Biologists have traditionally relied on temperature-sensitive mutants, transcriptional switches or degradation of coding transcripts by RNAi to regulate the function of a protein-of-interest (POI) in complex biological systems.[1, 2] Despite their utility, these systems offer less conditional control than small molecule inhibitors, a limitation that is exaggerated for long-lived and/or highly abundant proteins, and can be difficult to use in live animals. Unfortunately, although chemical genetic screening has successfully identified useful “probe” compounds with novel targets, many biologically interesting proteins (e.g., scaffolding proteins and transcription factors) lack specific, high-affinity small molecule ligands.[3, 4]
Alternatively, several groups have developed general methods to regulate protein abundance using cell-permeable small molecule ligands for a receptor genetically fused to a POI.[5] A set of interactions commonly used for this purpose is binding of the prolyl isomerase FKB12 or the FKB12-binding domain of mammalian target of rapamycin (FRB) by rapamycin, FK506 or derivatives of these natural products. For instance, this approach has been used to recruit a POI to the proteasome or mitochondrial outer membrane, thereby leading to its degradation or mislocalization, respectively.[6, 7] However, these methods require the introduction of two fusion proteins into cells. To address this issue, several labs have developed methods in which a POI is fused to an unstable protein fragment, termed a degradation domain (DD), that is rapidly degraded in the absence of bio-orthogonal “stabilizing” ligands.[8–11] Although DD-based systems have been used for in vivo studies, their utility for long-term experiments is compromised by the potential for fluctuations in POI abundance between injections of the stabilizing ligand.[12, 13] More recently, exposure of a cryptic DD has been used to degrade FKBP12-POI fusions in response to binding of a FK506 derivative.[14] However, preparation of the bio-orthogonal ligands for FKBP12 or FRB is synthetically challenging, making these systems prohibitively expensive for animal studies.
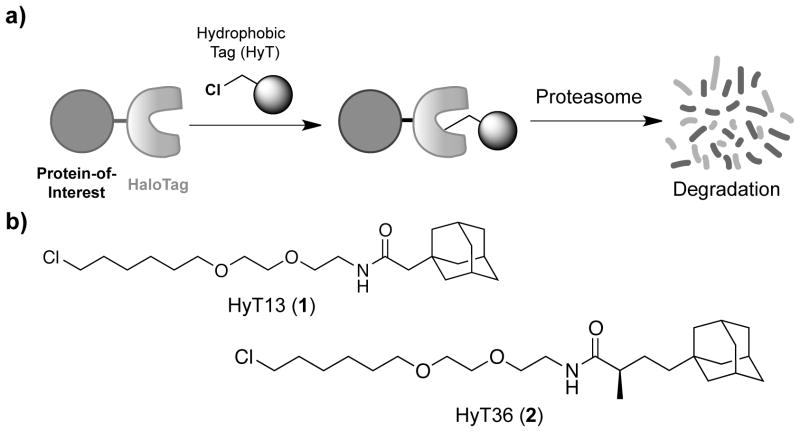
To develop a more accessible bio-orthogonal method to degrade any POI in response to a cell permeable small molecule we sought to co-opt the cellular quality control machinery. Specifically, we envisioned that labeling proteins with low molecular weight hydrophobic tags (HyTs) might promote their degradation because exposed hydrophobic regions are a hallmark of unfolded proteins leading to their elimination by the ubiquitin-proteasome system and/or autophagy.[15–17] As a proof-of-concept, we covalently labeled an early version of the HaloTag (HaloTag2) bacterial dehalogenase mutant with HyTs comprised of its cognate chloroalkane-reactive linker linked to a hydrophobic moiety, which promotes rapid, proteasome-dependent degradation of HaloTag2-fusion proteins (Figure 1a).[18] In particular, the adamantane-based HyT13 (1; Figure 1b) degraded a variety of fusion proteins in cell culture and zebrafish embryos in addition to inhibiting tumor progression driven by a HaloTag2-HRas1G12V fusion protein in mice. Of note, HyT13 (1) can be prepared using standard synthetic methods in four steps from commercially available starting materials with 63% overall yield.[18]
Figure 1.
Hydrophobic tagging-induced protein degradation. a) Fusion proteins composed of a protein-of-interest and HaloTag can be specifically targeted for proteasomal degradation with low molecular weight hydrophobic tags (HyTs). b) Chemical structures of HyT13 (1) and the more active HyT36 (2).
In the interim, HaloTag7, an improved HaloTag dehalogenase variant engineered to enhance stability for protein purification applications, has been developed.[19] Because of the burgeoning popularity of HaloTag7 and the commercial availability of expression constructs in which HaloTag7 is fused to nearly every mouse and human protein, we sought to determine whether HyT13 could efficiently de grade HaloTag7. Treating HEK 293 cells stably expressing a single copy of GFP-HaloTag2 or -HaloTag7 with HyT13 degraded the HaloTag2 fusion protein, but, unfortunately, had little effect on HaloTag7 as quantified by flow cytometry or immunoblot (Figure 2a and 2b; see Supporting Information for full experimental details).
Figure 2.
HyT13 does not significantly degrade the stabilized HaloTag7. a) and b) HEK 293 cells expressing GFP-HaloTag2 or GFP-HaloTag7 were treated with vehicle or HyT13 (1) (10 μM) for 24 hr followed by analysis of fusion protein levels by flow cytometry or immunoblot with specific antibodies as indicated. Images are representative of at least three independent experiments; band intensities were quantified and data presented as mean % degradation ± s.d.
To extend our hydrophobic tagging methodology to highly stabilized proteins like HaloTag7, we employed a chemical approach by conjugating a variety of hydrophobic moieties to the chloroalkane reactive linker (see Figure 1 in the Supporting Information). Hydrophobic moieties were selected in order to maximize hydrophobicity, minimize molecular weight and incorporate chemically diverse, commercially available scaffolds. Based on these parameters, 22 structurally distinct scaffolds were incorporated into approximately 40 HyTs by peptide coupling carboxylic acid derivatives of the hydrophobic moieties with the HaloTag chloroalkane reactive linker as described previously.[18]
We first evaluated the HyT library by treating HEK 293 cells expressing GFP-HaloTag2 since the greater dynamic range for degradation of HaloTag2 fusion protein better enables discrimination between small differences in activity. After 24 hr, GFP-HaloTag2 levels were assessed by flow cytometry followed by determination of the half-maximum inhibitory concentration (IC50) values by least-squares regression of the resulting concentration-response curves (see Table 1 in the Supporting Information). Consistent with our previous observations, HyT4 (30) and HyT5 (31), which lack large, hydrophobic moieties (CLogP ≤2) were completely inactive at all concentrations tested, indicating that covalent binding of chloroalkane linkers is insufficient to degrade HaloTag proteins. In contrast, appreciable reductions in GFP-HaloTag2 levels were detected with the remaining HyTs, which all have CLogP ≥2, indicating that a variety of hydrophobic moieties can degrade HaloTag proteins.
For the active HyTs, the maximum observed degradation did not correlate with CLogP (see Figure 2 in the Supporting Information) indicating that other factors such as cell permeability and/or stability contribute to observed activity. However, two trends did emerge from this data set. First, HyT activity decreases as the number of atoms between a particular hydrophobic moiety and the chloroalkane reactive linker increases. For example, HyT8 (9), HyT9 (25), HyT10 (26) and HyT7 (27) contain a triphenylmethyl group positioned 16, 22, 28 or 46 atoms from the reactive haloalkane, respectively, which correlates with a reduction in the maximum degradation from 76% for the shortest HyT to 28% for the longest. Similarly, increasing the distance of the adamantyl group from reactive haloalkane in HyT40 (23) is associated with a smaller maximum degradation (58%) compared to the 70% decrease observed with the shorter HyT13 (1). However, as observed for 38–44 reducing the distance between the chloroalkane linker and the hydrophobic moieties did not significantly decrease IC50 or maximum degradation beyond that observed with the corresponding longer linker (see Table 2 in the Supporting Information).
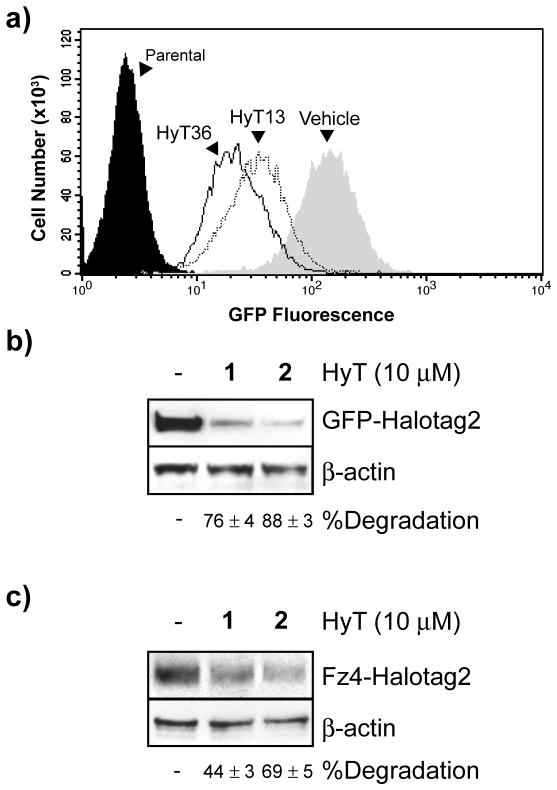
The more significant second trend is that incorporating branching hydrophobic groups at the α-carbon adjacent the peptide bond carbonyl increased potency as evidenced by greater maximum degradation observed for HyT36 (2; Figure 1b) and HyT39 (3), which bear a methyl or a phenyl methyl substituent at this position (see Figure 1 in the Supporting Information). Of these compounds, HyT36 (2) was selected for further evaluation because it displayed a reduced IC50 and greater maximum degradation of GFP-HaloTag2 relative to HyT13 (1). Flow cytometry and immunoblotting reconfirmed that treating HEK 293 cells with HyT36 (2) reduced GFP-HaloTag2 levels by ≈90% compared to the ≈75% decrease observed with HyT13 (1) (Figure 3a and 3b).
Figure 3.
HyT36 degrades HaloTag2 fusion proteins to a greater extent than HyT13. a) and b) HEK 293 cells expressing GFP-HaloTag2 were treated with vehicle, HyT13 (1) or HyT36 (2) for 24 hr followed by analysis of fusion protein levels by immunoblot or flow cytometry. c) HEK 293T cells expressing Fz4-HaloTag2 were treated with vehicle, HyT13 (1) or HyT36 (2) for 24 hr followed by detection of this fusion protein by immunoblot. Images are representative of at least three independent experiments; band intensities were quantified and data presented as mean degredation ± s.d.
Regulating transmembrane proteins is a challenge for existing technologies for small molecule control of protein function, which limits their applicability to many important classes of drug targets like G-protein coupled receptors (GPCRs).[20] Although HyT13 can efficiently degrade single-and 4-pass transmembrane proteins fused to HaloTag2, its effects on 7-pass transmembrane receptors, like GPCRs, is somewhat limited.[18] To determine whether HyT36 (2) can be used to degrade this class of proteins to a greater degree than HyT13 (1), we treated HEK 293T cells that had been infected with a retrovirus encoding the 7-pass transmembrane receptor Frizzled4 (Fz4) fused to the N-terminus of HaloTag2 with either compound. Similar to our results with GFP-HaloTag2, HyT36 (2) treatment decreased Fz4-HaloTag2 levels by ≈70%, relative to the ≈50% reduction observed with HyT13 (1) (Figure 3c). Hence, HyT36 (2) affords enhanced regulation of 7-pass transmembrane receptors.
Next, we sought to determine whether HyT36 (2) could be used to degrade the more stabilized HaloTag7 protein. For this experiment, HEK 293 cells expressing GFP-HaloTag7 were treated with HyT13 or HyT36 (10 μM, 24 hr) followed by evaluation of protein abundance by flow cytometry and immunoblotting. Both detection methods demonstrated a modest (ca. 30%) HyT13 (1)-induced degradation of GFP-HaloTag7, whereas HyT36 (2) was more than twice as effective, decreasing fusion protein abundance by ≈65% (Figure 4a and 4b). Significantly, concentrations of HyT36 used in this study were non-toxic in cell culture and did not degrade GFP when it was conjugated to a HaloTag protein (see Figures 3 and 4 in the Supporting Information). These results, along with our data for hydrophobic tagging-induced degradation of the 7-pass transmembrane receptor Fz4, indicate that HyT36 is better able to degrade stable proteins than HyT13.
Figure 4.
Effect of HyT36 on HaloTag7 fusion proteins. a) and b) HEK 293 cells expressing GFP-HaloTag7 were treated with vehicle, HyT13 (1) (10 μM) or HyT36 (2) (10 μM) for 24 hr followed by analysis of fusion protein levels by immunoblot or flow cytometry. c) Melting curves for HaloTag7 in the presence of vehicle (closed circles, solid line) or HyT13 (1) (10 μM, open circles, dashed line). d) Melting curves for HaloTag7 in the presence of vehicle (closed circles, solid line) or HyT36 (2) (10 μM, open circles, dashed line) or HyT36 (2) (40 μM, closed triangles, dotted line). Images are representative of at least three independent experiments; band intensities were quantified and data presented as mean % degredation ± s.d. Errors bars on thermal shift plots represent the mean ± s.d for each temperature.
The enhanced degradative activity of HyT36 (2) relative to HyT13 (1) could result from increased stability and/or cell permeability. Alternatively, HyT36 (2) might directly destabilize HaloTag proteins to a greater extent than HyT13 (1). We addressed the latter possibility using a fluorescence thermal shift assay in which a protein and analyte ligand are combined in the presence of SYPRO Orange, a dye that displays sharply increased fluorescence when bound to the hydrophobic interior of proteins.[21] Heating samples in a fluorimeter allows for protein unfolding to be measured in terms of increased fluorescence with the melting temperature (Tm) corresponding to the midpoint between the upper and lower limits of the fluorescence signal. In this assay, addition of an equimolar concentration of HyT13 (1) moderately reduced the Tm of purified HaloTag7 from 57.4 ± 0.4 to 56.4 ± 0.1 °C (p <0.01) indicating a very modest degree of destabilization (Figure 4c). By contrast, HaloTag7 was destabilized to a three-fold greater extent by equimolar HyT36 (2) as evidenced by a decrease in the Tm to 54.3 ± 0.2 °C (p <0.001) (Figure 4d). Significantly, incubation of HaloTag7 with a four-fold excess of HyT36 (2) did not significantly reduce the Tm further making it unlikely that the destabilization results from non-specific effects on protein stability (Figure 3d). While we cannot rule out a contribution of differences in stability and/or cell permeability, these in vitro data strongly suggest that the increased degradation of stabilized HaloTag fusion proteins observed with HyT36 in cells results from a greater, direct destabilizing effect on the HaloTag protein itself.
In summary, we synthesized and functionally evaluated a library of HyTs to identify members capable of more efficiently degrading stable proteins including HaloTag7, which is in wide use today.[22–24] The most active HyTs in this screen contained hydrophobic moieties with branching substituents at the a-carbon adjacent the peptide bond carbonyl. In particular, HyT36 (2), which bears a methyl substituent at this position, more robustly degraded both HaloTag2 and HaloTag7 fusion proteins, including the 7-pass transmembrane receptor Fz4, than the previously reported HyT13 (1). Although more structurally complex, HyT36 (2) can be prepared using standard synthetic methods in seven steps from commercially available starting materials with an overall 55% yield, making it amenable for general use as a hydrophobic tag. Finally, we have demonstrated a large destabilizing effect of HyT36 (2) on HaloTag7 in thermal shift assays, which suggests a mechanism whereby HyT binding directly destabilizes HaloTag proteins rather than the hydrophobic moieties serving as recognition elements for the cellular quality control machinery. As such, promoting protein unfolding by covalent linkage of destabilizing hydrophobic tags may represent a general strategy to induce targeted degradation of endogenous proteins including those often classified as “undruggable” using traditional methodologies.
Acknowledgments
We wish to acknowledge financial support from the US NIH (R01AI084140). T. Kirchhausen and G. Daley at Harvard Medical School kindly provided human Frizzled4 and the retroviral pEYK3.1 vector, respectively. D.J.N. is a Dept. of Defense NDSEG pre-doctoral fellow. A.G.R. is a Leopoldina - Nationale Akademie der Wissenschaften post-doctoral fellow. T.B.S. is supported by Post-Doctoral Fellowship 119708-PF-10-204-01-DDC from the American Cancer Society.
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
References
- 1.Banaszynski LA, Wandless TJ. Chem Biol. 2006;13(1):11–21. doi: 10.1016/j.chembiol.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Saghatelian A, Cravatt BF. Nat Chem Biol. 2005;1(3):130–142. doi: 10.1038/nchembio0805-130. [DOI] [PubMed] [Google Scholar]
- 3.Crews CM. Chem Biol. 2010;17(6):551–555. doi: 10.1016/j.chembiol.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schreiber SL. Proc Natl Acad Sci U S A. 2011;108(17):6699–6702. doi: 10.1073/pnas.1103205108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raina K, Crews CM. J Biol Chem. 2010;285(15):11057–11060. doi: 10.1074/jbc.R109.078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janse DM, Crosas B, Finley D, Church GM. J Biol Chem. 2004;279(20):21415–21420. doi: 10.1074/jbc.M402954200. [DOI] [PubMed] [Google Scholar]
- 7.Robinson MS, Sahlender DA, Foster SD. Dev Cell. 2010;18(2):324–331. doi: 10.1016/j.devcel.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau HD, Yaegashi J, Zaro BW, Pratt MR. Angew Chem Int Ed Engl. 2010;49(45):8458–8461. doi: 10.1002/anie.201003073. [DOI] [PubMed] [Google Scholar]
- 9.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. Cell. 2006;126(5):995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwamoto M, Bjorklund T, Lundberg C, Kirik D, Wandless TJ. Chem Biol. 2010;17(9):981–988. doi: 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu KJ, Arron JR, Stankunas K, Crabtree GR, Longaker MT. Nature. 2007;446(7131):79–82. doi: 10.1038/nature05557. [DOI] [PubMed] [Google Scholar]
- 12.Banaszynski LA, Sellmyer MA, Contag CH, Wandless TJ, Thorne SH. Nat Med. 2008;14(10):1123–1127. doi: 10.1038/nm.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dvorin JD, Martyn DC, Patel SD, Grimley JS, Collins CR, Hopp CS, Bright AT, Westenberger S, Winzeler E, Blackman MJ, Baker DA, Wandless TJ, Duraisingh MT. Science. 2010;328(5980):910–912. doi: 10.1126/science.1188191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonger KM, Chen LC, Liu CW, Wandless TJ. Nat Chem Biol. 2011;7(8):531–537. doi: 10.1038/nchembio.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredrickson EK, Rosenbaum JC, Locke MN, Milac TI, Gardner RG. Mol Biol Cell. 2011;22(13):2384–2395. doi: 10.1091/mbc.E11-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubota H. J Biochem. 2009;146(5):609–616. doi: 10.1093/jb/mvp139. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Liu Y, Soetandyo N, Baek K, Hegde R, Ye Y. Mol Cell. 2011;42(6):758–770. doi: 10.1016/j.molcel.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neklesa TK, Tae HS, Schneekloth AR, Stulberg MJ, Corson TW, Sundberg TB, Raina K, Holley SA, Crews CM. Nat Chem Biol. 2011;7(8):538–543. doi: 10.1038/nchembio.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohana RF, Encell LP, Zhao K, Simpson D, Slater MR, Urh M, Wood KV. Protein Expr Purif. 2009;68(1):110–120. doi: 10.1016/j.pep.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Schrader EK, Wilmington SR, Matouschek A. Chem Biol. 2010;17(9):917–918. doi: 10.1016/j.chembiol.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niesen FH, Berglund H, Vedadi M. Nat Protoc. 2007;2(9):2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 22.Strauch RC, Mastarone DJ, Sukerkar PA, Song Y, Ipsaro JJ, Meade TJ. J Am Chem Soc. 2011;133(41):16346–16349. doi: 10.1021/ja206134b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erkelenz M, Kuo CH, Niemeyer CM. J Am Chem Soc. 2011;133(40):16111–16118. doi: 10.1021/ja204993s. [DOI] [PubMed] [Google Scholar]
- 24.Zhou ZP, Shimizu Y, Tadakuma H, Taguchi H, Ito K, Ueda T. J Biochem. 2011;149(5):609–618. doi: 10.1093/jb/mvr010. [DOI] [PubMed] [Google Scholar]