Abstract
The advancement of neuroscience, perhaps one of the most information rich disciplines of all the life sciences, requires basic frameworks for organizing the vast amounts of data generated by the research community to promote novel insights and integrated understanding. Since Cajal, the neuron remains a fundamental unit of the nervous system, yet even with the explosion of information technology, we still have few comprehensive or systematic strategies for aggregating cell-level knowledge. Progress toward this goal is hampered by the multiplicity of names for cells and by lack of a consensus on the criteria for defining neuron types. However, through umbrella projects like the Neuroscience Information Framework (NIF) and the International Neuroinformatics Coordinating Facility (INCF), we have the opportunity to propose and implement an informatics infrastructure for establishing common tools and approaches to describe neurons through a standard terminology for nerve cells and a database (a Neuron Registry) where these descriptions can be deposited and compared. This article provides an overview of the problem and outlines a solution approach utilizing ontological characterizations. Based on illustrative implementation examples, we also discuss the need for consensus criteria to be adopted by the research community, and considerations on future developments. A scalable repository of neuron types will provide researchers with a resource that materially contributes to the advancement of neuroscience.
Keywords: neuron, property, type, part, relationship, value, ontology, characterization
Introduction
To understand the brain, one must first understand its component parts and their relationships to one another. A key component of the brain, from an information processing perspective, is the neuron. There are estimated to be 1011 neurons (Azevedo et al., 2009; Herculano-Houzel, 2009) in the human brain with 1015 connections (Sporns, 2011). It is uncontroversial that all neurons cannot simply be considered the same; at the same time, it is impractical and most likely of little value to consider all 1011 unique. Traditionally, neurons have been classified on the basis of one or more morphological, physiological, molecular, and/or developmental properties. Yet, there are relatively few clearly identified cell classes; rather the literature is rife with partially overlapping or irreconcilable classification schemes published for a particular brain region and usually based on a single technique. Trying to integrate these different classification schemes is bewildering at best and impossible at worst. Yet, interrelating different experimental findings requires common points of reference (Ref). Since the nineteenth century, the cell has provided such point of Ref for most tissues of the body. We believe it is time for the neuroscience community to come together and jointly devise a strategy for dealing with the cellular complexity of the nervous system. Specifically, we call for a common description framework to describe, identify, and name neuron types. In this perspective, we discuss current national and international efforts to begin to address the complexity of neuronal types within the Neuroscience Information Framework (NIF) and the International Neuroinformatics Coordinating Facility (INCF (http://incf.org) Neuron Registry initiative.
A way to deal with the cellular complexity of the nervous system is to group neurons into types based on their properties. The term property in this context means descriptive characterizations of neurons that include, but are not limited to, morphological, molecular, and physiological aspects. This approach is complementary to the emergent attempts to provide a classification purely based on molecular expression patterns and developmental transcription pathways (Bernard et al., 2009). A property-based characterization has clear empirical value, as it reflects the natural language used by scientists to record and communicate their observations. At the same time, the granularity of the description must be chosen carefully. Using too large a number of properties to maximize the resolution of neuron types might produce so many types as to hinder the understanding of basic mechanisms and common principles. Conversely, using too small a number of properties, while still possibly capturing substantive information, could potentially yield such general categories as to miss essential functional and computational consequences of neuronal diversity.
Describing neuron types by properties that refer to neuronal component parts (e.g., soma, axons, dendrites) and their interactions (e.g., connectivity) could enable an ideal balance between these extreme scenarios (Larson et al., 2007; Larson and Martone, 2009). Such an approach allows for the development of a powerful structured vocabulary for neuronal description. In addition, this style of vocabulary can be extended dynamically during property entry. In such a way, the level of detail of the property descriptors can be tailored to the most effective level of abstraction for any given context.
Ontologies are formal representations of domain knowledge leveraging normalized logical relationships (Larson and Martone, 2009). In recent years, ontological characterization has emerged as a promising mechanism to describe biological systems (http://www.obofoundry.org, http://bioportal.bioontology.org, http://www.ebi.ac.uk/ontology-lookup). As an example of a relevant ontology instantiation, we refer the reader to the Subcellular Anatomy Ontology (SAO; http://bioportal.bioontology.org/ontologies/1068). A particularly suitable entry mechanism for the descriptive semantics of neuronal properties can be based on the Resource Description Framework (RDF) approach (Decker et al., 2000). By relating descriptors (i.e., ontological terms), concepts can be formed much in the same way that sentences are constructed in natural language. Graphs of ontological relationships (Udrea et al., 2005) can be depicted, thereby illustratively defining the semantics of a property language. In this article we propose the use of subject-predicate-object triples (Horrocks et al., 2003) to form the basis of the ontological neuronal property descriptors. Founding a property language on these triples provides flexibility while maintaining specificity. This type of neuronal characterization has the advantage that it can accommodate the wide variety of informational variants associated with neuronal properties while facilitating unambiguous and computer readable scribing of those variants.
After briefly introducing the specific problem of representing and organizing the knowledge of neurons and their relations, we describe a methodology, some of the proposed properties to initiate an iterative refinement, and the infrastructure being developed to enable and encourage community interaction. Given the topic of this article, a note on terminology is appropriate. Anatomists tend to use “feature” for a morphological characteristic, whereas physiologists use “property” for most phenomena they record. We will use the terms “feature” and “property” interchangeably to signify descriptors of neuronal phenotypes.
The problem
The outward forms of nerve cells were first seen clearly using the silver impregnation method introduced by Camillo Golgi in the late nineteenth century. The Golgi method showed that, in any given brain region, cells are divided into several distinct types, which are characteristic for that region. A fundamental problem from the start was the haphazard naming of these cell types and their component parts. In the cerebellum, for example, some types were eponymous (Purkinje cell, named after its discoverer, Jan Purkinje; Golgi cell, named after Golgi). Some neurons had appellations based on size (granule cell, so-called because it appeared as small as a grain under the microscope). Other cells were named according to the form of their axonal endings (mossy fiber, because of its mossy-looking terminal; climbing fiber, because it appeared to climb over the Purkinje cell dendritic tree), on the basis of their dendritic trees (stellate cell, because of its star-shaped dendritic tree emanating in all directions). The cell type names in other regions were equally idiosyncratic.
By the early twentieth century, many types of nerve cells had been described by the classical histologists, which are summarized in Ramon y Cajal's great work on the histology of the nervous system (Cajal, 1911). His classification scheme was primarily based on the output connectivity, but this logic was not reflected in the terminology. Nevertheless, the names stuck, and people simply proceeded with their studies using the terms they preferred. The same lack of rules for a systematic terminology continues through the present day to hinder the inventorying of cells.
From a modern biological perspective, it is likely that neuronal classification will ultimately be based on, or at least relate to, developmental ontogeny (Wonders and Anderson, 2006). However, knowledge of lineage specification in mammals is still sparse and too many details are missing to attempt a systematic effort in that direction at this time. In addition, many properties of mature neurons will be dependent upon their local environment in the adult brain rather than their embryonic origin. A far more comprehensive identification of distinct neuron types in the literature depends on the phenotypic characterization of their properties.
What properties should be used to define neuron types? The most desirable categorization would be maximally informative of how each neuron type relates to others in the context of neuronal processing. For example, simply stating that our brain contains billions of neurons does little to advance our understanding of the brain. It is of higher value to describe the cellular composition of regions of the brain, such as the diencephalon, cerebellum, and cerebrum. More information is gleaned by further refining the parcels. For instance, the diencephalon contains the thalamus, hypothalamus, and epithalamus. The chosen properties should also reflect the myriad ways through which neuroscientists classify neurons in their studies. Most introductory text books provide an overview of these classification systems: by morphology, e.g., pyramidal or bipolar; by functional role (excitatory vs. inhibitory); by neurotransmitter(s) released; by physiological characteristics (fast-spiking, adapting, etc.,) and by the range of their connectivity (i.e., local circuit or projection neuron).
Combining these dimensions yields a combinatorial subdivision, with the danger of becoming lost in the detail while missing the context of overall function. Some descriptors, however, might seem too detailed to be useful only at first glance. Yet these same descriptors could be revealing when viewed in the proper perspective. For example, at the molecular level, DNA sequences could highlight the developmental tendencies of connectivity (Toni et al., 2007), which might lead to a prediction of all of the connections in the brain or a part of it, called a connectome (Lichtman et al., 2008) based on genetics. It is thus expected for approaches with a primary interest in distinct scales, such as subcellular neuroanatomy (Larson et al., 2007) or network connectivity (Bota and Swanson, 2008), and unique species, such as the fruit fly (Tweedie et al., 2009), to focus on different levels and types of detail. Given these different levels of focus, integration across scales is facilitated by use of a common framework.
An approach to a solution
A historically useful approach to characterization of types is that of ontology (Lorhard, 1606). Ontological characterizations are flexible yet structured, not too restrictive, and explicit enough to build logical inferences. Defining a neuronal ontology implies the formulation of universal descriptions from instances of neuron types and their associated properties. For example, published experimental evidence describes neurons with pyramidal-shaped somata in a single layer of the hippocampal CA1 region, with afferents from CA3 and efferents ending in the entorhinal cortex. These instances can then be generalized into universals defining a pyramidal-type neuron in region CA1 with like properties. The construction of a neuronal ontology could naturally leverage a relational approach. In this approach, relations such as “has part,” (in this example, soma), “has shape” (pyramidal), and “is located in” (principal layer of CA1) might constitute the properties that define neuron type universals along with “receives contact from” (afferents from CA3), and “makes contact to” (targets in the entorhinal cortex).
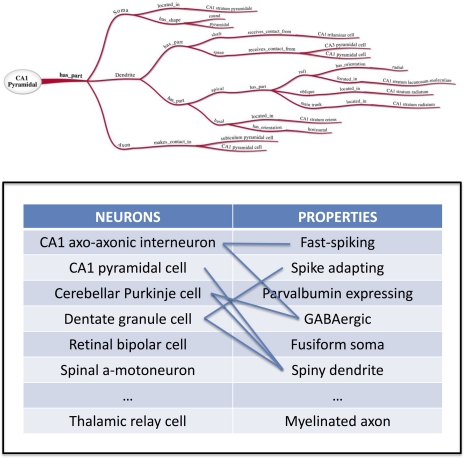
As illustrated in the above example, properties may thus be described as consisting of three fundamental aspects, namely a part descriptor, the relation itself, and a value target. In ontological terms, these are subject-predicate-object triples (Horrocks et al., 2003). For example, a neuron part, the soma, has a relation, shape, with a value of pyramidal. These facts are ontologically described as “part” = “soma,” “relation” = “has_shape,” and “value” = “pyramidal.” Figure 1 shows a graphical depiction of these types of ontological relationships, which define the properties of the neuronal classification. These properties could be extended to include neuronal location, axo-dendritic structure, brain subregions, local circuit connectivity, projection connectivity, neurotransmitters, other chemical markers, and membrane properties.
Figure 1.
Ontological Graphical Depiction (courtesy of the INCF Neuron Registry task force) and Neuron Registry Establishment → and illustration of the many-to-many relationship between neurons and their properties. The Neuron Registry will constitute a resource to browse and search neuron types based on their properties, and properties based on the neuron types in which they are found.
The advantage of a semantically consistent and formal representation of neuronal properties is the ability to automatically generate additional classifications of neurons through any single or multiple set of properties. For example, from the above relationships, we could automatically generate a set of all neurons with soma located in the principal layer of CA1 or that receive afferents from CA3. These are reasonable questions to ask, but currently this information is buried in the literature or scattered in databases requiring considerable human effort to assemble.
To formalize this approach to neuronal categorization, an effort is being sponsored by the INCF (http://incf.org) under the Program on Ontologies of Neural Structures (PONS; http://incf.org/core/programs/pons). The aim is to define a standard set of neuronal properties, and the associated ontologies required to define these properties for mammalian neurons. These standards will then be used to create a Neuron Registry that will serve as a knowledge base of neuronal types and the means to add definitions of new neurons based on their properties (Figure 1). Domain experts have knowledge acquired through years of practical experience, much of which is unique and often unpublished (Gardner et al., 2008). This knowledge must be gleaned from participating scientists and encoded into a searchable electronic form, using ontological techniques to formulate a structured, formalized description method.
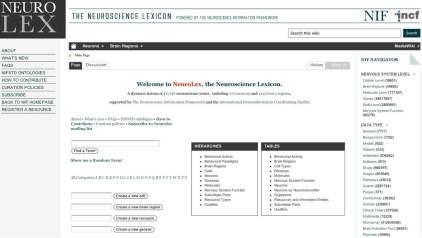
The Neuron Registry is building upon other prior and foundational efforts to employ a property-based approach to neuronal classification. Examples include BAMS (http://brancusi.usc.edu/bkms), SenseLab (http://senselab.med.yale.edu), SAO (http://bioportal.bioontology.org/ontologies/1068), and the NIF (http://www.neuinfo.org). Specifically, SAO is a formal ontology for the subcellular anatomy of the nervous system, covering multiple scales from neuron to its ultrastructural components, as well as the interactions between these components. The NIF standard ontology is an integration of different ontologies, including SAO. In SenseLab, the description of neurons is based on a predefined set of features, such as a schematic compartmental representation of the morphology, synaptic receptors, and membrane properties. The Neuron Registry aims to facilitate the expert definition of neuron types based on machine-readable descriptors of their properties. Thus, the scope of the Neuron Registry, as such, is aimed at providing semantic information about neurons in a community platform. The NIF project has taken steps toward providing the basic relations and entities for establishing a common neuronal knowledge base through the establishment of the NeuroLex (http://neurolex.org), a wiki facilitating the establishment of a lexicon of neuroscience concepts (Figure 2). The NeuroLex provides a semi-structured representation of neuroscience concepts necessary to construct statements about neurons and their properties. It assigns each of these concepts a unique identifier in the form of a numeric ID, to which multiple synonyms can be referenced. Thus, the NeuroLex provides a platform for knitting together the multiplicity of terms currently used to describe the nervous system. The NeuroLex also assigns each concept a preferred label. Although formal ontologies do not depend on the nomenclature choice, the systematic adoption of an agreed-upon terminology will benefit neuroscientists and automated agents (e.g., text processors) alike. Thus, a crucial contribution of the NeuroLex can be the establishment of a comprehensive controlled vocabulary for neuroscience. In particular, the NeuroLex and the Neuron Registry have created a consistent scheme for naming neuron types, which is described in section “Naming Conversion” below. The NeuroLex also selected several key properties that are useful for the classification of neuron types (e.g., their neurotransmitter), leading to the ability to infer broad classes such as “GABAergic neurons.”
Figure 2.
NeuroLex—Neuroscience Lexicon.
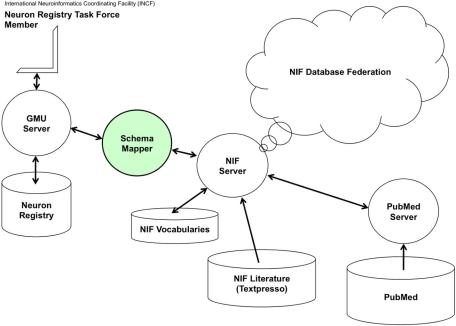
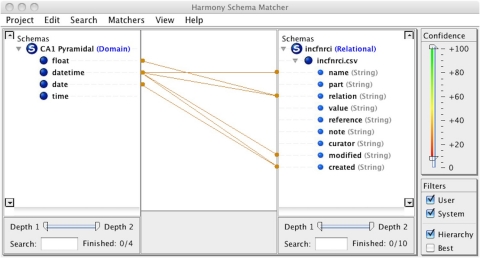
The NeuroLex definition of major “canonical” neuronal types, described based on a predetermined set of properties, is thus complementary to the goal of the Neuron Registry, which leaves the choice of defining properties up to the neuroscience expert. Such strategy provides a platform for describing any neuron that might be encountered during an experimental investigation, as is further expanded in sections “Implementation and a Representative Use Case Application” and “Guidelines for Describing a Neuronal Taxonomy based on Formal Ontologies” below. The Neuron Registry also more easily accommodates organism-specific characterizations of individual neuron types. At the same time, a synergistic collaboration is also in place (Figure 3): the Neuron Registry will define neurons using terms contained in the NeuroLex; and new terms defined in the Neuron Registry will be deposited into the NeuroLex (initially manually, but eventually automatically) so as to ensure continuous maintenance of a comprehensive repository of the entire neuroscience lexicon. Interoperability among the Neuron Registry, the NeuroLex, and other related neuroinformatics resources may be facilitated by recently developed tools such as Harmony (Smith et al., 2009), which provides interactive thresholding of schema correlation parameters, giving a researcher insight into prospective element matches (Figure 4). It is also planned to provide a SPARQL endpoint and RESTful web services for direct queries and data pulls from the Neuron Registry.
Figure 3.
Neuron Registry NIF Interface—This is a concept diagram depicting a planned capability not yet realized at the time of this writing.
Figure 4.
Schema Mapping using Harmony—Interoperability among neuroinformatic resources is aided through synchronization and sharing of data. Schema matching across disparate informatic repositories is critical to this sharing. Harmony assists the implementer in mapping data types properly so as to preserve integrity of meaning. Both automated (e.g., pattern recognition) and manual (e.g., thresholding) techniques are leveraged in this tool.
Implementation and a representative use case application
An information repository such as the Neuron Registry must be designed to house neuron properties, ontological depictions, type definitions, along with Refs, comments, and general additional related information. The design parameters of this database must include the facilitation of information importation and extraction, which are essential for information sharing and collaboration. Reports of data relations pertinent to answering the questions of investigators must be easily generated and quickly rendered. The database schema must also be flexible and adaptable, since the Neuron Registry is in its infancy and will evolve over time.
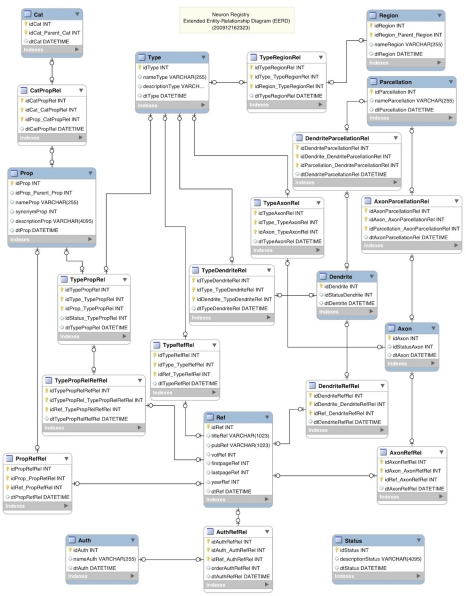
The RDF data model, with its subject-predicate-object triple form of representation (http://en.wikipedia.org/wiki/Resource_Description_Framework), is considered to be conceptually similar to the classic Entity-Relationship approach to data modeling (Chen, 1976). An extension to this classic modeling approach, the Extended Entity Relationship (EER) model, was subsequently proposed (Thalheim, 2007). An existing software tool called MySQL Workbench (http://wb.mysql.com) was leveraged to diagram the data models underlying the Neuron Registry as an Extended Entity-Relationship Diagram (EERD). An EERD depicts relations between objects in a database design (Figure 5). The key aspect of this design is to emphasize a highly indexed structure, which enables fast queries of nearly any combination of entity relationships. Objects are shown as labeled boxes containing the list of corresponding elements. Relations between the objects are shown with lines connecting the objects.
Figure 5.
Neuron Registry Extended Entity-Relationship Diagram.
In this design, the blue labeled boxes designate the primary informational objects such as category (Cat), property (Prop), and type. These primary objects are then related through relationship objects (Rel) such as CatPropRel and TypePropRel. Each record entered into the database is also tagged with a unique identity (id) along with the date/time stamp of entry. This allows for various manipulations such as cross-referencing by row id, multi-level indexing, sorting by date, and archival by date/time stamp, just to name a few. Being able to capture Refs and to correlate them across entries into the registry is also important. To facilitate this, a Ref object along with a related author (Auth) object is included as well in the database design. All other objects within the database, including the relationships themselves, can be related to the Ref object through the above-described cross-referencing mechanism (i.e., the relationship objects). This allows for Ref tagging of all information entered.
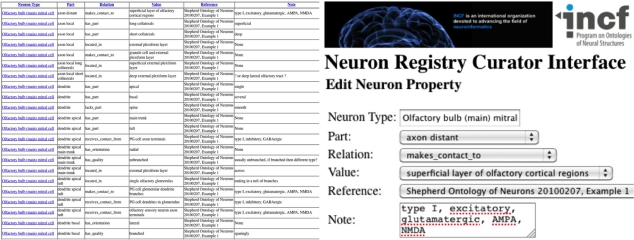
With a schema design in hand, the problem of populating the database becomes evident. If the task force members provide their information in freeform, translation into a usable structured electronic form is then dependent upon knowledge engineers. To avoid this dual-bottleneck workflow pipeline, the Neuron Registry has adopted a different approach that arms the task force with an interactive, focused entry portal (Figure 6; http://incfnrci.appspot.com/). Such a web-based curator interface eases information encoding and directly creates a framework to organize neuronal property descriptors and type definitions. In particular, a neuron type is defined by a set of properties, each consisting of ontological characterizations that include “part,” “relation,” and “value.” Other information fields house Ref and general notes along with the date/time that the curator entered the record. A folder view assists in the navigation of the repository and provides hierarchical insight into the registry content. Dropdown lists of terms help guide the curator by facilitating the adoption of structured vocabularies (Figure 6). To enter additional information, such as a new part value, a simple “other” option can be selected, which invokes an intuitive entry form. Thus, task force members can use the existing ontological descriptors or adapt and enhance the ontology to incrementally evolve the descriptive framework.
Figure 6.
Neuron Registry Curator Interface—List and Edit views (http://incfnrci.appspot.com/).
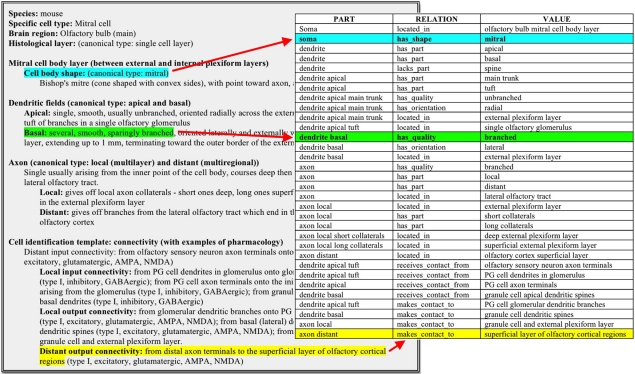
Using this approach, freeform descriptions of neuron types as typically reported in the scientific literature can be efficiently converted into a triple form of that same neuron type. A real-world example used during the early development of this conceptual framework is constituted by the olfactory bulb mitral cell. The freeform characterization provided by an olfactory neuroscientist was naturally organized into three major neuronal components: Soma, Dendrite, and Axon, which were logically mapped into corresponding “parts.” The translation from a freeform description to triples ensures computer readability of this same information (Figure 7). The Neuron Registry Curator Interface (http://incfnrci.appspot.com) facilitates direct entry in this form so as to avoid the need of a conversion altogether.
Figure 7.
Mapping of a freeform neuronal description of the defining properties of an Olfactory bulb (main) mitral cell into a part-relation-value ontological description.
Usage examples of the machine-readable information being entered into this Neuron Registry include searches of neurons based on properties (e.g., what neurons have axons located in a given region?), finding similarities and differences between two specific cell types (i.e., common and distinguishing properties), and checking to determine if a neuron with a given set of properties has already been characterized or might constitute a new type. Having described the rationale for the Neuron Registry implementation and a sample of representative use cases, we now turn attention to the guiding principles for associating neuron types with their defining properties.
Guidelines for describing a neuronal taxonomy based on formal ontologies
To maximize utility and consistency, the Neuron Registry task force has established a set of operating principles that guide contributor entries. Following the logic described above, each neuron type in the Neuron Registry is defined by a collection of properties, each expressed with a relation and a value referring to a specific neuron part. For example, one of the properties defining dentate gyrus granule cells is that their dendrites are in the molecular layer of the dentate gyrus. This is expressed in the Neuron Registry with the relation “located in,” the value “molecular layer,” and the assignment to the part “dendrites.” To build the Neuron Registry within the consistent semantic framework provided by the NIF project, each value comes from existing NeuroLex categories. When such categories are not available, they are added to the NeuroLex. By using common building blocks, the relationships among the entities specified in the Neuron Registry are by nature consistent with the entities utilized by the NeuroLex. If contradictory statements are made, these are more easily revealed because the id of entities referenced has been asserted. Perhaps of greater importance, because the Neuron Registry entries Ref their parts to anatomical structures and molecular entities within the Neurolex, the neuron descriptions are immediately interoperable with any other information artifact built from these same entities.
The defining properties are all necessary conditions for a neuron to be considered of a given type. This means that if a neuron is known or shown not to have one of these necessary properties, then it is a different type altogether. Taken together, the collection of properties, which defines a neuron type, also constitutes a set of sufficient conditions for a neuron to be of that type. This means that if a neuron has all of those properties, then it must be of that type. The adoption of “necessary and sufficient” properties is opposed to a “comprehensive description” of neuron types. The Neuron Registry aims at a minimal description of neuron types, in the sense of only including definitional information, rather than being generically descriptive. As such, there is no restriction on the range of neural property descriptors as long as the “necessary and sufficient” criterion is met.
As an example, having a relatively large soma located in a single cell body layer of the olfactory bulb, as well as one or more radial (“primary”) dendrites extending across the external plexiform layer to connect to one or more olfactory glomeruli, characterizes mitral cells of most vertebrate species. The number of radial dendrites varies in different species, from one in many mammals to two in turtles and up to 18 in some birds. These are, however, all considered to be mitral cells by virtue of their relatively large cell bodies in a single layer and smooth radial dendrite(s) connecting to the glomeruli. This means that if a neuron does not have these properties, it would not be considered a mitral cell. Likewise, if a neuron with these properties is found, then it is definitely a mitral cell. In contrast, one could argue that cell shape, although descriptive, is not truly defining of cell id. However, most, but not all, mitral cells have a cell body with a “mitral” shape, As another example, it might be tempting to state that a hippocampal neuron with a pyramidal-shaped soma located in the pyramidal layer must be a pyramidal neuron. However, this would in many cases be an incorrect assignment, as hippocampal basket cells are known whose somata are pyramidally shaped and lie in the pyramidal-cell layer.
At the same time, minimal descriptions must take into account that different experimenters, reports, and findings refer to different aspects of neurons. In the above described example, an olfactory bulb neuron whose soma is in the mitral cell body layer and has a single radial trunk across the external plexiform layer can only be a mitral cell. A researcher, however, might lack morphological observations on the neurites, and yet possess information on the connectivity (e.g., through the use of viral tracers). Indeed, mitral cells can be defined as neurons with somata in the mitral cell body layer and which make dendrodendritic synapses with granule cell spines; or else as neurons with somata in the mitral cell body layer and which send axons into the lateral olfactory tract to make synapses on dendrites in the olfactory cortex. Here is where other crucial aspects of information come into play, once they are expressed through relations such as “receives contact from” and “makes contact to” in the Neuron Registry.
Other complementary minimal descriptions can be considered based on pharmacological, developmental or physiological criteria (PING, 2008). The following examples each constitute viable definitions of mitral cells: neurons glutamatergic at their axonal or dendritic synapses, with somata in the mitral cell body layer; neurons not renewed by neurogenesis through the rostral migratory stream, with somata in the mitral cell body layer; neurons with input resistances of up to 100 megohms, with somata in the mitral cell body layer. In general, it is not required to show that a neuron has all of the properties defining a given type, in order to conclude that it is of that type. Usually, only a subset of the defining properties is observed in a given lab, time period, and research project. However, stating that a neuron belongs to a given type assumes that all of the defining properties would be verified if they were measured.
The Neuron Registry cannot list all of the known properties of a given type, nor is it expected that every neuron type entry in the Registry will have an exhaustive or even extensive list of properties. However, each neuron type entry should include all of its minimal defining properties. An illustrative example of this approach is provided by a proposal to define the “typical pyramidal neurons” in the mammalian neocortex based on eight specific features (Nieuwenhuys, 1994): (1) dendritic spines; (2) a radial apical dendrite; (3) terminating in the most superficial layer and; (4) distinct from basal dendrites; (5) descending axon to subcortical white matter; (6) intracortical axonal collaterals establishing; (7) asymmetric synapses with round vesicles using; (8) excitatory neurotransmitter glutamate and/or aspartate. Any cell with all of these features is a “typical pyramidal neuron.” However, several “atypical or aberrant pyramidal neurons” also exist, sharing most but not all features with their typical counterparts, such as aspiny pyramids and inverted pyramids (Nieuwenhuys, 1994). Because they lack at least one of the necessary defining properties, these other cell types constitute separate classes.
Last but not least, every assignment of a defining property to a neuron type must be accompanied by at least one citation of a peer-reviewed publication. Multiple citations may be necessary to properly attribute the source of information about particular neuronal properties. The choice of citation(s) may follow various criteria, such as historical credit to the original work or the first usage of the term, the high impact or wide availability of the source, and the perceived scholarly value of the publication and its authors. The responsibility to select the most appropriate criteria and thus citation ultimately rests with the expertise of the curator and is likely to vary on a case-by-case basis. The assignment of a property to a neuron type further may be accompanied by notes, e.g., of explanations or comments. This is particularly needed when there is controversy about a property, in which case annotation about the controversy can stimulate new experiments to resolve it. Annotation is also needed when a property changes during the course of development. Finally, physiological characterization of a neuron is highly dependent on a number of factors: in vivo vs. in vitro, tissue slice vs. cell culture, anesthesia depth and type, temperature, and many other factors. Thus, physiological properties will probably be the greatest challenge for inclusion in a neuron ontology (PING, 2008).
During the design phase of the Registry, only Task Force members actively entered neuron properties. Now, all neuroscientists are welcome to contribute. We are approaching this research with openness and rely on the entire scientific community to maintain quality and integrity. Specifically, we have initiated an Adopt-a-Neuron Campaign (adopt.a.neuron@incf.org) encouraging (self-)nomination of neuroscience experts to curate the properties of their specific neuron(s) of interest. A parallel Representation and Deployment Task Force (RDTF) is envisioned to support the activity of the Neuron Registry by ensuring that the relations linking the ontological classes and the values of instances are compliant with rigorous ontological principles (http://obofoundry.org). This separate curation step removes the necessity for the neuroscience experts to also possess and respect formal ontology expertise while trying to resolve open questions and/or integrate existing knowledge about neuron types. The end result will also be ready for incorporation into broader cell ontology efforts (http://cellontology.org).
Naming convention
Each neuron type in the Neuron Registry and in the NeuroLex is associated with a unique identifier (i.e., a code or string of characters not associated with a different type). The unique identifier is not to be confused with the common name of the neuron. Each neuron type is also given a preferred name and possibly one or more synonyms.
The preferred name for a neuron should adopt some standard naming conventions so that neurons are easy to organize for a human and easy to parse for an information system. Some defining properties are leveraged for incorporation into the name. For example, if the locations of the soma and/or dendrites are among the defining properties of the neuron type, then the name of that type should begin with the noun describing the region of the nervous system encompassing those locations (e.g., spinal cord motoneuron, dentate gyrus granule cell). If the name of the neuron includes additional terms to discriminate different subtypes, those should be appended at the end (e.g., spinal cord motor neuron alpha, spinal cord motor neuron gamma). We can uniquely identify a set of properties associated with a neuron population that resides in a particular brain region, regardless of the fact that it might be derived from the same precursor as a cell residing in another brain region.
The underlying rationale is that neuron cell bodies and surrounding dendrites are located in specific regions of the nervous system. Therefore, it is consistent for the name for all the cells in a given region to start with the name of that region. For example, a ganglion cell in the retina would have as its name Retina Ganglion Cell. It is not acceptable to combine this cell type with any other cell that happens to have the unhelpful name of “ganglion cell,” which has no meaning other than its discoverer calling it by a generic name signifying “large.” In this way, all of the different types of cells in the retina belong to the class “Retina.” The same logic applies to every other cell in the nervous system. Such a convention can be characterized by “Region Cell Name,” which avoids grouping cells haphazardly, such as granule cells of the cerebellum, olfactory bulb, dentate gyrus, etc., all falling under the name “granule” (Table 1).
Table 1.
Example of comparison between lists of common neuron names and structured neuron names as in NeuroLex.
| Unstructured Neuron Names | Structured Neuron Names |
|---|---|
| Basket cell in cerebellum | Cerebellum basket cell |
| Golgi cell in cerebellum | Cerebellum Golgi cell |
| Granule cell in cerebellum | Cerebellum granule cell |
| Purkinje cell in cerebellum | Cerebellum Purkinje cell |
| Stellate cell in cerebellum | Cerebellum stellate cell |
| Granule cell in main olfactory bulb | Olfactory bulb main granule cell |
| Mitral cell in main olfactory bulb | Olfactory bulb main mitral cell |
| Periglomerular cell in main olfactory bulb | Olfactory bulb main periglomerular cell |
| Deep short axon cell in main olfactory bulb | Olfactory bulb main deep short axon deep cell |
| Tufted cell in main olfactory bulb | Olfactory bulb main tufted cell |
The confusion of mixing neuron names from different regions is striking even for just two regions, compared with the ease of scanning and finding a specific name when structured by region (Table 1). The mixing problem is far worse with 200 neuron names, whereas the list remains clear when sorted by region. The structured list has the additional advantage of enabling the neuron names to be readily linked to brain atlases and databases of brain regional anatomy. This naming convention also provides the basis of logical definitions in ontologies such as the Neuroscience Information Framework Standard (NIFSTD) ontology (Bug et al., 2008). For example, restrictions may be derived based on the fact that a cerebellum neuron must be part of a cerebellum.
This convention of Region Cell Name has to subsume several possible complications. First, there may be several parts to the cell region. For example, the cochlear nucleus has dorsal and ventral parts, which are distinct in their cell types and functions. Although we refer to them colloquially as the dorsal cochlear nucleus and ventral cochlear nucleus, for the purposes of listing them, they must follow a parent-child convention to keep them classified together: Cochlear Nucleus Dorsal and Cochlear Nucleus Ventral. In each case, the cell name follows (i.e., Cochlear Nucleus Dorsal Fusiform Cell and Cochlear Nucleus Ventral Bushy Cell). Other examples are Olfactory Bulb Main Mitral Cell and Olfactory Bulb Accessory Mitral Cell. Thus, we have the expanded parent-child convention to “Region Subregion Cell Name.” This naming convention is useful for constructing an alphabetically arranged list of cell names. In addition, all neurons that belong to a single brain region will be immediately obvious in contrast to the current situation. Colloquially, we continue to use their common names in their common order, i.e., a “mitral cell of the main olfactory bulb.”
Further complexities apply to cortical regions because of their laminar characteristics. However, these are also accommodated by an expansion of the parent-child relation; thus a cortical pyramidal-cell in lamina 3 of the motor cortex is termed “Neocortex Motor Layer 3 Pyramidal-Cell.” Another level of complexity to be considered in naming will be encountered when characterizing neuron types by gene expression (Gong et al., 2003; Hatten and Heintz, 2005) in combination with or in addition to network connectivity (Polleux, 2005; Shepherd et al., 2005). In spite of these challenges, the utility of this approach has been tested on over 200 neuron types in the NeuroLex (available at http://neurolex.org/wiki/Category:Neuron), and provides the basis for future naming. Each of these neurons has been assigned a soma location of the brain region indicated by their name. All examples provided deal with the mammalian nervous system. The same approach can be used for any species, vertebrate or invertebrate, in which the cell type is unique or different from the general mammalian pattern. In those cases, an additional initial “species” modifier may be needed for clarity.
Concluding remarks
The Neurolex and Neuron Registry grew out of the recognition that neuroscience needs to begin developing and implementing common information frameworks to bring together data acquired across scales, organisms and techniques. In the genomic world, every sequence is registered within a standard format to a central database where it can be compared algorithmically to all other sequences. We do not have the luxury at the cellular level of a simple set of characters for defining neurons. However, having a standard grammar and centralized database where neuron descriptions can be deposited in a way that is amenable to machine-based processing and where unique identifiers can be assigned will be an important infrastructure for finally grappling with the cellular complexity of the nervous system. We anticipate that such a resource can be used, much like the genomic resources, for comparing neurons, determining functional or structural groupings and for building multiscale models of the nervous system.
There are multiple dimensions to the problem of neuronal classification. Neuronal properties can be described in a variety of ways (PING, 2008). Depending on the relation being described, the values can take on different meanings. For example, genetic markers in the context of development could be used to characterize neurons from a connectivity perspective. In addition to species dependence, metadata about the experimental models, such as age, strain, and sex, and experimental conditions, such as caging, diet, and behavior, might ultimately affect neuronal characterizations. Future considerations might also incorporate developmental/transcription factors in addition to the property-based approaches currently in use.
The number of neuron types in the mammalian brain ultimately depends on the resolution of the considered collection of properties. Too many neuronal properties will produce too large a number of neuron types to be useful. Too few properties will have the opposite effect. Ontologies provide a flexible characterization method that can be adapted to the resolution of choice to maximize resultant utility by neuroscientists toward advancement of their research. By using a standard set of properties and entities, we can automatically classify neurons along any single or combination of properties automatically through the definition of a rule. For example, in the Neurolex, we can generate the list of GABAergic neurons (any cell that is a member of the class neuron and uses GABA as a neurotransmitter), all spiny neurons, all cortical pyramidal neurons, etc.
The Neuron Registry and Neurolex use a lightweight semantic representation based on the RDF triple with the subject and objects drawn from community ontologies. As indicated, this representation is sufficiently flexible to generate multiple hierarchies based on single or multiple properties. A more semantically enriched representation is being pursued by the Cell Ontology consortium using OWL, to enable logical inferencing of cell classes (Bard et al., 2005). The Neuron Registry Task force is collaborating with this endeavor through the INCF PONS program. Thus, all three representations are being coordinated so that information can flow among them in a consistent manner while the pros and cons of each approach are determined. Given the complexity of the neuronal identification problem, we feel that multiple representations built from common entities currently constitute the best approach.
Although ontological characterization is a mature process, its application to neuroscience is relatively new. Neuron characterization with ontologies should yield important results and contribute significantly to neuroscience research. However, the approach described here is just in its infancy. It will take a concerted effort by teams of researchers throughout the neuroscience community to address all of the possible neuron types. The Neuron Registry task force will help define and refine this approach, and will hopefully catalyze a spiraling effort to encompass a wider and wider degree of community participation.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Stephen Larson, David Osumi-Sutherland, Alan Ruttenberg, and the members of the Neuron Registry task force for their ongoing contribution, valuable expertise, and critical dialog on many of the ideas discussed in this article. This work was supported in part by NIH R01 NS39600 and an INCF contract to Giorgio A. Ascoli, NIH R01 DC009977, NIDA SSSAB-2008, and R01 DC000086 to Gordon M. Shepherd, and NIDA DA016602 and R01 NS05829 to Maryann E. Martone. The NIF project is supported by the NIH Blueprint consortium through contract HHSN271200800035C administered by NIDA to Maryann E. Martone.
References
- Azevedo F. A. C., Carvalho L. R. B., Grinberg L. T., Farfel J. M., Ferretti R. E. L., Leite R. E. P., Filho W. J., Lent R., Herculano-Houzel S. (2009). Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 513, 532–541 10.1002/cne.21974 [DOI] [PubMed] [Google Scholar]
- Bard J., Rhee S. Y., Ashburner M. (2005). An ontology for cell types. Genome Biol. 6, R21 10.1186/gb-2005-6-2-r21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A., Sorensen S. A., Lein E. S. (2009). Shifting the paradigm: new approaches for characterizing and classifying neurons. Curr. Opin. Neurobiol. 19, 530–536 10.1016/j.conb.2009.09.010 [DOI] [PubMed] [Google Scholar]
- Bota M., Swanson L. W. (2008). BAMS Neuroanatomical Ontology: design and implementation. Front. Neuroinformatics 2:2 10.3389/neuro.11.002.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bug W., Ascoli G. A., Grethe J. S., Gupta A., Fennema-Notestine C., Laird A., Larson S., Rubin D., Shepherd G. M., Turner J. A., Martone M. E. (2008). The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics 6, 175–194 10.1007/s12021-008-9032-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal S. R. Y. (1911). Histologie du Système Nerveux de l'Homme et des Vertébrés. Trans. L. Azoulay. Paris: Maloine [Google Scholar]
- Chen P. P. (1976). The entity-relationship model: toward a unified view of data. ACM TODS 1, 9–36 [Google Scholar]
- Decker S., Melnik S., van Harmelen F., Fensel D., Klein M., Broekstra J., Erdmann M., Horrocks I. (2000). The Semantic Web: the roles of XML and RDF. IEEE Internet Comput. 4, 63–74 [Google Scholar]
- Gardner D., Akil H., Ascoli G. A., Bowden D. M., Bug W., Donohue D. E., Goldberg D. H., Grafstein B., Grethe J. S., Gupta A., Halavi M., Kennedy D. N., Marenco L., Martone M. E., Miller P. L., Müller H. M., Robert A., Shepherd G. M., Sternberg P. W., van Essen D. C., Williams R. W. (2008). The neuroscience information framework: a data and knowledge environment for neuroscience. Neuroinformatics 6, 149–160 10.1007/s12021-008-9024-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S., Zheng C., Doughty M. L., Losos K., Didkovsky N., Schambra U. B., Nowak N. J., Joyner A., Leblanc G., Hatten M. E., Heintz N. (2003). A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925 10.1038/nature02033 [DOI] [PubMed] [Google Scholar]
- Hatten M. E., Heintz N. (2005). Large-scale genomic approaches to brain development and circuitry. Annu. Rev. Neurosci. 28, 89–108 10.1146/annurev.neuro.26.041002.131436 [DOI] [PubMed] [Google Scholar]
- Herculano-Houzel S. (2009). The human brain in numbers: a linearly scaled-up primate brain. Front. Hum. Neurosci. 3:31 10.3389/neuro.09.031.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrocks I., Patel-Schneider P. F., van Harmelen F. (2003). From SHIQ and RDF to OWL: the making of a web ontology language. J. Web Semantics 1, 7–26 [Google Scholar]
- Larson S. D., Fong L. L., Gupta A., Condit C., Bug W. J., Martone M. E. (2007). A formal ontology of subcellular neuroanatomy. Front. Neuroinformatics 1:3 10.3389/neuro.11.003.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson S. D., Martone M. E. (2009). Ontologies for neuroscience: what are they and what are they good for? Front. Neurosci. 3:7 10.3389/neuro.01.007.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman J. W., Livet J., Sanes J. R. (2008). A technicolour approach to the connectome. Nat. Rev. Neurosci. 9, 417–422 10.1038/nrn2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorhard J. (1606). Diagraph of Metaphysic or Ontology. St. Gallen: Ogdoas scholastica Book 8. Published in Sangalli. Trans. Sara L. Uckelman. [Google Scholar]
- Nieuwenhuys R. (1994). The neocortex. An overview of its evolutionary development, structural organization and synaptology. Anat. Embryol. 190, 307–337 [DOI] [PubMed] [Google Scholar]
- PING (Petilla Interneuron Nomen-clature Group): Ascoli G. A., Alonso-Nanclares L., Anderson S. A., Barrionuevo G., Bena-vides-Piccione R., Burkhalter A., Buzsaìki G., Cauli B., DeFelipe J., Faireìn A., Feldmeyer D., Fishell G., Fregnac Y., Freund T. F., Gardner D., Gardner E. P., Goldberg J. H., Helmstaedter M., Hestrin S., Karube F., Kisvaìrday Z. F., Lambolez B., Lewis D. A., Marin O., Markram H., Muñoz A., Packer A., Petersen C. C. H., Rockland K. S., Rossier J., Rudy B., Somogyi P., Staiger J. F., Tamas G., Thomson A. M., Toledo-Rodriguez M., Wang Y., West D. C., Yuste R. (2008). Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat. Rev. Neurosci. 9, 557–568 10.1038/nrn2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polleux F. (2005). Genetic mechanisms specifyingcortical connectivity: let's make some projections together. Neuron 46, 395–400 10.1016/j.neuron.2005.04.017 [DOI] [PubMed] [Google Scholar]
- Shepherd G. M. G., Stepanyants A., Bureau I., Chklovskii D., Svoboda K. (2005). Geometric and functional organization of cortical circuits. Nat. Neurosci. 8, 782–790 10.1038/nn1447 [DOI] [PubMed] [Google Scholar]
- Smith K., Mork P., Seligman L., Rosenthal A., Morse M., Wolf C., Allen D., Li M. (2009). The Role of Schema Matching in Large Enterprises. The MITRE Corporation, CIDR Perspectives. [Google Scholar]
- Sporns O. (2011). Networks of the Brain. Cambridge, MA: The MIT Press [Google Scholar]
- Thalheim B. (2007). Extended Entity-Relationship Model. (Kiel: Christian-Albrechts University; ), 1–8 [Google Scholar]
- Toni N., Teng E. M., Bushong E. A., Aimone J. B., Zhao C., Consiglio A., van Praag H., Martone M. E., Ellisman M. H., Gage F. H. (2007). Synapse formation on neurons born in the adult hippocampus. Nat. Neurosci. 10, 727–734 10.1038/nn1908 [DOI] [PubMed] [Google Scholar]
- Tweedie S., Ashburner M., Falls K., Leyland P., McQuilton P., Marygold S., Millburn G., Osumi-Sutherland D., Schroeder A., Seal R., Zhang H. (2009). FlyBase Consortium. Nucleic Acids Res. 37, D555–D559 10.1093/nar/gkn788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udrea O., Deng Y., Ruckhaus E., Subrahmanian V. S. (2005). “A graph theoretical foundation for integrating RDF ontologies,” in Proceedings of AAAI'05, (Pittsburgh, PA: AAAI Press; ), 1442–1447 [Google Scholar]
- Wonders C. P., Anderson S. A. (2006). The origin and specification of cortical interneurons. Nat. Rev. Neurosci. 7, 687–696 10.1038/nrn1954 [DOI] [PubMed] [Google Scholar]