of various mechanisms. By choosing appropriate mechanisms, the user can duplicate an existing model or build an entirely new one in a modular way.
3 CREATING A TOOLKIT
To build a toolkit in the ‘Create Toolkit’ menu, users have to create mechanisms first (in the menu ‘Create Mechanism’). As described above, toolkits are collections of components, to each of which is associated any number of mechanisms. Different mechanisms may reflect cell-type-specific differences or differences in the level of mathematical description. Mechanisms are defined in terms of compartments, species and reactions in a way familiar to anyone using compartmentalized ordinary differential equations or SBML (Hucka et al., 2004). Compartments typically correspond to 3D volumes: a dish containing cells or the nucleus or the cytoplasm of a single cell. They can also represent 2D surfaces, such as the inner face of the plasma membrane. Compartment sizes may be specified when the compartment is defined. Species are chemical entities that participate in reactions. They are localized to a specific compartment, specified upon definition, so if the same chemical entity is found in several compartments, a different species is defined for each of these. Reactions are treated as a link between substrate species and product species with specified stoichiometry. The user provides a formula for the rate of the reaction, specified in Matlab syntax. Any names in the formula that do not correspond to defined species are treated as parameters, which are extracted and made available independently. The values of parameters or the initial concentrations of species are only needed at simulation time; Proteus gives them default values unless the user specifies them. Since mechanisms may be at any level of granularity, the entire network or pathway that is being studied may be defined as a mechanism and compiled into a working model. Conversely, any existing SBML file may be uploaded and its entities made available. This provides several useful facilities. For instance, any model in the Biomodels database may be uploaded to Proteus, modified in terms of its compartments, species, reactions or parameters and then saved as SBML or compiled into Matlab or Fortran. Cannibalizing existing models is often the easiest way to make new mechanisms.
Once a library of mechanisms has been established, users may combine mechanisms and assign them to components in the ‘Combine Mechanisms’ menu. The creation of components along with the assignment of mechanisms to components is the final step in creating a toolkit. A more detailed guide on how to use and create toolkits is provided in the online help menu as well as in the Supplementary Materials.
We have created a prototype Ca2+-signalling toolkit that includes mechanisms described in eight papers. The toolkit has been uploaded to Proteus and is available to any user. A significant feature of Ca2+-signalling is that many hormones elicit repetitive Ca2+ spikes and this has been widely studied experimentally and mathematically (Schuster et al., 2002; Woods et al., 1986). We were interested to know how spiking depended on the particular assumptions behind each model. We used the toolkit to build a range of ‘Frankensteinian’ models, which mixed mechanisms from different papers. This often abolished spiking at the parameter values used in the original papers but other parameter values could be found at which spiking was restored. This suggests that spiking is robust to different mechanistic assumptions and, consequently, that the mere occurrence of spiking does not help to discriminate between different models. A full discussion will appear elsewhere. The Matlab program used for parameter searching is also available from the Proteus website.
4 OUTLOOK
We envisage Proteus being used to incrementally build up toolkits for individual molecular networks like ‘calcium’, ‘NF-κB’, ‘Wnt’, etc. Different models of a network could then be brought together under a common format and namespace, comparisons more easily made and new models more easily constructed and tested. Context-sensitive biochemical and dynamical knowledge about a network could thereby evolve through collective interactions between independent researchers. Such a facility would not only help to integrate experiment and modelling but also to nucleate new research communities. We hope this application note will increase the user community and help in further improving Proteus. We believe that tools of this kind will become increasingly important in tackling the molecular complexity underlying cellular behaviour.
Funding: NIH under R01-GM081578.
Conflict of Interest: none declared.
Supplementary Material
Footnotes
†Present address: Department of Bioinformatics and Computational Biology, Genentech Inc., South San Francisco, California 94080, USA.
REFERENCES
- Berridge M.J., et al. Calcium signalling: dynamics, homeostasis and remodelling. Nature Rev. Mol. Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- Faeder J.R., et al. Rule-based modeling of biochemical systems with BioNetGen. Methods Mol. Biol. 2009;500:113–167. doi: 10.1007/978-1-59745-525-1_5. [DOI] [PubMed] [Google Scholar]
- Ginkel A., et al. Modular modelling of cellular systems with ProMot/Diva. Bioinformatics. 2003;19:1169–1176. doi: 10.1093/bioinformatics/btg128. [DOI] [PubMed] [Google Scholar]
- Hlavacek W.S., et al. Rules for modeling signal-transduction systems. Sci. STKE. 2006;344:re6. doi: 10.1126/stke.3442006re6. [DOI] [PubMed] [Google Scholar]
- Hucka M., et al. Evolving a lingua franca and associated software infrastructure for computational systems biology: the Systems Biology Markup language (SBML) project. Syst. Biol. (Stevenage) 2004;1:41–53. doi: 10.1049/sb:20045008. [DOI] [PubMed] [Google Scholar]
- Mallavarapu A., et al. Programming with models: modularity and abstraction provide powerful capabilities for systems biology. J. R. Soc. Interface. 2009;6:257–270. doi: 10.1098/rsif.2008.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novére N.L., et al. Biomodels database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res. 2006;34:D689–D691. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S., et al. Modeling of simplex and complex calcium oscillations: from single-cell responses to intercellular signaling. Eur. J. Biochem. 2002;269:1333–1355. doi: 10.1046/j.0014-2956.2001.02720.x. [DOI] [PubMed] [Google Scholar]
- Woods N.M., et al. Repetitive transient rises in cytoplasmic free calcium in hormone-stimulated hepatocytes. Nature. 1986;319:600–602. doi: 10.1038/319600a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.