Abstract
Neural stem cells (NSCs) generate neurons throughout life in the hippocampal dentate gyrus (DG). How gene expression signatures differ among NSCs and immature neurons remains largely unknown. We isolated NSCs and their progeny in the adult DG using transgenic mice expressing a GFP reporter under the control of the Sox2 promoter (labeling NSCs) and transgenic mice expressing a DsRed reporter under the control of the doublecortin (DCX) promoter (labeling immature neurons). Transcriptome analyses revealed distinct gene expression profiles between NSCs and immature neurons. Among the genes that were expressed at significantly higher levels in DG NSCs than in immature neurons was the growth factor insulin-like growth factor 2 (IGF2). We show that IGF2 selectively controls proliferation of DG NSCs in vitro and in vivo through AKT-dependent signaling. Thus, by gene expression profiling of NSCs and their progeny, we have identified IGF2 as a novel regulator of adult neurogenesis.
Introduction
A number of neuropsychiatric diseases such as epilepsy, stroke, age-related cognitive decline, and depression result in neuronal loss or dysfunction (Zhao et al., 2008). The discovery of neural stem cells (NSCs) in the adult brain that are proliferating and able to generate functional neurons gave rise to the idea that neuronal loss could be ameliorated by harnessing endogenous NSCs for brain repair (Lie et al., 2004; Suh et al., 2009). To achieve this goal, NSCs first have to be characterized in detail and their developmental programming must be better understood.
Under physiological conditions, the generation of substantial amounts of new neurons is restricted to two brain areas: the subventricular zone (SVZ) lining the lateral ventricles (Lois and Alvarez-Buylla, 1994) and the subgranular zone (SGZ) of the dentate gyrus (DG) in the hippocampus (Kuhn et al., 1996). NSCs of both neurogenic regions share intrinsic stem cell properties: when isolated and cultured in vitro, adult NSCs from the SVZ and DG are self-renewing, multipotent, and express several stem cell-associated proteins, such as the transcription factor SRY-box2 (SOX2) (Palmer et al., 1995; Doetsch et al., 1999; Seaberg and van der Kooy, 2002; Shi et al., 2004; Babu et al., 2007; Suh et al., 2007; Walker et al., 2008). In addition, newborn neurons express the microtubuli-associated protein Doublecortin (DCX), which is expressed for ∼3 weeks after cells are born, before they become functionally incorporated into the hippocampal or olfactory circuitry (van Praag et al., 2002; Carleton et al., 2003; Jessberger and Kempermann, 2003; Kempermann et al., 2004; Couillard-Despres et al., 2005).
Over the last decade, substantial progress has been made in identifying regulators of stem cell activity and neuronal differentiation. It appears that a number of extrinsic and intrinsic factors such as Wnt, Shh, BMP, and Notch control stem cell proliferation (Lai et al., 2003; Lie et al., 2005; Breunig et al., 2007; Lugert et al., 2010; Mira et al., 2010). Similarly, neuronal differentiation and maturation are regulated by intrinsic programs orchestrated through transcription factors such as NeuroD1 and a number of external cues, among them GABA and glutamate signaling (Ge et al., 2006; Tashiro et al., 2006; Gao et al., 2009).
Gene expression profiling of NSCs and their neuronal progeny holds the potential of identifying novel and niche-specific regulators of neurogenesis; however, previous attempts have failed to do so, either due to the sparseness of NSCs and newborn neurons within the adult brain or because these studies have focused exclusively on NSCs or immature neuronal populations (Pennartz et al., 2004; Beckervordersandforth et al., 2010).
In this study, we sought to identify novel regulators of adult neurogenesis by comparing gene expression profiles of discrete cell populations representing either NSCs or immature neurons. NSCs were isolated from transgenic mice expressing a GFP reporter under the control of the Sox2 promoter (hereafter called SOX2+ cells), and immature neurons were isolated from mice expressing a DsRed reporter under the control of the DCX promoter to isolate immature neurons (hereafter called DCX+ cells; Couillard-Despres et al., 2006; Suh et al., 2007). Using this approach, we identified novel and selective regulators of distinct steps in the developmental course of adult hippocampal neurogenesis, thereby providing the first gene expression-based analysis of adult hippocampal neurogenesis.
Materials and Methods
Fluorescence-activated cell sorting.
Two previously described transgenic mouse lines were used for fluorescence-activated cell sorting (FACS) of SOX2+ and DCX+ cells: one line expressed GFP under the Sox2 promoter and the other line expressed DsRed under the Dcx promoter (Couillard-Despres et al., 2006; Suh et al., 2007). For each experiment, the DGs of 10 hippocampi from 6- to 8-week-old mice were dissected and pooled. Dissected tissue was chopped into ∼1 mm3 pieces using sterile razor blades and dissociated by incubation for 30 min in a solution containing 0.01% papain (25 U/mg, Worthington Biochemicals), 0.1% neutral protease (0.5 U/mg, Roche), and 0.01% DNaseI (2788 U/mg, Worthington Biochemicals). The cell suspension was mixed with an equal volume of DMEM:F12 media (containing 1 mm l-glutamine and 10% fetal bovine serum) and filtered through a 70 μm nylon mesh, mixed with an equal amount of Percoll solution and pelleted (20,000 g for 30 min). Cellular debris was removed, and cells were spun down and dissolved in 2 ml of DMEM:F12 plus N2 supplement. For FACS, GFP was excited with a 488 nm water-cooled argon laser; DsRed was excited with a 561 nm solid-state laser. The filters used were 530/30 nm for GFP and 630/22 nm for DsRed using a BD FACSvantage Diva system (BD Biosciences). Three independent FACS experiments were performed for each cell type.
Microarray analysis.
RNA from 1000 cells from each cell population was isolated with TRIzol reagent (Invitrogen), amplified and reverse transcribed using the SuperAmp protocol (Miltenyi). cDNA was hybridized on 44 K mouse whole genome arrays (Agilent). The integrity of the cDNA was validated with the Agilent 2100 Bioanalyzer. Raw Cy3 data were normalized to the median and all values that were smaller than 1 were set to zero. All genes that had zero values in at least 5 of 6 arrays were deleted from the list, leaving 29,148 genes for analysis. Data were analyzed using the CARMA software suite (https://carmaweb.genome.tugraz.at/carma/; Rainer et al., 2006). The values were log2 transformed and quantile normalized. Expression differences were calculated over three replicates between two groups of experiments with the LIMMA package using a modified t-statistic. The Benjamini and Hochberg false discovery rate was used to calculate p values. The final gene lists contain all genes with a p value ≤0.05. Expression values of genes that were represented more than once on the array were averaged. Heatmaps were generated using the statistics software R (R Development Core Team, 2008) using the heatmap.2 method implemented in the gplots package (www.rosettabio.com/products/resolver). Array data are available online at the Gene Expression Omnibus (GSE21208; www.ncbi.nlm.nih.gov/geo). Gene Ontology classification and pathway analyses were performed using the MetaCore software package (www.genego.com/metacore).
Quantitative PCR.
Two methods for quantitative PCR (qPCR) experiments were used: SYBR Green and TaqMan. cDNA was synthesized using the SuperAmp protocol (Miltenyi). qPCR was performed either with a Power SYBR Green PCR Master Mix on an ABI Prism 7000 Sequence Detection System (Applied Biosystems) or with a TaqMan PCR Master Mix (Applied Biosystems) on an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). All qPCRs were performed using the same protocol (50°C, 2 min; 95°C, 10 min for 1 cycle; 95°C, 30 s; 60°C, 1 min for 40 cycles). All qPCRs were done in biological and technical triplicates. Data were analyzed using the SDS2.1 software package (Applied Biosystems) and Microsoft Excel. GAPDH was used as an endogenous normalization control and the fold expression relative to GAPDH was determined by the ΔΔCt method. All primer sequences used are available upon request. We used the Student's t test to test for significance.
Cell culture and constructs.
We isolated NSCs from micro-dissected DG and SVZ of 6- to 8-week-old adult mice as described previously (Ray and Gage, 2006). shRNA oligos were cloned into the original LentiLox3.7 vector (Addgene 11795), which contains an U6-driven cloning site and a CMV-driven GFP or a modified vector containing an U6-driven cloning site and a CMV-driven mCherry. Target sequences were CCAAAGAGCTCAAAGAGTT and GGACCGCGGCTTCTACTTCAGT (Zhu et al., 2007). As a control, we used vectors expressing an shRNA that does not have a murine target (nontargeting shRNA, CCTAAGGTTAAGTCGCC). Coding sequences for AKT (Addgene plasmid #9021) and dominant-active AKT (AKTdPH, Addgene plasmid #9009) were cloned into retroviral vectors as described previously (Jessberger et al., 2008). To test the efficiency of shRNAs to knockdown IGF2 protein levels, 293T cells were collected in standard RIPA buffer 48 h after transfection with Igf2-overexpressing and shRNA constructs using Lipofectamine 2000 (Invitrogen). Knockdown of IGF2 protein was analyzed by Western blots as described previously (Jessberger et al., 2008). For rescue experiments, IGF2 (20 ng/ml, Biotrend) was added to the culture medium 1 d after lentiviral transduction for 2 d before proliferation analyses. The peptide analog of IGF-I, JB1 (Bachem Biosciences), was used to block IG1R function. JB1 (1 μg/ml) was added to the culture medium 1 d after virus transduction. Two d after JB1 application, EdU was added to the culture medium for 1 h to measure proliferation rate.
For phosphoprotein Western blots, the RIPA buffer contained a mix of phosphatase inhibitors (Phosphatase Inhibitor Cocktail 1 and Phosphatase Inhibitor Cocktail 2, Sigma). Protein was isolated from mouse DG and mouse SVZ stem cell cultures either under proliferation or differentiation conditions (2 d for proliferation; 2 weeks for differentiation by growth factor withdrawal). Cells were infected with lentiviruses 1 d after plating and lysed using RIPA buffer containing protease inhibitors (Complete mini EDTA-free, Roche). Proteins were separated by SDS-PAGE on a 4–12% Bis-Tris gradient gel (Invitrogen) and transferred to a nitrocellulose membrane (Bio-Rad). Membranes were probed overnight using mouse α-pAKT (Cell Signaling Technology), rabbit α-AKT (Cell Signaling Technology), rabbit α-pIRS-1 (Cell Signaling Technology), rabbit α-IRS-1 (Cell Signaling Technology), or goat α-IGF-2 (Santa Cruz Biotechnology). HRP-conjugated secondary antibodies or AP-conjugated antibodies (Promega) were used at 1:4000 dilutions. Bands were detected by enhanced chemiluminescence substrate (Millipore) or CDS-Star substrate (Roche).
The levels of IGF2 in murine NSC cultures were measured using the Mouse IGF-II DuoSet (R&D Systems) ELISA kit, according to the manufacturer's instructions. IGF2 samples were taken from mDG/mSVZ and differentiated mDG stem cell cultures. Lysates were extracted using RIPA buffer and medium was first concentrated using Amicon Ultra centrifugal filter devices (Millipore) before measurement. Serial dilutions of IGF2 standard were assayed in parallel with lysates and culture medium samples. IGF2 concentration was expressed as pg/ml lysate or culture medium. The optical density was plotted against standard IGF2 concentrations to generate the standard curve, according to the manufacturer's protocol. Samples were read at 450 nm using a SpectraMax 190 Absorbance Microplate Reader (Molecular Devices).
For proliferation experiments, cells were pulsed for 1 h with 10 μm EdU (Invitrogen) and then fixed. EdU was visualized according to the manufacturer's protocol (Invitrogen). To analyze the mode of IGF2 action, we transduced NSCs using lentiviruses expressing shRNA against Igf2 with varying virus titers, thus leading to the infection of NSC cultures in different concentrations with transduced and nontransduced cells in the same culture dish. For differentiation, growth factors were withdrawn and cells were kept in DMEM:F12 plus heparin for 14 d. For all virus-mediated Igf2 knockdown experiments in vitro, 25 fields per coverslip (n ≥ 3) were imaged using a 40× objective on an inverted microscope (Zeiss Axiovert Observer-D1). GFP+ and EdU+ cells were counted using ImageJ (colocalization and cell counter plug-ins, NIH). Approximately 2000 GFP+ cells were counted per condition in the DG NSC proliferation assay and in the rescue experiment using 20 ng/ml IGF2 as a supplement. Approximately 1500 cells were counted for the proliferation assay of SVZ NSCs. Approximately 1000 cells per condition were analyzed for the differentiation experiments. In the rescue experiment, using a virus to overexpress AKT together with Igf2 knockdown or control viruses, ∼400 DG NSCs were counted per condition. In the IGF1R inhibition experiment using the pharmacological inhibitor JB1 (1 μg/ml), ∼800 GFP+ cells were counted. For the dilution series experiments, ∼6000 DAPI+ cells were counted per condition. GFP+ cells were calculated by subtracting the GFP− cells from the total DAPI+ cells. The GFP+EdU+ cells were calculated by subtracting the total GFP−EdU+ cells from the total EdU cell number.
Animals and immunostaining.
Female mice were used for all immunohistochemical experiments: WT (C57BL/6) were 6–8 weeks old and nestinGFP transgenic mice were 8 weeks old (Yamaguchi et al., 2000). All protocols were approved by the Salk Institute's Institutional Animal Care and Use Committee or by the veterinary office of the Canton of Zurich. For knockdown of IGF2, animals were injected with lentiviruses (two shRNAs targeting Igf2 mRNA, one nontargeting, control shRNA) into the DG (coordinates from bregma anteroposterior (AP) −2.0, ML, ±1.5, DV −2.3 from skull or for SVZ AP −1.0, ML ± 1.0, DV −2.8 from skull). Three weeks after viral injections, animals received one injection of IdU (87 mg/kg bodyweight) and were killed 24 h later. Tissue was processed for immunostaining as described earlier (Jessberger and Kempermann, 2003). Briefly, brains were cut into 40-μm-thick sections and distributed into 12 series. One series from each brain (n ≥ 3) was stained and used for counting cell numbers. All countings were done using serial scanning microscopy. For all combinations analyzed, ≥50 cells per animal (with n ≥ 3) were phenotyped for the expression of the respective markers. For intensity measurement, images were taken with a Leica SP2 confocal fluorescence microscope, using the same settings for all conditions. All sections were stained and imaged in parallel. IGF2 staining fluorescence intensity was measured and calculated using ImageJ (NIH). Immunostaining intensity was measured over the virus-infected area and normalized to the intensity of the nontargeting shRNA. Integrated density measurements of the control were set to 1.
Primary antibodies used were rat α-BrdU (Harlan Seralab), mouse α-BrdU (Becton Dickinson; for IdU detection), rabbit α-GFP (Invitrogen), chicken α-GFP (Aves Labs), rabbit α-SOX2 (Millipore Bioscience Research Reagents), goat α-SOX2 (Santa Cruz Biotechnology), goat α-DCX (Santa Cruz Biotechnology), rabbit α-DCX (Cell Signaling Technology), mouse α-GFAP (Millipore Bioscience Research Reagents), goat α-IGF2 (Santa Cruz Biotechnology), and rabbit α-Ki67 (Novocastra). All secondary antibodies were from Jackson ImmunoResearch. Fluorescent signals were detected using two different spectral confocal microscopes (Radiance 2100, Bio-Rad, and Leica SP2). Nuclei of cells and tissues were counterstained with 4–6-diamidino-2-phenylindole (DAPI, Sigma). Images were processed in Adobe Photoshop (CS5 for Mac; Adobe Systems) for general contrast enhancements and color adjustments.
In situ hybridization.
Single-stranded RNA probes against Igf2 mRNA were generated by in vitro transcription of a full-length Igf2 cDNA (Open Biosystems plasmid MMM1013–9201704) using T7 or Sp6 RNA polymerase in the presence of digoxigenin (DIG)-UTP. The hybridization procedure was performed according to standard procedures (Wilkinson, 1998).
Statistical analyses.
All numerical analyses were performed using Excel (Microsoft) or SPSS (IBM). Two sample t tests or ANOVA (if applicable) were used for all comparisons. Differences were considered significant at p < 0.05.
Results
FACS-based isolation and transcriptome analyses of SOX2+ and DCX+ cells from the adult DG
We microdissected the DG of transgenic mice that were expressing either GFP under the control of the Sox2 promoter or DsRed under the control of the Dcx promoter (Couillard-Despres et al., 2006; Suh et al., 2007), and we subsequently performed FACS for SOX2+ and DCX+ cells (Fig. 1A,B). We used the resultant cell populations for RNA preparation and performed microarray hybridization experiments. To identify differentially regulated genes, we used statistical analysis of microarray data (see Materials and Methods, Microarray analysis). Only genes that were found to be significant after multiple testing corrections (Benjamini and Hochberg; p < 0.05) were included in the analyses. We validated a set of differentially regulated genes with quantitative PCR (qPCR; Fig. 1C,D). The array and qPCR expression data were in agreement for all tested genes, with a generally high concordance between fold changes calculated from the array and from qPCR analyses (data not shown). Cluster analyses of differentially expressed genes revealed the high reproducibility of the biological triplicates (Fig. 1E).
Figure 1.
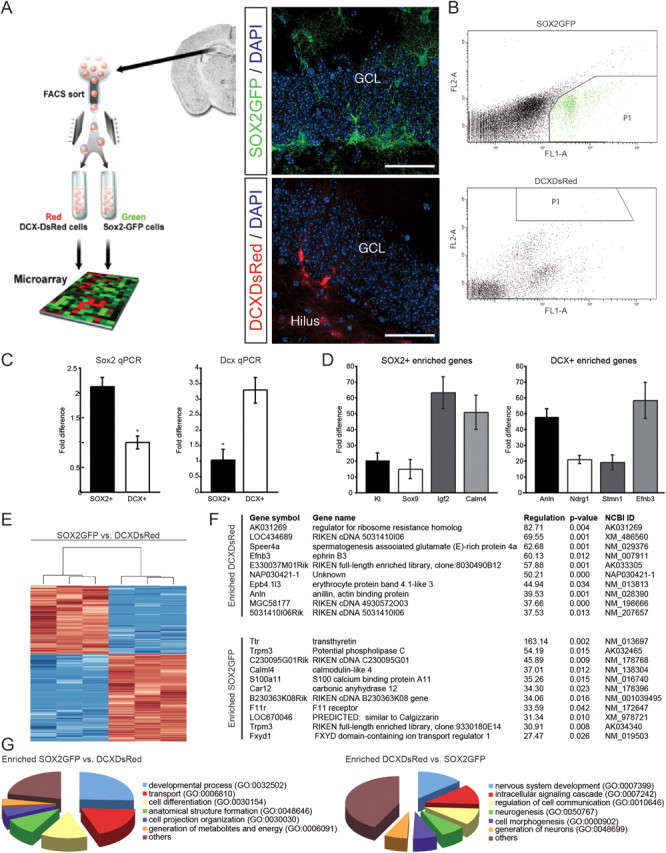
Experimental strategy and expression of genes differentially regulated between NSCs and their progeny. A, NSCs and their neuronal progeny were directly isolated from the adult DG of SOX2GFP or DCXDsRed transgenic mice using FACS, followed by RNA isolation and subsequent array hybridization (left). GFP and DsRed reporter expression in the adult DG (insets, right). Scale bars, 100 μm. GCL, granule cell layer. B, FACS of adult hippocampal NSCs and their neuronal progeny. Representative examples of FACS experiments using transgenic mice labeling SOX2GFP- or DCXDsRed-expressing cells. Top, SOX2GFP-expressing cells isolated from the DG. Bottom, DCXDsRed-expressing cells isolated from the DG. FL1-A represents the GFP signal intensity, FL2-A represents the DsRed signal intensity. Boxed areas show the cell population that was selected for RNA isolation. C, Verification of Sox2 and Dcx enrichment using qPCR. To verify the enriched expression of Sox2 and Dcx in SOX2GFP- and DCXDsRed-expressing cells, we measured their expression levels using quantitative PCR. Shown are the fold differences between SOX2+ and DCX+ cells (left, Sox2 qPCR; right, Dcx qPCR). Data are presented as mean ± SEM. D, Verification of array data using qPCR. To verify the array data, we picked four candidates of the top regulated genes from all comparisons and measured their expression levels using quantitative PCR. Shown are the fold differences either to SOX2GFP- or DCXDsRed-expressing cells within DG. Data are presented as mean ± SEM. Kl, Klotho; Sox9, SRY (sex determining region Y)-box 9; Igf2, insulin-like growth factor 2; Calm4, Calmodulin 4; Anln, Anilin; Ndrg1, N-myc downstream regulated gene 1; Stmn1, Stathmin 1; Efnb3, EphrinB3. E, Cluster analysis of differentially regulated genes expressed in SOX2+ versus DCX+ cells indicates the high reproducibility of biological triplicates. Red and blue denote high and low expression levels respectively. F, Top 10 upregulated and downregulated genes between SOX2+ and DCX+ cells of the DG. Regulation is expressed as the relative difference in gene expression levels between SOX2+ and DCX+ cells. G, GO analysis of differentially regulated genes. All classes shown were significantly enriched in their respective categories (p < 0.05).
Analyses of gene expression data showed that 1494 genes (3.4% of all transcripts interrogated) were differentially regulated between SOX2+ and DCX+ cells in the DG. Of these, 840 genes (∼56%) were upregulated and 654 genes (∼44%) were downregulated in DCX+ cells compared with SOX2+ cells. The top 10 regulated genes are shown in Figure 1F. We used gene annotation analysis of gene ontology (GO) terms to identify pathways and processes of regulated genes that had a significant enrichment of regulated genes within the DG. We found that, among others, the GO classes of developmental processes and cell differentiation were significantly enriched in SOX2+ cells (Fig. 1G). In DCX+ cells, we found GO terms of genes enriched that are associated with nervous system development, neurogenesis, generation of neurons, and neurite development. Among the highly expressed genes in DCX+ cells were previously identified regulators of differentiation such as Stathmin 1 (Stmn1), a cytosolic phosphoprotein that plays an important role in regulating microtubule dynamics in the context of mitosis as well as neuronal migration (Peschanski et al., 1993). Another example is Prox1, which has recently been identified to be critical for neuronal differentiation of hippocampal granule cells (Lavado et al., 2010; Karalay et al., 2011). In total, we found that ∼31% of the genes that were expressed more highly in DCX+ cells had been previously associated with neurogenesis or differentiation, thus supporting the validity of the gene array data. In analogy, we found that ∼29% of all genes overexpressed in SOX2+ cells had been previously implicated in stem cell activity or cell proliferation.
Surprisingly, we also found many genes enriched in SOX2+ cells enriched in GO classes associated with generation of energy, lipid metabolism and catabolic processes, which might point toward a previously unrecognized metabolic program of DG NSCs. For example, we found 12 genes enriched in SOX2+ cells that were annotated in the GO term regulation of lipid metabolic process (GO 0019216), among them Bmp6, Apoe, Acacb, Igfbp7, Edf1, Irs, Lrp1, Cpt1a, Pparc1a, Acaa2, and Acsl6.
IGF2 is highly expressed in NSCs in the DG
We next sought to characterize the functional significance of a gene that is highly expressed in the SOX2+ cell population in the DG. We focused our analyses on IGF2, which we identified to be highly expressed in SOX2+ cells compared with DCX+ cells (∼60-fold; Fig. 2A). This expression profile, together with previous data showing that the growth factor IGF2 plays an important role during embryonic development and has been implicated in sonic hedgehog (Shh)-mediated cell proliferation and neuronal survival (Toretsky and Helman, 1996; Hartmann et al., 2005; Agis-Balboa et al., 2011), made IGF2 an intriguing candidate for a regulator of hippocampal neurogenesis. After confirming the array data by qPCR (Fig. 2A), we performed in situ hybridization using probes directed against Igf2 mRNA and found an enrichment of Igf2 mRNA in the SGZ, the site within the DG that harbors NSCs and immature neurons (Fig. 2B and data not shown). In support of the mRNA expression, we detected high expression levels of IGF2 protein in the SGZ with higher expression in SOX2+ cells compared with DCX+ cells in the DG (Fig. 2C,D), corroborating the array- and mRNA-based Igf2 gene expression results. This finding also confirmed previous reports that showed Igf2 expression in the postnatal DG (Zhang et al., 2007). We next characterized the expression of IGF2 in more detail based on previously described markers of NSCs in the adult DG (Seri et al., 2001; Kronenberg et al., 2003; Suh et al., 2007). We found that IGF2 was highly expressed in radial glia-like NSCs expressing SOX2GFP, NestinGFP, or glial fibrillary acidic protein (GFAP) and that a substantial fraction of dividing (Ki67+) cells colabeled with IGF2 (Fig. 2E,F). Thus, the gene expression profiling strategy we used correctly identified IGF2 to be highly enriched in a NSC population within the adult DG.
Figure 2.
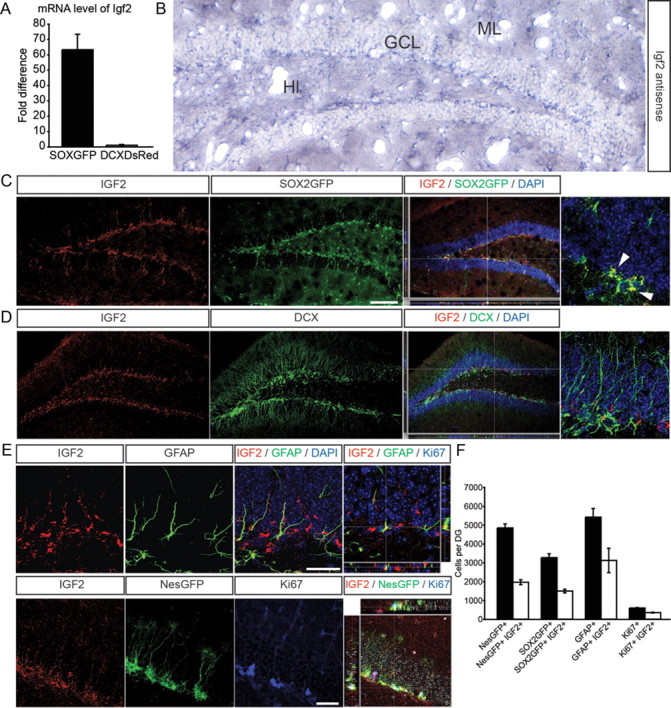
IGF2 is expressed in the SGZ of the DG. A, Quantitative RT-PCR shows strong enrichment of Igf2 in SOX2+ cells of the DG compared with DCX+ cells. B, In situ hybridization using a riboprobe against Igf2 mRNA shows expression in the granule cell layer (GCL) and an enrichment of Igf2 mRNA in the SGZ of the DG. Hl, hilus; ML, molecular layer. C, IGF2 (red) shows high expression in SOX2+ cells (green) in the DG. A high-power view of a SOX2GFP/IGF2 coexpressing cell is shown on the far right (arrowheads). D, IGF2 (red) shows only low expression in DCX+ cells (green). A high-power view of a DCX+ cell (green) that does not colabel with IGF2 is shown on the far right. E, Representative images of IGF2 expression (red) in costainings with GFAP (top, green), NestinGFP (bottom, green), and Ki67 (bottom, blue). Z-stacks show 3-dimensional reconstruction of IGF2/GFAP colabeled cells (top) and IGF2/NestinGFP colabeled cells (bottom). F, Using confocal microscopy, we quantified the number of NestinGFP+, SOX2GFP+, GFAP+, and Ki67+ cells colabeling with IGF2. We found that a substantial fraction of radial-glia like cells in the DG expressed IGF2. Nuclei were counterstained with DAPI. Data are presented as mean ± SEM. Scale bars (in C) C, D, 100 μm; E, 50 μm.
IGF2 regulates proliferation of hippocampal NSCs in vitro
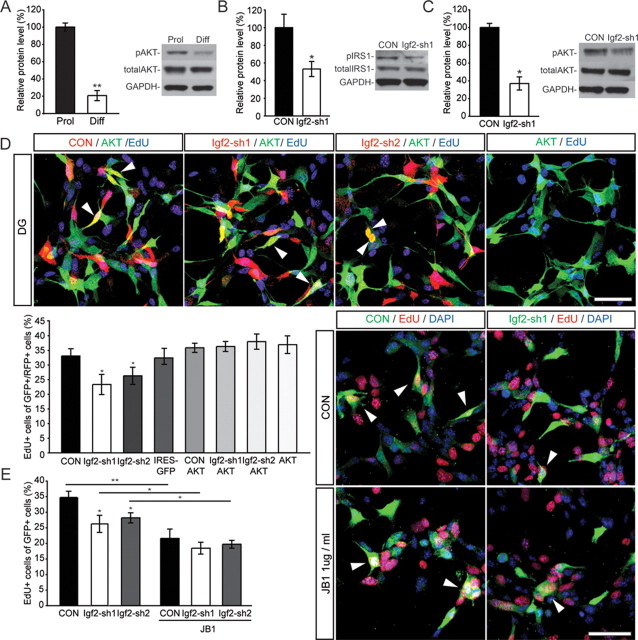
To test the functional importance of Igf2, we isolated NSCs from the adult DG and analyzed IGF2 protein levels in vitro. Independent array data obtained from SOX2+ cells from the SVZ indicated that IGF2 was only enriched in SOX2+ cells from the DG but not in SOX2+ cells isolated from the SVZ. Thus, we isolated in parallel NSCs from the adult SVZ. Confirming the gene expression profile, Western blots and ELISA showed that IGF2 protein was strongly enriched in DG-derived NSCs compared with NSCs derived from the SVZ (Fig. 3A,B). Further, IGF2 was substantially downregulated with differentiation of NSC cultures, as measured by total protein amount and protein levels in the supernatant by ELISA (Fig. 3A,C).
Figure 3.
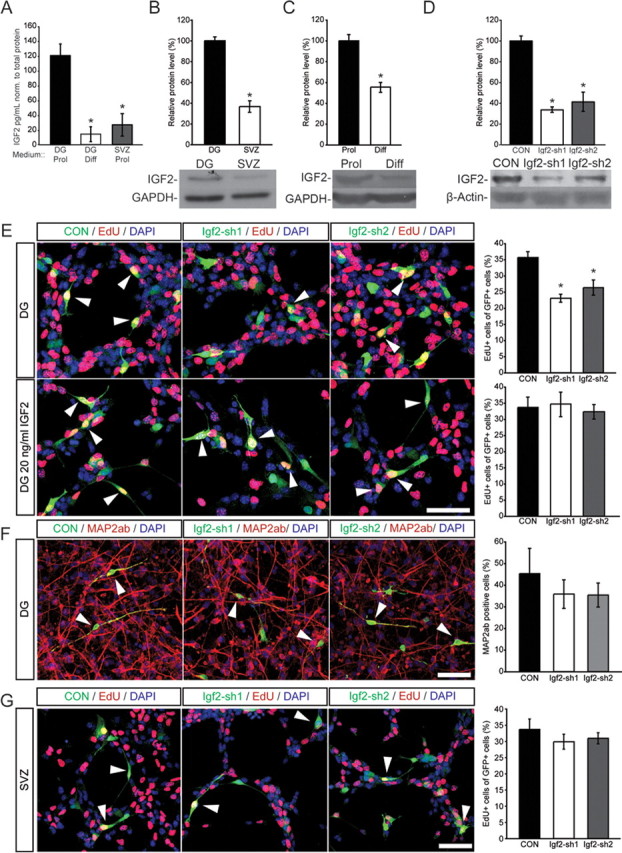
IGF2 regulates proliferation of DG NSCs in vitro. A, ELISAs for IGF2 were performed. Medium from proliferating DG and SVZ NSCs and from 14 d differentiated DG NSCs was collected, concentrated and analyzed. Graph shows the amount of Igf2 in the medium in pg/ml normalized to total protein (mg/ml). B, IGF2 protein levels are higher in DG-derived NSCs than in SVZ-derived NSCs. Total protein was isolated from proliferating NSCs isolated from DG or SVZ. Western blot analysis showed higher protein levels of IGF2 in DG- than in SVZ-derived cultures. C, IGF2 is downregulated with differentiation of DG-derived NSCs. Shown is a representative Western blot and quantification of relative protein levels. D, DG cells were infected with lentiviruses expressing different shRNA sequences targeting Igf2 mRNA. shRNA sequences sh1 and sh2 resulted in ∼75% and 60% decreases in IGF2 protein levels, respectively. Shown are representative Western blots with nontargeting shRNA (CON) or shRNA against Igf2. E, Knockdown of IGF2 using two independent lentiviruses expressing shRNAs (Igf2-sh1 and Igf2-sh2) targeting the murine Igf2 mRNA resulted in a robust decrease in the number of proliferating, EdU-labeled, DG-derived NSCs in vitro (top, red). Exogenous IGF2 rescued the proliferation defect caused by knockdown of endogenous IGF2 knockdown. IGF2 supplementation (20 ng/ml IGF2) rescued the proliferation phenotype (as measured by EdU incorporation, bottom, red) of IGF2 knockdown in DG-derived NSCs. Images show representative pictures of cells transduced with a lentivirus expressing nontargeting shRNA (CON, green, left) or expressing shRNA sequence 1 (Igf2-sh1) or 2 (Igf2-sh2). Arrowheads point to GFP-expressing, EdU-labeled cells. Data are presented as mean ± SEM. Scale bar, 50 μm. F, After transduction with viruses expressing shRNAs directed against Igf2 or nontargeting control shRNA, NSCs were differentiated for 14 d. There was no effect on the number of MAP2ab/GFP+ cells compared with control. Shown are examples and the quantification of GFP (virus-expressing, green) and MAP2ab (red) colabeled cells in DG NSC cultures. Nuclei were counterstained with DAPI (blue). Data are presented as mean ± SEM. Scale bar, 100 μm. G, IGF2 knockdown did not affect the number of EdU-labeled cells in NSC cultures derived from the adult SVZ. Right, representative pictures of cells transduced with a nontargeting shRNA expressing lentivirus (CON, green, left) or expressing shRNA sequence 1 (Igf2-sh1) or 2 (Igf2-sh2) (green, middle and right). Arrowheads point toward GFP-expressing, EdU-labeled cells. Scale bar, 50 μm. *p < 0.05.
We next tested whether knockdown of Igf2 would affect NSC behavior in a cell type-specific manner. We transduced cells in vitro with lentiviruses expressing two different silencing short-hairpin RNAs (shRNAs) directed against Igf2 mRNA (Fig. 3D). When we measured proliferation rates of DG-derived NSCs with 5-ethynyl-2-deoxyuridine (EdU) pulse labeling or by staining for Ki67, we found that IGF2 knockdown significantly decreased proliferation (Fig. 3E and data not shown). Notably, Igf2 knockdown was not associated with enhanced apoptosis of NSCs in vitro, as measured by TUNEL assay (data not shown). Further, we found that reduced proliferation after Igf2 knockdown of DG-derived NSCs was fully rescued by the addition of exogenous IGF2 (Fig. 3E), whereas addition of IGF2 to control cells had only a minor effect on NSC proliferation (data not shown). As expected from the robust downregulation of IGF2 with differentiation, we did not observe an effect of IGF2 knockdown on neuronal differentiation of either DG- or SVZ-derived cells (Fig. 3F). However, we did not find an effect on proliferation in SVZ-derived NSCs after knockdown of Igf2 suggesting that production of IGF2 is only critical for SOX2+ NSCs derived from the adult DG (Fig. 3G).
In conclusion, these data suggest that IGF2 plays a role in the proliferation of NSCs derived from the adult DG in vitro.
IGF2 regulates NSC proliferation via AKT signaling in a largely autocrine manner
We next sought to characterize the signaling pathways through which IGF2 might regulate proliferation of DG-derived NSCs. Pathway analyses of our gene expression profiles (data not shown) and previously published reports suggested that IGF2 might act via v-akt murine thymoma viral oncogene homolog 1 (AKT) to regulate NSC proliferation (Hartmann et al., 2005; Peltier et al., 2007). To dissect the molecular mechanisms underlying the effects of IGF2 on stem cell behavior, we focused on the role of AKT-dependent signaling. We found that levels of phospho-AKT (pAKT) were significantly downregulated with neuronal differentiation, supporting a role for AKT signaling in NSC proliferation (Fig. 4A). ShRNA-mediated knockdown of Igf2 led to a reduction in insulin receptor substrate 1 (IRS1) phosphorylation, an intracellular adaptor protein that becomes phosphorylated upon IGF-receptor activation (Sun et al., 1991), and a reduction in pAKT levels (Fig. 4B,C). In addition, we found that the phenotype of Igf2 knockdown (i.e., impaired proliferation) was rescued by overexpression of a dominant-active AKT (Fig. 4D). Together, these data suggest that IGF2 acts via AKT-dependent signaling to regulate proliferation of hippocampal NSCs.
Figure 4.
IGF2 acts via AKT signaling to regulate NSC proliferation. A, Levels of pAKT are dynamically regulated with differentiation of NSCs, suggesting a role for pAKT signaling in NSC proliferation. B, Knockdown of IGF2 decreases IRS1 phosphorylation. C, Knockdown of IGF2 results in decreased AKT phosphorylation levels. A–C show representative Western blots and densitometric quantification. Data are normalized to total AKT (A, C) or total IRS1 (B). D, Virus-mediated overexpression of AKT (green) in NSCs rescues the proliferation defect as measured by EdU labeling (blue) caused by IGF2 knockdown (control and shRNA-expressing virus in red). Arrowheads point to EdU/GFP/mCherry-labeled cells. Bars show quantification of NSC proliferation. E, Virus-mediated knockdown of IGF2 (control and shRNA-expressing viruses in green) together with blocking IGF1R function (using JB1) indicates that inhibition of IGF1R strongly reduces NSC proliferation, exceeding the effect of IGF2 knockdown alone. Arrowheads point to EdU/GFP-labeled cells. Data are presented as mean ± SEM. Scale bar, 50 μm. **p < 0.01; *p < 0.05.
We next analyzed whether IGF2 functions via an IGF1 receptor (IGF1R) or IGF2 receptor (IGF2R). Although most data suggest that IGF2R does not transduce a signal but rather sequesters the activity of IGF2 away from the IGF1R (for a review see Pollak, 2008), recently published evidence has suggested that IGF2R might be able to mediate IGF2-dependent signaling (Chen et al., 2011). We used a IGF1R-specific blocking peptide (JB1; Pietrzkowski et al., 1992) and found that inhibition of IGF1R strongly reduced NSC proliferation, exceeding the effect of IGF2 knockdown alone (Fig. 4E). In addition, IGF2 knockdown did not further decrease NSC proliferation after JB1 treatment, suggesting that IGF2 functions via the IGF1R but also that additional growth factors (such as IGF1) may act on the IGF1R to regulate NSC proliferation.
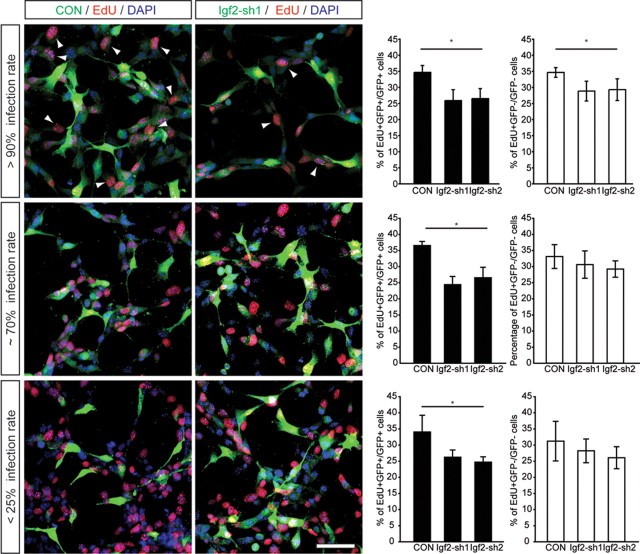
To test whether IGF2 functions predominantly in an autocrine manner, we cocultured NSCs transduced with shRNA-expressing viruses in distinct ratios together with nontransduced cells. Notably, we found that only high ratios of IGF2-knockdown cells (>90% of all cells) affected the proliferation on nontransduced NSCs, as measured with EdU pulse labeling (Fig. 5). In contrast, low ratios of IGF2-knockdown cells (<25% of all cells) did not show any effect on the proliferation on nontransduced NSCs (Fig. 5). This finding suggests that IGF2 largely functions in an autocrine manner but also exerts paracrine effects to regulate hippocampal NSC proliferation.
Figure 5.
IGF2 functions predominantly in an autocrine manner. Cocultured DG NSCs transduced with shRNA-expressing viruses (control and shRNA-expressing virus in green) were plated in three ratios (>90%, top; ∼70%, middle; <25%, bottom) together with nontransduced DG-derived NSCs. Proliferation of transduced and nontransduced cells was measured by EdU labeling (red). High ratios of IGF2-knockdown cells (>90% of all cells) affected the proliferation rate of nontransduced cells, whereas at lower rates no differences could be detected compared with control. Arrowheads point to GFP-expressing, EdU-labeled cells. Data are presented as mean ± SEM. Scale bar, 50 μm. *p < 0.05.
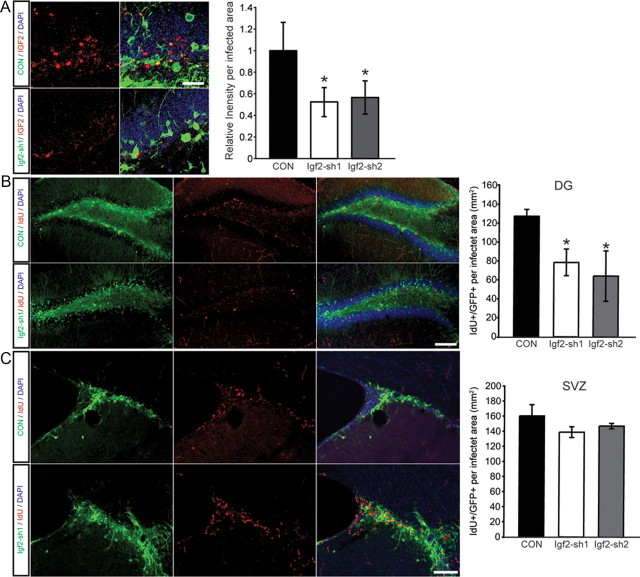
Regulation of NSC proliferation by IGF2 in the adult brain
Next we analyzed whether the effects of IGF2 on NSCs that we observed in vitro also occurred within the adult brain. We injected shRNA-expressing lentiviruses stereotactically into the DG of adult mice and observed a significant drop in IGF2 protein levels within the adult DG (Fig. 6A,D). We next analyzed cell proliferation using the thymidine analog 5-iodo-2-deoxy-uridine (IdU) 3 weeks after virus injections. In analogy to the in vitro experiments, we found that IGF2 knockdown reduced proliferation of hippocampal NSCs (Fig. 6B). Corroborating the in vitro data, we found no effect of IGF2 knockdown on cell proliferation in the adult SVZ (Fig. 6C). Together, these results suggest that IGF2 acts as a regulator of adult hippocampal neurogenesis. Furthermore, this finding highlights the usefulness of an unbiased gene expression profiling approach to reveal new molecular mechanisms underlying life-long neurogenesis in the mammalian brain.
Figure 6.
IGF2 knockdown reduces NSC proliferation in the adult DG. A, Fluorescence intensity measurements of IGF2 staining in either nontargeting shRNA or shRNA against Igf2-infected cells. Shown are confocal images of the DG after injection of a control virus expressing a nontargeting shRNA (CON, green, top) or virus expressing shRNA against Igf2 (Igf2-sh1, green, bottom) and stained for IGF2 (red, left). Quantification of the IGF2 staining in virus-transfected cells of the DG, showing a reduction in the relative fluorescence level in the Igf2 knockdown cells compared with the CON shRNA cells. B, Shown are confocal images of the DG after injection of a control virus expressing a nontargeting shRNA (green, top) or virus expressing shRNA against Igf2 (Igf2-sh1, green, bottom). Proliferating cells in the DG were visualized by IdU labeling (red). Note the reduction in IdU-labeled cells in the DG. Right, an overlay of single channels. C, Shown are confocal images of the SVZ after injection of a control virus (green, top) or virus-expressing shRNA against Igf2 (Igf2-sh1, green, bottom). Right, an overlay of single channels. In contrast to the DG, IGF2 knockdown does not impair proliferation in the SVZ. Data are presented as mean ± SEM. Scale bars, 100 μm. *p < 0.05.
Discussion
In this study we used reporter gene-assisted cell sorting of NSCs and immature neurons isolated from the DG, a neurogenic zone in adult mouse brain, and subsequently performed genome-wide transcriptome analysis of the different cell populations. We used the cell type-specific markers SOX2+ and DCX+ to discern between NSCs and their neuronal progeny, respectively. With this approach, we identified IGF2 as a novel niche-specific regulator of neurogenesis that controls the proliferation of NSCs in the adult DG.
In addition, we identified a large number of genes that were previously implicated in stem cell maintenance, proliferative activity, or neuronal differentiation [for example: Sox9, Cheng et al., 2009; reversion-inducing-cysteine-rich protein with kazal motifs (Reck), Muraguchi et al., 2007; TAG-1, Furley et al., 1990; Smad interacting protein 1 (Sip1), Seuntjens et al., 2009].
Although these findings validate our strategy of combining reporter-based cell isolation with microarray analysis, this approach is limited by the cell-type specificity of the markers we used. Although SOX2 expression is a hallmark of NSCs, a substantial fraction of classical astrocytes is also positive for SOX2 (Couillard-Despres et al., 2006; Suh et al., 2007). In fact, we found that ∼30% of all SOX2GFP-expressing cells in the DG colabeled with S100β, which is not expressed in NSCs (data not shown). Furthermore, DCX marks maturing cells ranging from neuroblasts to rather mature neurons of ∼3 weeks of age (Couillard-Despres et al., 2006; Suh et al., 2007); therefore, DCX+ cells also represent a somewhat heterogeneous population. These limitations may have diminished or even masked some differences in gene expression that might have been found with access to perfectly homogeneous cell populations. Despite these limitations, we believe that our data represent a useful resource for identifying and functionally testing genes in novel pathways involved in neurogenesis. For example, we also found that EfnB3, one of the genes identified to be upregulated in DCX+ cells of the DG, is functionally involved in the survival of newborn hippocampal neurons (data not shown).
Gene expression analyses identify IGF2 as a regulator of DG neurogenesis
One of the genes we found to be highly expressed in the stem cells of the DG was Igf2. IGF signaling has been shown to be involved in organ growth during embryonic development (DeChiara et al., 1990; Sim et al., 2006; Ye and D'Ercole, 2006), and recent data suggest that IGF1 is critically involved in brain development and NSC behavior (for review, see D'Ercole and Ye, 2008). Furthermore, IGF2 acts in concert with Shh signaling to regulate proliferation of cerebellar progenitors (Fernandez et al., 2010). IGF2 appears to also be important for the proliferation of medulloblastoma cells (Corcoran et al., 2008).
Our validation experiments confirmed high levels of Igf2 mRNA and protein in the DG, and we found that proliferation of DG-derived NSCs is regulated by IGF2 via IGF1R and AKT-dependent signaling. Notably, our data indicate that IGF2 is mainly produced by SOX2+ NSCs and largely acts in an autocrine manner. This mode of IGF2 signaling seems to be different from the second neurogenic area of the postnatal brain, where it has been recently shown that Igf2 is expressed in the choroid plexus of the lateral ventricles and that IGF2 appears to modulate embryonic neurogenesis by being diffused through the CSF and by signaling through the IGF1R on SVZ NSCs (Lehtinen et al., 2011). Thus, it appears that the same signaling factor, IGF2, although derived from different sources (choroid plexus in the SVZ, SOX2+ NSCs in the DG), has similar effects on NSC proliferation upon activation of IGF1R. This hypothesis is also supported by our findings that FACS-based isolation of SOX2+ cells from the SVZ showed that SVZ-derived NSCs expressed very low levels of IGF2 compared with SOX2+ cells in the DG. Furthermore, knockdown of IGF2 in vitro or within the adult SVZ did not affect NSC proliferation.
Gene expression analyses followed by pathway mapping and our experimental data suggest that IGF2 acts via AKT signaling to regulate proliferation of DG-derived NSCs. AKT appears to be critically involved in cell survival and anti-apoptotic signaling throughout the brain (Brazil et al., 2004). In addition, AKT has previously been implicated in precursor proliferation of adult NSCs (Peltier et al., 2007). The molecular components downstream of AKT activation that mediate NSC proliferation in the context of IGF2 signaling remain unclear. However, it seems reasonable to speculate that AKT may serve as a hub to integrate pro-proliferative signals (such as IGF2) with pathways that induce quiescence, such as BMP or PTEN signaling (Groszer et al., 2001; Mathieu et al., 2008). Further studies are required to understand the interplay between these signaling cascades in regulating the proliferative behavior of hippocampal NSCs.
Harnessing endogenous NSCs for brain repair
Surveying gene expression profiles of NSCs of the adult brain and their progeny is an important step in understanding the basic biology of adult neurogenesis. Notably, ongoing neurogenesis in the adult brain has been implicated in a variety of diseases, and the continuous generation of new neurons appears to play important roles for brain tissue maintenance and brain function (Scharfman and Hen, 2007; Kempermann et al., 2008; Zhao et al., 2008). A thorough understanding of the molecular mechanisms underlying the neurogenic process in the mature brain is therefore an important prerequisite to devising strategies aimed at targeting NSCs for brain repair. Thus, the identification of mechanisms regulating the activity of NSCs in the adult brain might prove a critical step for targeted manipulation of NSC populations in certain diseases. By providing the molecular framework for a deeper understanding of neurogenesis in the adult brain, the data provided in this work will bring us closer to using adult NSCs for endogenous brain repair.
Notes
Supplemental Tables 1 and 2 for this article are available at www.files.ethz.ch/cb/tables.xls. This material has not been peer reviewed.
Footnotes
This study was supported by the National Centres of Competence in Research–Neural Plasticity and Repair, Swiss National Science Foundation, TH Grant ETH-01 08–1, Zurich Neuroscience Center, Novartis Foundation, and Theodore Ott Foundation (to S.J.), and by grants from the U.S. National Institute of Neurological Disorders and Stroke, the Lookout Fund, the Christopher and Dana Reeve Foundation, the Picower Foundation, and James S. McDonnell Foundation (to F.H.G.). O.B. was supported by a German Academic Exchange Service student exchange fellowship. S.A. was a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-1859–05). M.K. was supported by the Janggen-Pöhn Foundation. We thank Mary Lynn Gage for editorial comments, Ueli Suter for conceptual input, members of the Jessberger and Gage laboratories for helpful discussions, David Chambers and Jonna Barrie for essential help with the FACS experiments, the Swiss Federal Institute of Technology Zurich Light Microscopy Center for help with imaging, and Stefan Zoller (Functional Genomics Center Zurich) for bioinformatic support.
References
- Agis-Balboa RC, Arcos-Diaz D, Wittnam J, Govindarajan N, Blom K, Burkhardt S, Haladyniak U, Agbemenyah HY, Zovoilis A, Salinas-Riester G, Opitz L, Sananbenesi F, Fischer A. A hippocampal insulin-growth factor 2 pathway regulates the extinction of fear memories. EMBO J. 2011;30:4071–4083. doi: 10.1038/emboj.2011.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu H, Cheung G, Kettenmann H, Palmer TD, Kempermann G. Enriched monolayer precursor cell cultures from micro-dissected adult mouse dentate gyrus yield functional granule cell-like neurons. PLoS ONE. 2007;2:e388. doi: 10.1371/journal.pone.0000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth R, Tripathi P, Ninkovic J, Bayam E, Lepier A, Stempfhuber B, Kirchhoff F, Hirrlinger J, Haslinger A, Lie DC, Beckers J, Yoder B, Irmler M, Götz M. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell [Erratum (2011) 8: 119] 2010;7:744–758. doi: 10.1016/j.stem.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Brazil DP, Yang ZZ, Hemmings BA. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem Sci. 2004;29:233–242. doi: 10.1016/j.tibs.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Breunig JJ, Silbereis J, Vaccarino FM, Sestan N, Rakic P. Notch regulates cell fate and dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proc Natl Acad Sci U S A. 2007;104:20558–20563. doi: 10.1073/pnas.0710156104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton A, Petreanu LT, Lansford R, Alvarez-Buylla A, Lledo PM. Becoming a new neuron in the adult olfactory bulb. Nat Neurosci. 2003;6:507–518. doi: 10.1038/nn1048. [DOI] [PubMed] [Google Scholar]
- Chen DY, Stern SA, Garcia-Osta A, Saunier-Rebori B, Pollonini G, Bambah-Mukku D, Blitzer RD, Alberini CM. A critical role for IGF-II in memory consolidation and enhancement. Nature. 2011;469:491–497. doi: 10.1038/nature09667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Bachar Raveh T, Barakat MT, Lee EY, Scott MP. Insulin-like growth factor 2 is required for progression to advanced medulloblastoma in patched1 heterozygous mice. Cancer Res. 2008;68:8788–8795. doi: 10.1158/0008-5472.CAN-08-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillard-Despres S, Winner B, Schaubeck S, Aigner R, Vroemen M, Weidner N, Bogdahn U, Winkler J, Kuhn HG, Aigner L. Doublecortin expression levels in adult brain reflect neurogenesis. Eur J Neurosci. 2005;21:1–14. doi: 10.1111/j.1460-9568.2004.03813.x. [DOI] [PubMed] [Google Scholar]
- Couillard-Despres S, Winner B, Karl C, Lindemann G, Schmid P, Aigner R, Laemke J, Bogdahn U, Winkler J, Bischofberger J, Aigner L. Targeted transgene expression in neuronal precursors: watching young neurons in the old brain. Eur J Neurosci. 2006;24:1535–1545. doi: 10.1111/j.1460-9568.2006.05039.x. [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Efstratiadis A, Robertson EJ. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature. 1990;345:78–80. doi: 10.1038/345078a0. [DOI] [PubMed] [Google Scholar]
- D'Ercole AJ, Ye P. Expanding the mind: insulin-like growth factor I and brain development. Endocrinology. 2008;149:5958–5962. doi: 10.1210/en.2008-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Fernandez C, Tatard VM, Bertrand N, Dahmane N. Differential modulation of Sonic-hedgehog-induced cerebellar granule cell precursor proliferation by the IGF signaling network. Dev Neurosci. 2010;32:59–70. doi: 10.1159/000274458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furley AJ, Morton SB, Manalo D, Karagogeos D, Dodd J, Jessell TM. The axonal glycoprotein TAG-1 is an immunoglobulin superfamily member with neurite outgrowth-promoting activity. Cell. 1990;61:157–170. doi: 10.1016/0092-8674(90)90223-2. [DOI] [PubMed] [Google Scholar]
- Gao Z, Ure K, Ables JL, Lagace DC, Nave KA, Goebbels S, Eisch AJ, Hsieh J. Neurod1 is essential for the survival and maturation of adult-born neurons. Nat Neurosci. 2009;12:1090–1092. doi: 10.1038/nn.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Goh EL, Sailor KA, Kitabatake Y, Ming GL, Song H. GABA regulates synaptic integration of newly generated neurons in the adult brain. Nature. 2006;439:589–593. doi: 10.1038/nature04404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- Hartmann W, Koch A, Brune H, Waha A, Schüller U, Dani I, Denkhaus D, Langmann W, Bode U, Wiestler OD, Schilling K, Pietsch T. Insulin-like growth factor II is involved in the proliferation control of medulloblastoma and its cerebellar precursor cells. Am J Pathol. 2005;166:1153–1162. doi: 10.1016/S0002-9440(10)62335-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger S, Kempermann G. Adult-born hippocampal neurons mature into activity-dependent responsiveness. Eur J Neurosci. 2003;18:2707–2712. doi: 10.1111/j.1460-9568.2003.02986.x. [DOI] [PubMed] [Google Scholar]
- Jessberger S, Aigner S, Clemenson GD, Jr, Toni N, Lie DC, Karalay O, Overall R, Kempermann G, Gage FH. Cdk5 Regulates Accurate Maturation of Newborn Granule Cells in the Adult Hippocampus. PLoS Biol. 2008;6:e272. doi: 10.1371/journal.pbio.0060272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karalay O, Doberauer K, Vadodaria KC, Knobloch M, Berti L, Miquelajauregui A, Schwark M, Jagasia R, Taketo MM, Tarabykin V, Lie DC, Jessberger S. Prospero-related homeobox 1 gene (Prox1) is regulated by canonical Wnt signaling and has a stage-specific role in adult hippocampal neurogenesis. Proc Natl Acad Sci U S A. 2011;108:5807–5812. doi: 10.1073/pnas.1013456108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G, Jessberger S, Steiner B, Kronenberg G. Milestones of neuronal development in the adult hippocampus. Trends Neurosci. 2004;27:447–452. doi: 10.1016/j.tins.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Kempermann G, Krebs J, Fabel K. The contribution of failing adult hippocampal neurogenesis to psychiatric disorders. Curr Opin Psychiatry. 2008;21:290–295. doi: 10.1097/YCO.0b013e3282fad375. [DOI] [PubMed] [Google Scholar]
- Kronenberg G, Reuter K, Steiner B, Brandt MD, Jessberger S, Yamaguchi M, Kempermann G. Subpopulations of proliferating cells of the adult hippocampus respond differently to physiologic neurogenic stimuli. J Comp Neurol. 2003;467:455–463. doi: 10.1002/cne.10945. [DOI] [PubMed] [Google Scholar]
- Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J Neurosci. 1996;16:2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K, Kaspar BK, Gage FH, Schaffer DV. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat Neurosci. 2003;6:21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
- Lavado A, Lagutin OV, Chow LM, Baker SJ, Oliver G. Prox1 is required for granule cell maturation and intermediate progenitor maintenance during brain neurogenesis. PLoS Biol. 2010;8:e1000460. doi: 10.1371/journal.pbio.1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, D'Ercole AJ, Wong ET, LaMantia AS, Walsh CA. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie DC, Song H, Colamarino SA, Ming GL, Gage FH. Neurogenesis in the adult brain: new strategies for central nervous system diseases. Annu Rev Pharmacol Toxicol. 2004;44:399–421. doi: 10.1146/annurev.pharmtox.44.101802.121631. [DOI] [PubMed] [Google Scholar]
- Lie DC, Colamarino SA, Song HJ, Désiré L, Mira H, Consiglio A, Lein ES, Jessberger S, Lansford H, Dearie AR, Gage FH. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science. 1994;264:1145–1148. doi: 10.1126/science.8178174. [DOI] [PubMed] [Google Scholar]
- Lugert S, Basak O, Knuckles P, Haussler U, Fabel K, Götz M, Haas CA, Kempermann G, Taylor V, Giachino C. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell. 2010;6:445–456. doi: 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Mathieu C, Sii-Felice K, Fouchet P, Etienne O, Haton C, Mabondzo A, Boussin FD, Mouthon MA. Endothelial cell-derived bone morphogenetic proteins control proliferation of neural stem/progenitor cells. Mol Cell Neurosci. 2008;38:569–577. doi: 10.1016/j.mcn.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Mira H, Andreu Z, Suh H, Lie DC, Jessberger S, Consiglio A, San Emeterio J, Hortigüela R, Marqués-Torrejón MA, Nakashima K, Colak D, Götz M, Fariñas I, Gage FH. Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell. 2010;7:78–89. doi: 10.1016/j.stem.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Muraguchi T, Takegami Y, Ohtsuka T, Kitajima S, Chandana EP, Omura A, Miki T, Takahashi R, Matsumoto N, Ludwig A, Noda M, Takahashi C. RECK modulates Notch signaling during cortical neurogenesis by regulating ADAM10 activity. Nat Neurosci. 2007;10:838–845. doi: 10.1038/nn1922. [DOI] [PubMed] [Google Scholar]
- Palmer TD, Ray J, Gage FH. FGF-2-responsive neuronal progenitors reside in proliferative and quiescent regions of the adult rodent brain. Mol Cell Neurosci. 1995;6:474–486. doi: 10.1006/mcne.1995.1035. [DOI] [PubMed] [Google Scholar]
- Peltier J, O'Neill A, Schaffer DV. PI3K/Akt and CREB regulate adult neural hippocampal progenitor proliferation and differentiation. Dev Neurobiol. 2007;67:1348–1361. doi: 10.1002/dneu.20506. [DOI] [PubMed] [Google Scholar]
- Pennartz S, Belvindrah R, Tomiuk S, Zimmer C, Hofmann K, Conradt M, Bosio A, Cremer H. Purification of neuronal precursors from the adult mouse brain: comprehensive gene expression analysis provides new insights into the control of cell migration, differentiation, and homeostasis. Mol Cell Neurosci. 2004;25:692–706. doi: 10.1016/j.mcn.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Peschanski M, Hirsch E, Dusart I, Doye V, Marty S, Manceau V, Sobel A. Stathmin: cellular localization of a major phosphoprotein in the adult rat and human CNS. J Comp Neurol. 1993;337:655–668. doi: 10.1002/cne.903370410. [DOI] [PubMed] [Google Scholar]
- Pietrzkowski Z, Wernicke D, Porcu P, Jameson BA, Baserga R. Inhibition of cellular proliferation by peptide analogues of insulin-like growth factor 1. Cancer Res. 1992;52:6447–6451. [PubMed] [Google Scholar]
- Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8:915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, Trajanoski Z. CARMAweb: comprehensive R- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 2006;34:W498–W503. doi: 10.1093/nar/gkl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray J, Gage FH. Differential properties of adult rat and mouse brain-derived neural stem/progenitor cells. Mol Cell Neurosci. 2006;31:560–573. doi: 10.1016/j.mcn.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Scharfman HE, Hen R. Neuroscience. Is more neurogenesis always better? Science. 2007;315:336–338. doi: 10.1126/science.1138711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaberg RM, van der Kooy D. Adult rodent neurogenic regions: the ventricular subependyma contains neural stem cells, but the dentate gyrus contains restricted progenitors. J Neurosci. 2002;22:1784–1793. doi: 10.1523/JNEUROSCI.22-05-01784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seri B, García-Verdugo JM, McEwen BS, Alvarez-Buylla A. Astrocytes give rise to new neurons in the adult mammalian hippocampus. J Neurosci. 2001;21:7153–7160. doi: 10.1523/JNEUROSCI.21-18-07153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, Nave KA, Huylebroeck D, Tarabykin V. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat Neurosci. 2009;12:1373–1380. doi: 10.1038/nn.2409. [DOI] [PubMed] [Google Scholar]
- Shi Y, Chichung Lie D, Taupin P, Nakashima K, Ray J, Yu RT, Gage FH, Evans RM. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427:78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- Sim FJ, Keyoung HM, Goldman JE, Kim DK, Jung HW, Roy NS, Goldman SA. Neurocytoma is a tumor of adult neuronal progenitor cells. J Neurosci. 2006;26:12544–12555. doi: 10.1523/JNEUROSCI.0829-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H, Consiglio A, Ray J, Sawai T, D'Amour KA, Gage FH. In vivo fate analysis reveals the multipotent and self-renewal capacities of SOX2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H, Deng W, Gage FH. Signaling in adult neurogenesis. Annu Rev Cell Dev Biol. 2009;25:253–275. doi: 10.1146/annurev.cellbio.042308.113256. [DOI] [PubMed] [Google Scholar]
- Sun XJ, Rothenberg P, Kahn CR, Backer JM, Araki E, Wilden PA, Cahill DA, Goldstein BJ, White MF. Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature. 1991;352:73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Sandler VM, Toni N, Zhao C, Gage FH. NMDA-receptor-mediated, cell-specific integration of new neurons in adult dentate gyrus. Nature. 2006;442:929–933. doi: 10.1038/nature05028. [DOI] [PubMed] [Google Scholar]
- Toretsky JA, Helman LJ. Involvement of IGF-II in human cancer. J Endocrinol. 1996;149:367–372. doi: 10.1677/joe.0.1490367. [DOI] [PubMed] [Google Scholar]
- van Praag H, Schinder AF, Christie BR, Toni N, Palmer TD, Gage FH. Functional neurogenesis in the adult hippocampus. Nature. 2002;415:1030–1034. doi: 10.1038/4151030a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker TL, White A, Black DM, Wallace RH, Sah P, Bartlett PF. Latent stem and progenitor cells in the hippocampus are activated by neural excitation. J Neurosci. 2008;28:5240–5247. doi: 10.1523/JNEUROSCI.0344-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG. In situ hybridization: a practical approach. Oxford: Oxford UP; 1998. [Google Scholar]
- Yamaguchi M, Saito H, Suzuki M, Mori K. Visualization of neurogenesis in the central nervous system using nestin promoter-GFP transgenic mice. NeuroReport. 2000;11:1991–1996. doi: 10.1097/00001756-200006260-00037. [DOI] [PubMed] [Google Scholar]
- Ye P, D'Ercole AJ. Insulin-like growth factor actions during development of neural stem cells and progenitors in the central nervous system. J Neurosci Res. 2006;83:1–6. doi: 10.1002/jnr.20688. [DOI] [PubMed] [Google Scholar]
- Zhang J, Moats-Staats BM, Ye P, D'Ercole AJ. Expression of insulin-like growth factor system genes during the early postnatal neurogenesis in the mouse hippocampus. J Neurosci Res. 2007;85:1618–1627. doi: 10.1002/jnr.21289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Zhu CQ, Popova SN, Brown ER, Barsyte-Lovejoy D, Navab R, Shih W, Li M, Lu M, Jurisica I, Penn LZ, Gullberg D, Tsao MS. Integrin alpha 11 regulates IGF2 expression in fibroblasts to enhance tumorigenicity of human non-small-cell lung cancer cells. Proc Natl Acad Sci U S A. 2007;104:11754–11759. doi: 10.1073/pnas.0703040104. [DOI] [PMC free article] [PubMed] [Google Scholar]