Abstract
Glycosaminoglycans (GAGs) interact with a number of cytokines and growth factors thereby playing an essential role in the regulation of many physiological processes. These interactions are important for both normal signal transduction and the regulation of the tissue distribution of cytokines/growth factors. In the present study, we employed surface plasmon resonance (SPR) spectroscopy to dissect the binding interactions between GAGs and murine and human forms of interleukin-7 (IL-7). SPR results revealed that heparin binds with higher affinity to human IL-7 than murine IL-7 through a different kinetic mechanism. The optimal oligosaccharide length of heparin for the interactions to human and murine IL-7 involves a sequence larger than a tetrasaccharide. These results further demonstrate that while IL-7 is principally a heparin/heparan sulfate binding protein, it also interacts with dermatan sulfate, chondroitin sulfates C, D, and E, indicating that this cytokine preferentially interacts with GAGs having a higher degree of sulfation.
Keywords: interleukin-7, glycosaminoglycans, heparin, surface plasmon resonance
1. Introduction
Glycosaminoglycans (GAGs), a family of polyanionic, polydisperse, linear polysaccharides perform a variety of critical biological functions [1]. Interactions between heparin, heparan sulfate (HS), and other GAGs with proteins mediate diverse biological processes including: blood coagulation, cell growth and differentiation, host defense and viral infection, lipid transport and metabolism, cell-to-cell and cell-to-matrix signaling, inflammation and cancer [2–6]. Thus, an understanding of GAG-protein interactions at the molecular level is of fundamental importance to biology and will aid in the development of highly specific therapeutic agents to these pathways [2].
One of the major binding partners of GAGs involves numerous cytokine and growth factors and these interactions lead to a myriad of functions. Numerous hematopoietic cytokines and growth factors have been shown to interact to GAGs, in particular heparin and HS, including: interleukins (IL)-1, -2, -6, -7, 8, -12, and fibroblast growth factors (FGFs) [7–14]. The interactions of GAGs with these proteins may localize them to specific environments or niches, regulate degradation of these proteins during transport, and provide an immobilized matrix to present these cytokines to receptors on targeted cells [11]. The GAG interactions with cytokines/growth factors may serve as co-receptors enhancing or inhibiting interactions with the soluble cytokines with their transmembrane anchored cell surface receptors [5,12].
The IL-7 signaling pathway is fundamental in the development, proliferation, and homeostasis of B- and T-cells depending on the organism [15–17]. The biological responses of B-cells occurs only in mice; whereas, the biological responses of T-cells occurs in both mice and humans [15–17]. The IL-7 signal is activated upon the interaction of soluble IL-7 with its own cell surface α-receptor (IL-7Rα, CD127) and the common gamma chain (γc, CD132) receptor [18]. The bringing together of these two cytokine receptors by IL-7 activates intracellular pathways up-regulating gene targets. IL-7 from human (hIL-7) and murine (mIL-7) are basic proteins of 154 and 129 amino acids, respectively, and share 60% sequence identity [19, 20]. The gene for hIL-7 encodes another exon of 19 amino acids extending the loop connecting helix C and D [19]. However, the additional amino acids in hIL-7 appears not to be critical for its function as deletion of it resulted in similar biological response relative to the full-length protein [19, 20]. Structural studies are ongoing on the IL-7 signaling pathway relying on crystal structure determinations of hIL-7 bound to the human IL-7Rα extracellular domain (ECD) [21].
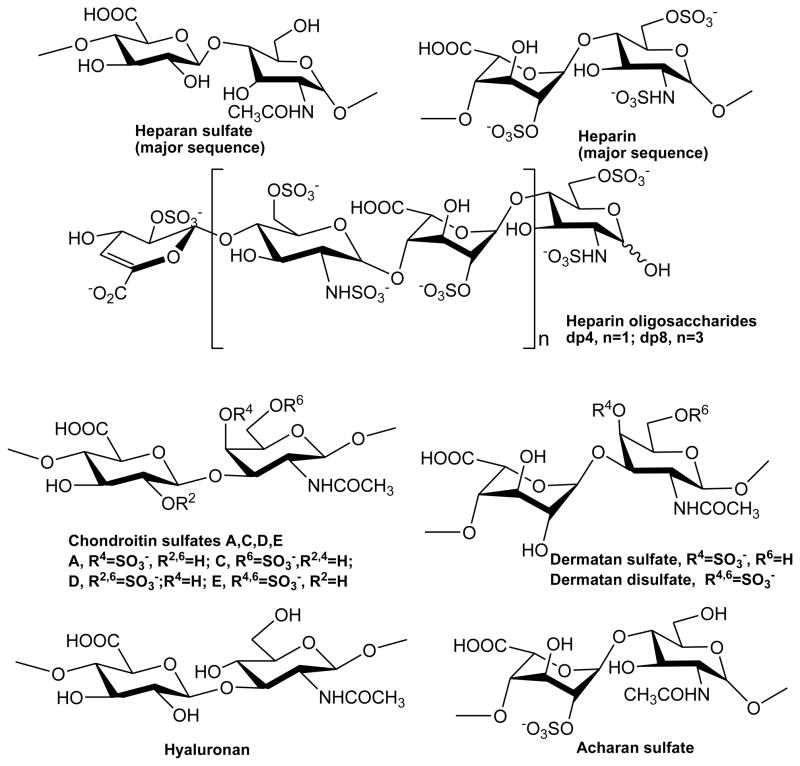
Previous studies reported IL-7 as a heparin/HS-binding cytokine that can directly regulate bioactivity of this protein [11,12, 22, 23]. The addition of heparin to mIL-7 abrogated lymphopoiesis, but not myelopoiesis in cultured B-cells [11]. Similarly, addition of heparin to human IL-7 inhibited an IL-7-dependent B-cell line [12]. Removal of B-cells of heparan sulfate using enzymes blocks mIL-7 binding and cells lacking heparan sulfate displayed significantly diminished proliferative growth factors to IL-7 [11]. Human IL-7 was found to bind to the extracellular matrix (ECM) and to fibronectin and resulted in the adhesion of T-cells in a dose- and integrin-dependent manner [23]. Human IL-7 was shown to be more protease resistant when bound to heparin than unbound [12]. It appears from these limited studies that IL-7 may be circulating bound to heparin enabling it to be more protease resistant and larger molecule escaping clearance through the kidneys or liver. Further, IL-7 may localize to specific microenvironments by interacting with cellular or extracellular matrix bound heparan sulfate molecules. However, a binding competition has to occur of IL-7 binding to its cognate cytokine receptors (IL-7Rα and γc) over heparin/HS to trigger the signaling cascade. While these studies have provided a general functional understanding of GAG-IL-7 interaction, there lacks structural and biophysical data of these interactions. Therefore, in this manuscript we report key structural and energetic features of the interactions of GAGs (Figure 1) to murine and human forms of IL-7 using surface plasmon resonance (SPR) spectroscopy.
Figure 1.
Chemical structures of GAGs and heparin-derived oligosacchrides.
2. Materials and methods
2.1. Materials
Human IL-7 was expressed and purified as described previously [24]. An Escherichia coli codon optimized gene of murine IL-7 was purchased from Bioclone Inc (San Diego, CA). The mIL-7 gene (residues 1-129 of the mature form; accession code P10168) was cloned into the NcoI and BamHI restriction sites of the expression vector pET-15b (Novagen Inc. Madison, WI) and subsequently confirmed by DNA sequencing. Murine IL-7 was expressed in E. coli cell line BL21(DE3) CodonPlus RP (Stratagene Inc. La Jolla, CA), refolded from inclusion bodies, and purified to homogeneity using the same protocol as for hIL-7 [24]. Wavelength scans using circular dichroism displayed classical α-helical secondary structure features for both forms of IL-7 as described earlier [24]. Mass spectrometry confirmed the molecular weights of both forms of IL-7. Both murine and human IL-7 contains an extra Met-Gly at the N-terminal end of the protein sequence. A molar absorption coefficient at 280 nm (ε280) of 6.9 mM−1 cm−1 was used to determine protein concentrations [25]. Purified human and murine IL-7 was buffer exchanged into 10 mM Tris-HCl (pH 7.0) and 150 mM NaCl.
Several different GAGs were utilized in this study. One set included porcine intestinal heparin (13 kDa), heparan sulfate (from porcine intestinal mucosa) (HS, 14.8 kDa, Celsus Inc., Cincinnati, OH), a low molecular weight heparin (LMWH, 4.8 kDa, Celsus Inc.), chondroitin sulfate A from porcine rib cartilage (CSA, 20 kDa, Sigma, St. Louis, MO), dermatan sulfate from porcine intestine (DS, also known as chondroitin sulfate B, 30 kDa, Sigma), and dermatan disulfate (Dis-DS, 4,6 disulfo DS, 33 kDa, Celsus Inc.). The other GAG set included chondroitin sulfate C from shark cartilage (CSC, 20 kDa, Sigma), chondroitin sulfate D from whale cartilage (CSD, 20 kDa, Seikagaku, Tokyo, Japan), chondroitin sulfate E from squid cartilage (CSE, 20 kDa from squid cartilage, Seikagaku), and hyaluronan from Streptococus zooepidemicus (HA, 100 kDa, Sigma). Acharan sulfate from Achatina fulica (AS, 29 kDa) was provided by Dr. Yeong-Shik Kim (Seoul National University). A disaccharide (dp2), tetrasaccharide (dp4), hexasaccharide (dp6), and octasaccharide (dp8) form of heparin was prepared from controlled partial heparin lyase 1 treatment of bovine lung heparin (Sigma) [26].
2.2. Preparation of a heparin sensor chip
SPR measurements were performed using a GE Biacore 3000 SPR instrument. Biotinylated heparin was prepared by reaction of sulfo-N-hydroxysuccinimide long-chain biotin (Pierce, Rockford, IL) with the free amino groups of unsubstituted glucosamine residues in the polysaccharide chain following a published procedure [27]. Biotinylated heparin (MW ~ 13 kD) was immobilized to streptavidin (SA) coated CM5 sensor chip (GE Heathcare, Uppsala, Sweden) using the manufacturer’s protocol. In brief, a 20 μL solution of the heparin-biotin conjugate (0.1 mg/mL) in HBS-EP (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.005% surfactant P20) buffer was injected over flow-cell 2 (FC2) of a SA sensor chip at a flow-rate of 10 μL/min. Approximately 100 resonance units (RU) of heparin were coupled to the sensor chip. A control flow-cell (FC) was prepared with a 1 min injection with saturated biotin in HBS-EP buffer.
2.3. SPR binding kinetics of IL-7-heparin interactions
The binding kinetics of the IL-7 interactions over the heparin sensor chip were assayed using HBS-EP buffer at 25 °C. Two-fold serial dilutions of IL-7 were injected over the sensor chip at a flow-rate of 30 μL/min for a period of 3 minutes followed by a 3 minutes dissociation period. The sensor chip was regenerated for subsequent runs using a 30 μL injection of 2 M NaCl. SPR experiments were performed in duplicate or triplicate at each concentration, confirming reproducibility. The binding sensorgrams (RU versus time) were pooled, trimmed, double referenced, and experimental fit to different kinetic models using BIAevaluation software v4.0.1.
2.4. SPR solution competition study of heparin oligosaccharides
A SPR solution competition study was performed by pre-equilibrating different heparin oligosaccharides with IL-7 followed by a subsequent injection of this solution over the heparin immobilized sensor chip. For the different chain lengths of heparin, IL-7 at 500 nM was pre-mixed with 1,000 nM of heparin disaccharide (dp2), tetrasaccharide (dp4), hexasaccharide (dp6), or octasaccharide (dp8) in HBS-EP buffer and injected over heparin chip at a flow-rate of 30 μl/min. Similarly, for the 10 other GAGS, IL-7 at 500 nM was pre-mixed with 1,000 nM of GAG and injected over heparin chip at a flow-rate of 30 μl/min. After each run, a dissociation period and regeneration protocol was performed as described above. The SPR experiments were performed in duplicate.
2.5. SPR solution competition study of different GAGs
For testing of inhibition of other GAGs to the IL-7/heparin interaction, IL-7 at 500 nM was pre-mixed with 1,000 nM of GAG and injected over the heparin chip at a flow-rate of 30 μl/min. After each run, a dissociation period and regeneration protocol was performed as described above. To measure the IC50 (concentration of competing GAGs resulting in a 50% decrease in IL-7 binding to heparin on the chip surface), IL-7 (500 nM) was mixed with different concentrations of GAGs in HBS-EP buffer and injected over the heparin chip. The IC50 values were calculated from plots of the binding signal as a function of GAG concentrations in solution.
3. Results
3.1. Binding kinetics of IL-7/heparin interactions
Binding kinetics were measured for murine and human IL-7 to immobilized heparin using SPR spectroscopy to provide a quantitative picture of these interactions. The SPR binding sensorgrams of the mIL-7/heparin and hIL-7/heparin interactions are displayed in Figure 2. The two binding sensorgrams fit reasonable well to a 1:1 Langmuir biomolecular reaction model. The interaction of mIL-7 to heparin displays an on-rate (kon) of 177 (± 12) M−1 s−1 and an off-rate (koff) of 1.10 ×10−2 (±8.5 ×10−5) s−1 (± standard errors were from the global fitting of different protein concentrations). The binding equilibrium dissociation constant (Kd = koff/kon) for the mIL-7/heparin interaction is 62 μM. The interaction of hIL-7 to heparin displays a kon of 6.10 × 104 (±900) M−1s−1 and a koff of 2.70 ×10−2 (± 2.6 × 10−4) s−1. The Kd for the hIL-7/heparin interaction is 0.44 μM (440 nM). The interaction of hIL-7 to heparin displayed a 345-fold and 2.5-fold faster on- and off-rates compared with the mIL-7/heparin interaction. The significantly faster kinetic rates for the hIL-7/heparin interaction translate into a 140-fold higher binding affinity than the mIL-7/heparin interaction. Possible reasons for the differences in the binding properties of murine and human IL-7 to heparin will be discussed below. A previous heparin-hIL-7 interaction study by Clarke et al. (12) reported the affinity of IL-7 for heparin measured using affinity co-electrophoresis method was 25 nM, lower than our SPR measurement of 440 nM. The disparity between these results might result from differences in binding conditions, such as buffer and temperature or from differences between a solution phase and solid phase binding measurement.
Figure 2.
Binding kinetics of murine (A) and human (B) IL-7 to heparin using SPR. SPR sensorgrams of IL-7/heparin interaction. IL-7 concentrations of 63, 125, 250, 500, and 1,000 nM were assayed over the immobilized heparin surface. The IL-7/heparin binding kinetics were determined by globally fitting the curves to a 1:1 biomolecular reaction model (black lines) using the BIAevaluation software package.
3.2. Binding preference of heparin chain size to IL-7 interactions
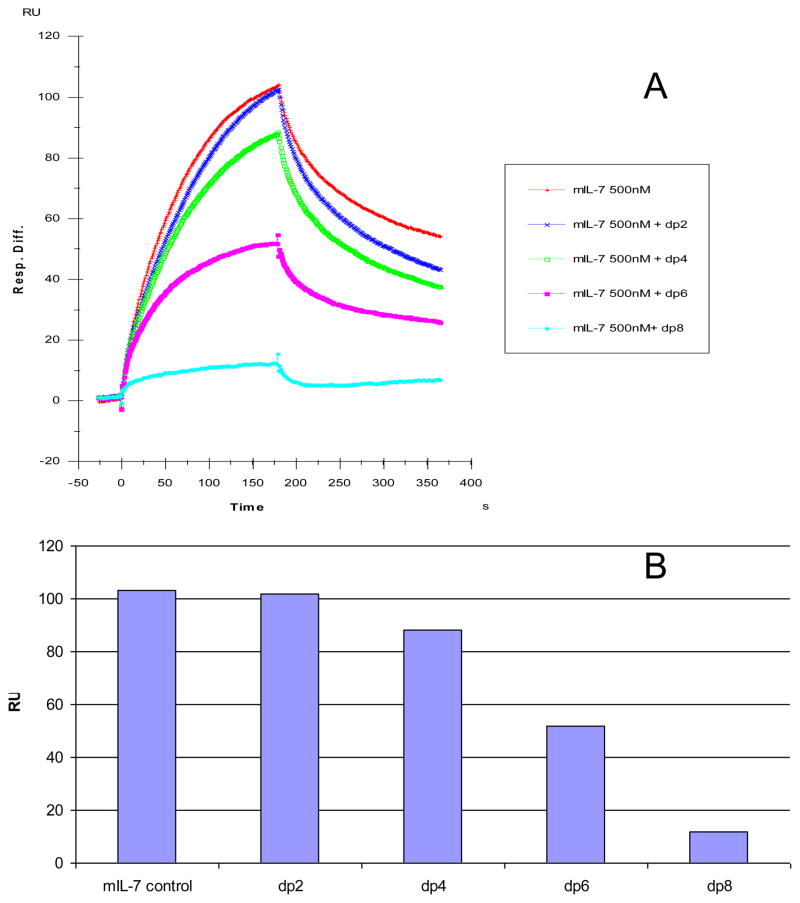
The chain length dependence of the heparin molecule critical to the binding interactions of both forms of IL-7 was determined using a SPR competition assay [31]. Different size heparin-derived oligosaccharides named for the degree of polymerization (dp2 to dp8) were assayed. A fixed concentration of each heparin oligosaccharide (1,000 nM) was incubated with a fixed concentration of IL-7 (500 nM). Subsequently, this solution was injected over the heparin immobilized sensor chip and the change in the maximal response was monitored relative to an IL-7 control containing no oligosaccharide of heparin. The SPR competition sensorgrams and the levels of competition of the different chain lengths of heparin for murine and human IL-7 interactions are illustrated in Figures 3 and 4, respectively. For both murine and human IL-7, little competition was observed when 1,000 nM of the disaccharide (dp2) to tetrasaccharide (dp4) heparin molecules were present with the protein solution (15% and 12% competition levels for mIL-7 and hIL-7, respectively). The dp6 heparin chain competed to a level of 48% for mIL-7/heparin immobilized interaction and this chain competed to a level of 42% for the hIL-7/heparin immobilized interaction. For both the mIL-7 and hIL-7 interactions with immobilized heparin, the dp8 heparin chain competed to a level of ~90%. These results suggest that the optimal binding interactions of both murine and human IL-7 to heparin require oligosaccharides larger than a tetrasaccharide (dp4).
Figure 3.
(A) Binding preferences of mIL-7 to various chain lengths of heparin using a SPR competition assay. Heparin disaccharide (dp2), tetrasaccharide (dp4), hexasaccharide (dp6), and octasaccharide (dp8) were each pre-equilibrated at a concentration of 1,000 nM with 500 nM of mIL-7. (B) Bar graphs (based on duplicated experiments with less than 5% errors) displaying the level of inhibition with the addition of different sized heparin chains to the mIL-7/immobilized heparin interaction.
Figure 4.
(A) Binding preferences of human IL-7 to various chain lengths of heparin using a SPR competition assay. A disaccharide (dp2), a tetrasaccharide (dp4), a hexasaccharide (dp6), and an octasaccarharide (dp8) of heparin was pre-equilibrated at a concentration of 1,000 nM with 500 nM of hIL-7. (B) Bar graphs (based on duplicated experiments with less than 5% errors) displaying the level of inhibition with the addition of different sized heparin chains to the hIL-7/immobilized heparin interaction.
3.3. Binding preferences of IL-7 to various GAGs
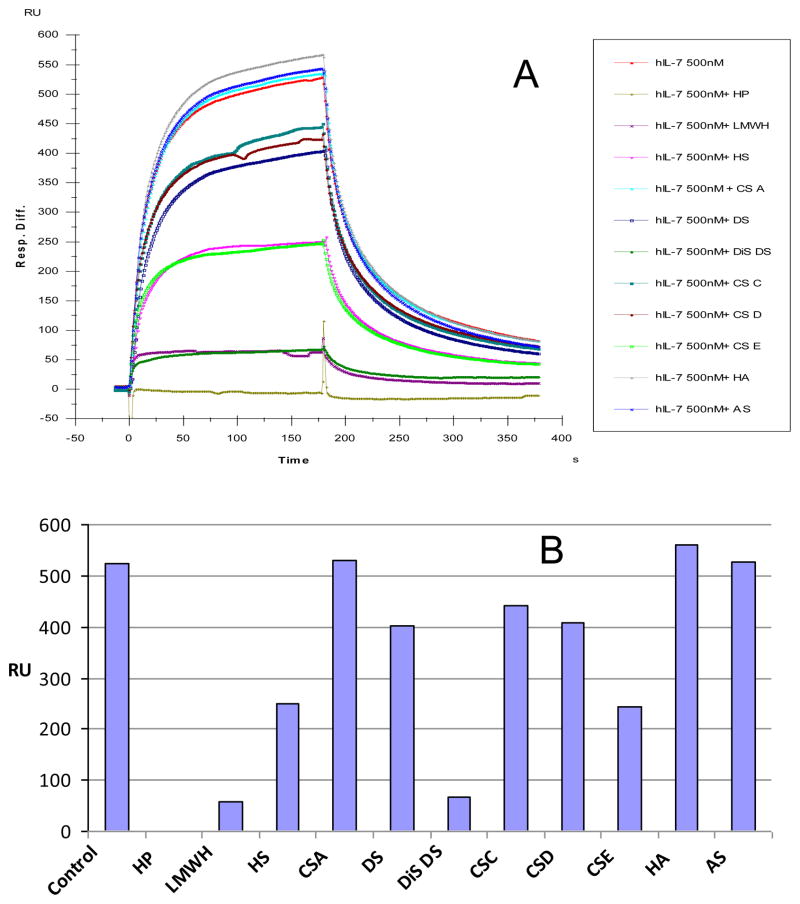
The SPR competition assay was also utilized to determine the binding preference of hIL-7 or mIL-7 to a panel of various GAGs (Figure 1). As above, the same concentration of each GAG (1,000 nM) was pre-equilibrated with hIL-7 or mIL-7. SPR competition sensorgrams and bar graphs of the GAG competition levels are displayed in Figure 5. Heparin (HP), low molecular weight heparin (LMWH), and dermatan disulfate (Dis-DS) produced the strongest inhibition by competing >90% of the IL-7/immobilized signal (heparin > LMWH > Dis-DS). Modest inhibitory activities were observed for chondroitin sulfate E (CSE) and heparan sulfate. Weak inhibitory activities were observed for chondroitin sulfates C, D (CSD and CSE), and dermatan sulfate (DS). Finally, chondroitin sulfate A (CSA), hyaluronan (HA), and acharan sulfate (AS) were unable to compete with surface heparin/hIL-7 interaction at the concentrations examined. The binding interactions of IL-7 to GAGs appear structure-dependent and highly influenced by the level of GAG sulfation. The inhibition behavior of each GAG towards hIL-7 and mIL-7 binding to surface heparin was very similar.
Figure 5.
(A) Binding preferences of various GAGs to the interaction with human IL-7 using a SPR competition assay. Various GAGs at 1,000 nM were pre-equilibrated with 500 nM of hIL-7 before injection over heparin immobilized sensor chip. (B) Bar graphs (based on duplicated experiments with less than 5% errors) displaying the level of inhibition with the addition of various GAGs to the hIL-7/immobilized heparin interaction.
Next, IC50 values were measured to see the inhibition activity of these GAGs to IL-7 heparin binding. IC50 values are summarized in Table 1. These data agree with the results described above.
Table 1.
The IC50 values of GAGs to inhibit hIL-7 and mIL-7 binding to surface heparin (based on duplicated experiments with less than 5% errors)
| GAGs | IC50 (nM) for hIL-7 | IC50 (nM) for mIL-7 |
|---|---|---|
| Heparin | 50 | 50 |
| LMWH | 250 | 200 |
| Chondroitin Sulfate A | ni | ni |
| Chondroitin Sulfate C | >2000 | >2000 |
| Chondroitin Sulfate E | 1,000 | 500 |
| Chondroitin Sulfate D | >2000 | >2000 |
| Hyaluronan | ni | ni |
| Dermatan Sulfate | 2000 | 500 |
| Dermatan Disulfate | 150 | 100 |
| Heparan Sulfate | 1,000 | 1,000 |
| Acharan Sulfate | ni | ni |
ni – no inhibition
4. Discussion
Heparin sulfate proteoglycans (HSPGs) consist of several families (e.g., syndecans and perlecan) of core proteins covalently linked to large GAG chains that are found on the cell surface and in the ECM. Intensive biochemical and genetic studies have shown that HSPGs play crucial roles in regulating key developmental signaling pathways, such as the pathways for Wnt, Hedgehog (Hh), transforming growth factor-β (TGF-β), and fibroblast growth factors (FGFs) [30]. In the FGF signaling pathway, FGF receptor (FGFR) dimerization with FGF requires the association of heparin or highly sulfated HS polysaccharide chains of HSPGs. For example, FGF1 signaling is transmitted across the cell membrane through the formation of a ternary complex of FGF1, FGFR1, and HS [28]. In the Hh signaling pathway, HSPGs are essential for proper Hh distribution, stabilization, and signaling activity [29]. HSPGs are also believed to facilitate Hh ligand presentation to responding cells, and participate as part of a larger receptor complex [30]. Numerous cytokines including IL-7, the focus of this study, have been shown to interact with GAGs (particularly to heparin and HS) resulting in cytokine bioavailability and localization of hematopoietic cells to specific microenvironments [7–14].
Previous studies established an association of IL-7 with heparin/GAGs and these interactions play important roles in the IL-7 signaling cascade [11,12, 22, 23]. However, quantitative biophysical binding studies exploring IL-7 species differences (murine and human) to a broad range of GAGs have not been examined. SPR competition experiments were performed to: 1) determine the binding effects of heparin saccharide chain lengths to IL-7 interactions; 2) test the specificities of the heparin and HS GAGs in promoting IL-7 interactions; and 3) delineate the impact of GAG structure (e.g. sulfation level and spacing) to IL-7 interactions. The binding studies revealed a heparin chain-length dependence for the interactions to murine and human IL-7 and demonstrated a minimum heparin oligosaccharide length greater than a tetrasaccharide (dp4). Similar chain length requirements for heparin/HS protein interactions were reported for binding to other proteins including: Shh-heparin and FGF-heparin interactions [31, 32].
The SPR competition experiments with GAGs and chemically modified GAGs clearly showed an inhibitory activity that was dependent on the level of GAG sulfation and fine structure. The inhibitory effects of soluble GAGs to the hIL-7/immobilized heparin interaction was greatest for heparin with 2.8 moles of sulfate per disaccharide, followed by CSE with 1.5–2 moles of sulfate per disaccharide, then HS with 1.2 moles of sulfate per disaccharide, and finally by DS and CSC that both consist of 1 mole of sulfate per disaccharide. There was no inhibition of the hIL-7/immobilized heparin interaction for the GAGs of AS (1 mole of sulfate per disaccharide), CSA (< 1 mole of sulfate per disaccharide), or HA (no sulfates) at the concentrations assayed. These results are similar to the GAG sulfation preference of other signaling proteins of Shh, Ihog, FGF1, and FGF2 by interacting with more highly sulfated heparin than the less sulfated HS [29, 30]. The GAG fine structure also appears to play a prominent role in the IL-7 interaction. CSD, CSE, and Dis-DS are composed of disulfate disaccharide repeating units, yet displayed differing inhibitory activities to the IL-7/immobilized heparin interaction with greater activity of Dis-DS followed by CSE, and then CSD. CSE and CSD differ in the positions of their sulfo groups attached to the 4,6 versus the 2,6 positions, respectively (Figure 1). Dis-DS and CSE both contain disulfation at the 4, 6 position, but differ in the chirality of their uronic and glucuronic acids versus iduronic acid, respectively. Future studies determining a complex structure of IL-7 bound to a heparin derivative will be essential to map the specific interactions among the basic residues of IL-7 and the stereochemistry of the GAG sulfo groups.
An unanswered question from this study involves the binding stoichiometry of the interactions between IL-7 and GAGs. Unfortunately, SPR has limitations in conclusively determining the stoichiometry of molecular interactions. A simple 1:1 Langmuir biomolecular reaction model is most commonly selected to process the binding data of molecular interactions when other information is available on binding stoichiometry. Since heparin is heterogeneous and contains sufficiently long chains to interact with multiple protein molecules, the interaction between heparin and IL-7 may not be simple 1:1 binding. Future experiments such as isothermal titration calorimetry will be useful in determining the stoichiometry of the heparin-IL-7 interaction.
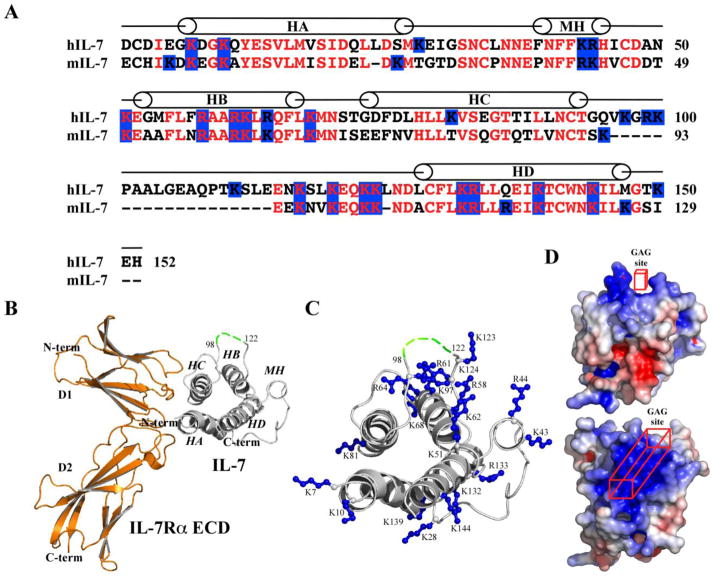
SPR analysis demonstrated that heparin bound to murine and human forms of IL-7 with different binding kinetics and affinities (Figure 2). A 140-fold tighter Kd was observed for the interaction of hIL-7 to heparin than the mIL-7/heparin interaction. The tighter binding affinity for the hIL-7/heparin interaction to mIL-7 resulted primarily through a faster on-rate (104 versus 102 M−1s−1). While both sequences of IL-7 are cationic, hIL-7 has a higher isoelectric point (pI) of 9.0 in comparison of 8.7 for mIL-7. The primary sequence of hIL-7 contains additional arginine (6 total) and lysine (19 total) residues relative to mIL-7 (Figure 6A). While the human and murine IL-7 sequences share 60% sequence identity, the tracks of lysine and arginine residues are highly conserved between the two species. With determinations of crystal structures of human IL-7 bound to unglycosylated and glycosylated forms of the human IL-7Rα ECD, structural insights may be gleaned into the potential GAG binding region on IL-7 (Figure 6B and C).
Figure 6.
Primary and tertiary structures of IL-7 and the potential IL-7-GAG binding site. (A) Primary sequence alignment of human and murine IL-7. The mini-helix (MH) and the 4-helix bundle (HA-HD) of hIL-7 are labeled. The second cross-over loop occurs from residues 96-114 using the hIL-7 numbering scheme. (B) Ribbon diagram of a crystal structure of hIL-7 to the hIL-7Rα ECD (3di3.pdb) [21]. The α-helices of IL-7 are labeled. The green dashed line connecting residues 98-122 represents the unobserved residues of hIL-7 in the crystal structure. (C) The basic residues of hIL-7 are displayed in ball-and-stick representation. The figure is roughly oriented in the same position as in B with HA and HC, the interaction site with the IL-7Rα ECD, positioned to the left. (D) Two views of electrostatic surfaces of hIL-7 contoured at levels of ± 5 kT/e (blue = positive and red = negative) by solving the linear Poisson-Boltzmann equation using ABPS [38] with 0.15 salt at 25 °C. The top view of hIL-7 is similar as in B while the bottom view is rotated 90° into the paper highlighting the top part of the molecule and the predicted GAG binding site. Structural figures were rendered using PyMOL [39].
The hIL-7 structure provides a model to predict the localization of the GAG binding site for both murine and human IL-7. There are three regions where the basic residues group on the hIL-7 structure (presumably also mIL-7 based on sequence alignment, Figure 6). The first region involves basic residues of helix A (K7 and K10) and C (K81) that are involved in the binding interface with the IL-7Rα ECD. Another region contains basic residues of the first loop (K28) and helix D (K132, R133, and K139) that localize to the predicted binding site for the interaction with the γc receptor ECD [21, 33-35]. The final region of basic residues involves residues of the mini-helix (K43 and R44), parts of the second cross-over loop (K123 and K124), and helix B (K51, R58, R61, K62, R64, and K68). Helix B contains a modified primary GAG binding motif of BXXBBXB (B = basic residue and X = any amino acid) [36]. Examination of the electropotential surface of hIL-7 indicates this region encompassing the mini-helix, helix B, and parts of the cross-over loop forms a large positive surface area that may specifically interact with GAGs such as heparin and HS. A cavity of basic residues is formed on IL-7 with distant residues in sequence space from the mini-helix and helix B (Figure 6D). A similar discontinuous GAG binding epitope has been reported for the lymphotactin-heparin interaction [37]. The second cross-over loop of hIL-7 from residues 98–122 was not observed in the electron density maps indicating a high degree of mobility [21]. This region consists of the additional exon of hIL-7 versus mIL-7. The additional exon of hIL-7 encodes three more lysine residues than the mIL-7 and may offer a rationale for the enhanced on-rate to heparin seen for hIL-7 over mIL-7. We posit that the GAG binding site on IL-7 involves residues localized to helix B, the mini-helix, and the second cross-over loop. Further site-directed mutagenesis studies of the basic IL-7 residues and structural determination of an IL-7/heparin complex will nail down the precise molecular mechanism of the interactions of GAGs with IL-7.
Highlights.
SPR was used to dissect the interactions between GAGs and murine/ human forms of IL-7
Heparin binds with higher affinity to human IL-7 than murine IL-7
The results provide key structural and energetic features of the interactions of GAGs to murine/human IL-7
Acknowledgments
This work was supported by grants from the National Institutes of Health: GM-38060 to R.J.L and AI-72142 to S.T.R.W.
Abbreviations
- IL-7
interleukin-7
- m
murine
- h
human
- SPR
surface plasmon resonance
- GAG
glycosaminoglycan
- HS
heparan sulfate
- HP
heparin
- LMWH
low molecular weight heparin
- CSA
chondroitin sulfate A
- DS
dermatan sulfate
- Dis-DS
dermatan disulfate
- CSC
chondroitin sulfate C
- CSD
chondroitin sulfate D
- CSE
chondroitin sulfate E
- HA
hyaluronic acid
- AS
acharan sulfate
- ECM
extracellular matrix
- SA
streptavidin
- FC
flow-cell
- RU
resonance unit
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Linhardt RJ. Heparin: structure and activity. Journal of Medicinal Chemistry. 2003;46:2551–2564. doi: 10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- 2.Capila I, Linhardt RJ. Heparin-protein interactions. Angewandte Chemie-International Edition. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 3.Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nature Reviews Molecular Cell Biology. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- 4.Parish CR. The role of heparan sulphate in inflammation. Nature Reviews Immunology. 2006;6:633–643. doi: 10.1038/nri1918. [DOI] [PubMed] [Google Scholar]
- 5.Powell AK, Yates EA, Fernig DG, Turnbull JE. Interactions of heparin/heparan sulfate with proteins: appraisal of structural factors and experimental approaches. Glycobiology. 2004;14:17R–30R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- 6.Sasisekharan R, Raman R, Prabhakar V. Glycomics approach to structure-function relationships of glycosaminoglycans. Annual Review of Biomedical Engineering. 2006;8:181–231. doi: 10.1146/annurev.bioeng.8.061505.095745. [DOI] [PubMed] [Google Scholar]
- 7.Roberts R, Gallagher J, Spooncer E, Allen TD, Bloomfield F, Dexter TM. Heparan sulphate bound growth factors: a mechanism for stromal cell mediated haemopoiesis. Nature. 1988;332:376–378. doi: 10.1038/332376a0. [DOI] [PubMed] [Google Scholar]
- 8.Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- 9.Turnbull JE, Fernig DG, Ke YQ, Wilkinson MC, Gallagher JT. Identification of the basic fibroblast growth factor binding sequence in fibroblast heparan sulfate. Journal of Biological Chemistry. 1992;267:10337–10341. [PubMed] [Google Scholar]
- 10.Ramsden L, Rider CC. Selective and differential binding of interleukin (IL)-1 alpha, IL-1 beta, IL-2 and IL-6 to glycosaminoglycans. European Journal of Immunology. 1992;22:3027–3031. doi: 10.1002/eji.1830221139. [DOI] [PubMed] [Google Scholar]
- 11.Borghesi LA, Yamashita Y, Kincade PW. Heparan sulfate proteoglycans mediate interleukin-7-dependent B lymphopoiesis. Blood. 1999;93:140–148. [PubMed] [Google Scholar]
- 12.Clarke D, Katoh O, Gibbs RV, Griffiths SD, Gordon MY. Interaction of interleukin 7 (IL-7) with glycosaminoglycans and its biological relevance. Cytokine. 1995;7:325–330. doi: 10.1006/cyto.1995.0041. [DOI] [PubMed] [Google Scholar]
- 13.Hasan M, Najjam S, Gordon MY, Gibbs RV, Rider CC. IL-12 is a heparin-binding cytokine. Journal of Immunology. 1999;162:1064–1070. [PubMed] [Google Scholar]
- 14.Spillmann D, Witt D, Lindahl U. Defining the interleukin-8-binding domain of heparan sulfate. Journal of Biological Chemistry. 1998;273:15487–15493. doi: 10.1074/jbc.273.25.15487. [DOI] [PubMed] [Google Scholar]
- 15.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nature Reviews Immunology. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Q, Li WQ, Aiello FB, Mazzucchelli R, Asefa B, Khaled AR, Durum SK. Cell biology of IL-7, a key lymphotrophin. Cytokine & Growth Factor Reviews. 2005;16:513–533. doi: 10.1016/j.cytogfr.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Mazzucchelli R, Durum SK. Interleukin-7 receptor expression: intelligent design. Nature Reviews Immunology. 2007;7:144–154. doi: 10.1038/nri2023. [DOI] [PubMed] [Google Scholar]
- 18.Noguchi M, Nakamura Y, Russell SM, Ziegler SF, Tsang M, Cao XQ, Leonard WJ. Interleukin-2 receptor gamma chain: a functional component of the interleukin-7 receptor. Science. 1993;262:1877–1880. doi: 10.1126/science.8266077. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin RG, Lupton S, Schmierer A, Hjerrild KJ, Jerzy R, Clevenger W, Gillis S, Cosman D, Namen AE. Human interleukin 7: molecular cloning and growth factor activity on human and murine B-lineage cells. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:302–306. doi: 10.1073/pnas.86.1.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lupton SD, Gimpel S, Jerzy R, Brunton LL, Hjerrild KA, Cosman D, Goodwin RG. Characterization of the human and murine IL-7 genes. Journal of Immunology. 1990;144:3592–3601. [PubMed] [Google Scholar]
- 21.McElroy CA, Dohm JA, Walsh STR. Structural and biophysical studies of the human IL-7/IL-7Ralpha complex. Structure. 2009;17:54–65. doi: 10.1016/j.str.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ariel A, Hershkoviz R, Cahalon L, Williams DE, Akiyama SK, Yamada KM, Chen C, Alon R, Lapidot T, Lider O. Induction of T cell adhesion to extracellular matrix or endothelial cell ligands by soluble or matrix-bound interleukin-7. European Journal of Immunology. 1997;27:2562–2570. doi: 10.1002/eji.1830271015. [DOI] [PubMed] [Google Scholar]
- 23.Kimura K, Matsubara H, Sogoh S, Kita Y, Sakata T, Nishitani Y, Watanabe S, Hamaoka T, Fujiwara H. Role of glycosaminoglycans in the regulation of T cell proliferation induced by thymic stroma-derived T cell growth factor. Journal of Immunology. 1991;146:2618–2624. [PubMed] [Google Scholar]
- 24.Wickham J, Walsh STR. Crystallization and preliminary X-ray diffraction of human interleukin-7 bound to unglycosylated and glycosylated forms of its alpha receptor. Acta Crystallographica Section F-Structural Biology and Crystallization Communications. 2007;63:865–869. doi: 10.1107/S1744309107042807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Science. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pervin A, Gallo C, Jandik KA, Han XJ, Linhardt RJ. Preparation and structural characterization of large heparin-derived oligosaccharides. Glycobiology. 1995;5:83–95. doi: 10.1093/glycob/5.1.83. [DOI] [PubMed] [Google Scholar]
- 27.Hernaiz M, Liu J, Rosenberg RD, Linhardt RJ. Enzymatic modification of heparan sulfate on a biochip promotes its interaction with antithrombin III. Biochemical and Biophysical Research Communications. 2000;276:292–297. doi: 10.1006/bbrc.2000.3453. [DOI] [PubMed] [Google Scholar]
- 28.Wu ZLL, Zhang LJ, Yabe T, Kuberan B, Beeler DL, Love A, Rosenberg RD. The involvement of heparan sulfate (HS) in FGF1/HS/FGFR1 signaling complex. Journal of Biological Chemistry. 2003;278:17121–17129. doi: 10.1074/jbc.M212590200. [DOI] [PubMed] [Google Scholar]
- 29.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes & Development. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 30.Lin X. Functions of Heparan Sulfate Proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- 31.Zhang FM, McLellan JS, Ayala AM, Leahy DJ, Linhardt RJ. Kinetic and structural studies on interactions between heparin or heparan sulfate and proteins of the hedgehog signaling pathway. Biochemistry. 2007;46:3933–3941. doi: 10.1021/bi6025424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang FM, Zhang ZQ, Lin XF, Beenken A, Eliseenkova AV, Mohammadi M, Linhardt RJ. Compositional analysis of heparin/heparan sulfate interacting with fibroblast growth factor.fibroblast growth factor receptor complexes. Biochemistry. 2009;48:8379–8386. doi: 10.1021/bi9006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olosz F, Malek TR. Three loops of the common gamma chain ectodomain required for the binding of interleukin-2 and interleukin-7. Journal of Biological Chemistry. 2000;275:30100–30105. doi: 10.1074/jbc.M004976200. [DOI] [PubMed] [Google Scholar]
- 34.Wang XQ, Rickert M, Garcia KC. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science. 2005;310:1159–1163. doi: 10.1126/science.1117893. [DOI] [PubMed] [Google Scholar]
- 35.LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi XL, Heller NM, Keegan AD, Garcia KC. Molecular and Structural Basis of Cytokine Receptor Pleiotropy in the Interleukin-4/13 System. Cell. 2008;132:259–272. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fromm JR, Hileman RE, Caldwell EEO, Weiler JM, Linhardt RJ. Pattern and spacing of basic amino acids in heparin binding sites. Archives of Biochemistry and Biophysics. 1997;343:92–100. doi: 10.1006/abbi.1997.0147. [DOI] [PubMed] [Google Scholar]
- 37.Peterson FC, Elgin ES, Nelson TJ, Zhang FM, Hoeger TJ, Linhardt RJ, Volkman BF. Identification and characterization of a glycosaminoglycan recognition element of the C chemokine lymphotactin. Journal of Biological Chemistry. 2004;279:12598–12604. doi: 10.1074/jbc.M311633200. [DOI] [PubMed] [Google Scholar]
- 38.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schrodinger L. The PyMOL Molecular Graphics System. 1.3 [Google Scholar]