Abstract
Summary: The affordability of high-throughput sequencing has created an unprecedented surge in the use of genomic data in basic, translational and clinical research. The rapid evolution of sequencing technology, coupled with its broad adoption across biology and medicine, necessitates fast, collaborative interdisciplinary discussion. SEQanswers provides a real-time knowledge-sharing resource to address this need, covering experimental and computational aspects of sequencing and sequence analysis. Developers of popular analysis tools are among the >4000 active members, and ~40 peer-reviewed publications have referenced SEQanswers.
Availability: The SEQanswers community is freely accessible at http://SEQanswers.com/
Contact: david.mittelman@vt.edu; ecolivares@gmail.com
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The Human Genome Project represents one of the greatest concerted achievements of the life sciences. This massive global effort jump-started the genomics era and enabled more ambitious and collaborative projects such as the Cancer Genome Atlas (Cancer Genome Atlas Research Network, 2008), 1000 Genome Project (1000 Genomes Project Consortium, 2010) and Human Microbiome Project (The NIH HMP Working Group et al., 2009). These large population-scale studies, powered by high-throughput sequencing (HTS) technologies, have generated massive amounts of genomic data with the potential to revolutionize genetics and medicine.
The translation of these data to actionable medicine, however, is complicated by the challenges of extracting meaningful information from HTS data (Mardis, 2010). The challenge is not purely computational, as bioinformatics is bound by the experimental methods employed to produce genomic data (Alkan et al., 2011). A successful experiment minimizes false positives and depends on the optimization of an entire pipeline, from sample preparation to computational analysis.
As HTS begins to transform nearly all aspects of biological and medical science, more labs will incorporate the production and analysis of genomic data into their studies. However, these experimental and computational methods are evolving at an incredible pace and it is increasingly challenging for smaller research groups outside of major genome centers to stay current. Real-time, interdisciplinary collaboration helps large genome centers optimize analysis pipelines and methods, and allows smaller groups to exploit them, even if they did not have resources to facilitate the initial development.
2 THE SEQANSWERS COMMUNITY
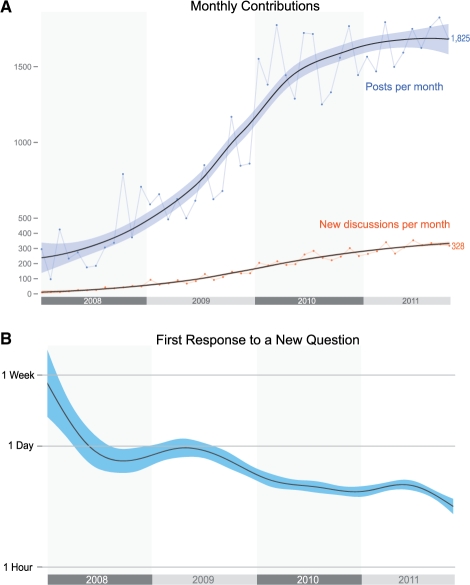
SEQanswers was launched in 2007 as an open forum to enable scientists across disciplines to collaboratively advance genomics and, particularly, HTS technologies. To date, there are >4000 active users visiting the online community each month. There is a rapidly growing number of discussion threads (currently >10 000) that span topics from sequencing platforms, experimental design, data analysis and biological interpretation (Fig. 1A). The SEQanswers community is truly global (Supplementary Fig. S1) and includes members from major genome centers and individual groups, as well as key developers of popular data analysis tools and methods. The community currently hosts >300 new questions, and 1800 new responses per month. This incredibly high rate of participation has led to rapid responses to questions, shortening initial response time from a week in early 2008 to less than a day in 2011 (Fig. 1B). Collaborative and transparent discussion on SEQanswers has triggered the development of new experimental techniques, data analysis methods and pipelines, as well as collaborative assessment of analysis standards (Supplementary Table S1). This innovation is captured in part by >30 peer-reviewed publications that cite SEQanswers so far (http://seqanswers.com/wiki/Papers_Referencing_SEQanswers).
Fig. 1.
SEQanswers is an active and fast growing community. (A) Monthly contributions to SEQanswers measured by the number of new posts (blue points/line) and discussions (orange points/line). Discussion counts include threads with at least two posts and exclude those with no answers. Also excluded are automated publication announcements. (B) The average response time to a new forum thread.
SEQanswers is not the only online resource for knowledge sharing and collaboration: major sequencing technology companies have platform-centric user communities, but these are often restricted to customers and exclude the greater scientific community. In contrast, BioStar (Parnell et al., 2011), an open, community-driven bioinformatics resource, currently hosts >2300 threads, which far exceeds the sum of discussions found on communities operated by sequencing companies (Supplementary Table S2). BioStar's principle feature is to enable researchers to ask questions and obtain brief answers, ranked by community vote, to bioinformatics-related problems. BioStar's success can be attributed to its well-defined scope and focus on a simple question and answer format for bioinformatics. However, this format precludes other forms of collaboration, discussion and debate.
SEQanswers differs from BioStar both in format and scope. In an almost complementary capacity, SEQanswers eschews the Q&A format in favor of a more traditional forum format to facilitate collective discussion of technologies, methods and standards of practice. The traditional forum format emphasizes the chronology and evolution of collective thought, rather than focusing on identifying a single, best answer. The scope of SEQanswers differs from BioStar's exclusive bioinformatics focus, including all aspects of genomics, experimental and computational. Finally, in recognition of the sometimes lengthy and tediously detailed threads that can emerge from sequential discussion, we have developed a manually curated database, SEQwiki (Li et al., 2012), that consists of frequently asked questions, analysis methods, tutorials and sequencing service providers.
3 CONCLUSION
The massive amounts of data and rapid pace of genome technology development necessitates innovations in scientific communication. The current standard for scientific communication between disparate research groups focuses on peer-reviewed research published in traditional scientific journals. These journals have evolved for the Internet age, especially with the new emphasis on open access and fast publishing from both new, exclusively open access journals to traditional journals that have created new outlets for open access publication. While scientific journals will continue to have important roles as curators of research and referees for the peer-review process, there is an opportunity for open, internet-based platforms to supplement traditional journals by enabling the rapid exchange of results, techniques and data, the latter two being crucial for advancing research,1 but notoriously difficult to access. SEQanswers was designed to address this need for genomics. The community has since developed into a thriving community that offers a wealth of information, including discussions that have facilitated the construction of analysis pipelines and consensus on standards in the genomics community.
Supplementary Material
ACKNOWLEDGEMENT
The authors would like to thank the members of the SEQanswers community for helpful suggestions on the manuscript.
Funding: This work was supported by an award through the NVIDIA Foundation's ‘Compute the Cure’ program to D.M.
Conflict of Interest: none declared.
Footnotes
1In a global survey, 62% of respondents claimed data accessibility is very important among all information to their research (Elsevier, 2010).
REFERENCES
- 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C., et al. Limitations of next-generation genome sequence assembly. Nat. Methods. 2011;8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsevier. Access vs. Importance – A Global Study Assessing the Importance of and Ease of Access to Professional and Academic Information – Phase I Results. Publishing Research Consortium; 2010. [Google Scholar]
- Li J.W., et al. The SEQanswers wiki: a wiki database of tools for high-throughput sequencing analysis. Nucleic Acids Res. 2012;40:D1313–D1317. doi: 10.1093/nar/gkr1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis E.R. The $1,000 genome, the $100,000 analysis? Genome Med. 2010;2:84. doi: 10.1186/gm205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell L.D., et al. BioStar: an Online Question & Answer Resource for the Bioinformatics Community. PLoS. Comput. Biol. 2011;7:e1002216. doi: 10.1371/journal.pcbi.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The NIH HMP Working Group et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.