Abstract
QRS interval on the electrocardiogram reflects ventricular depolarization and conduction time, and is a risk factor for mortality, sudden death, and heart failure. We performed a genome-wide association meta-analysis in 40,407 European-descent individuals from 14 studies, with further genotyping in 7170 additional Europeans, and identified 22 loci associated with QRS duration (P < 5 × 10−8). These loci map in or near genes in pathways with established roles in ventricular conduction such as sodium channels, transcription factors, and calcium-handling proteins, but also point to novel biologic processes, such as kinase inhibitors and genes related to tumorigenesis. We demonstrate that SCN10A, a gene at our most significant locus, is expressed in the mouse ventricular conduction system, and treatment with a selective SCN10A blocker prolongs QRS duration. These findings extend our current knowledge of ventricular depolarization and conduction.
Search Terms: QRS interval, ECG, quantitative trait, genome-wide association study
The electrocardiographic QRS interval reflects ventricular depolarization and its duration is a function of electrophysiological properties within the His-Purkinje system and the ventricular myocardium. A diseased ventricular conduction system can lead to life-threatening bradyarrhythmias, such as heart block, and tachyarrhythmias, such as ventricular fibrillation. Longer QRS duration is a predictor of mortality and sudden death in the general population and in cohorts with hypertension and coronary artery disease.1–3 In a population-based study, prolonged baseline QRS was associated with incident heart failure.4
Twin and family studies suggest a genetic contribution to QRS duration, with heritability estimates of up to 40%.5, 6 Prior candidate gene and smaller genome-wide studies identified a limited number of loci associated with QRS duration, supporting the hypothesis of the contribution of common genetic variation to QRS duration.7–9 To identify additional loci and highlight physiologic processes associated with ventricular conduction, we performed a meta-analysis of 14 genome-wide association studies (GWAS) of QRS duration in a total of 40,407 individuals of European descent, where we adjusted the analyses for age, sex, height, and body mass index after appropriate sample exclusions (Methods). After an initial discovery phase, we further genotyped selected variants representing nine loci with P-values ranging from 1 × 10−6 to 5 × 10−9 in an additional cohort of 7170 European individuals.
Results
Meta-analysis of genome-wide association results
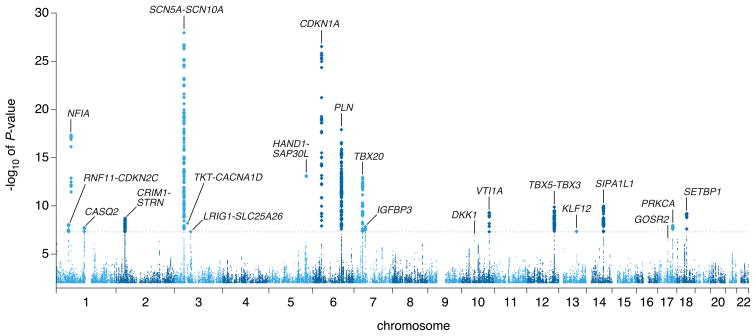
We conducted meta-analyses for approximately 2.5 million single nucleotide polymorphisms (SNPs) in 40,407 individuals of European ancestry from 14 GWAS (Supplementary Tables 1a and 1b). Overall, 612 variants in 20 loci exceeded our genome-wide significance P-value threshold of 5 × 10−8 after adjusting for modest genomic inflation (λGC = 1.059) (Figure 1 and Supplementary Figure 1). The loci associated with QRS interval duration are detailed in Table 1 and Supplementary Figure 2, with the index SNP (representing the most significant association) labeled for each independent signal.
Figure 1. Manhattan plot.
Manhattan plot showing the association of SNPs with QRS interval duration in a GWAS of 40,407 individuals. The dashed horizontal line marks the threshold for genome-wide significance (P = 5 × 10−8). Twenty loci (labeled) reached genome-wide significance. Two additional loci, GOSR2 and DKK1, reached significance after genotyping of select SNPs in an additional sample of 7170 individuals (see Results).
Table 1.
Significant loci at P<5×10−08 in combined GWAS and candidate SNP meta-analysis
| Locus | Chr | Index SNP | Coded/Non-coded Allele | AF | GWAS β | GWAS SEGC | GWAS PGC | I2 | Prevend β | Prevend P | POverall | Multi SNP β | Multi SNP P | Nearest Gene | SNP Annotation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | rs6801957 | T/C | 0.41 | 0.77 | 0.07 | 1.10×10−28 | 45.3 | - | - | 1.10×10−28 | 0.54 | 3.43×10−14 | SCN10A | intron |
| 3 | rs9851724 | C/T | 0.33 | −0.66 | 0.07 | 1.91×10−20 | 57.1 | - | - | 1.91×10−20 | −0.60 | 5.78×10−16 | SCN10A/SCN5A | intergenic | |
| 3 | rs10865879 | T/C | 0.41 | 0.77 | 0.07 | 1.10×10−28 | 53.6 | - | - | 1.10×10−28 | 0.33 | 1.67×10−04 | SCN5A/EXOG | intergenic | |
| 3 | rs11710077 | T/A | 0.21 | −0.84 | 0.09 | 5.74×10−22 | 23.8 | - | - | 5.74×10−22 | −0.44 | 1.33×10−06 | SCN5A | intron | |
| 3 | rs11708996 | C/G | 0.16 | 0.79 | 0.10 | 1.26×10−16 | 0.0 | - | - | 1.26×10−16 | 0.47 | 7.23×10−06 | SCN5A | intron | |
| 3 | rs2051211 | G/A | 0.26 | −0.44 | 0.08 | 1.57×10−08 | 0.0 | - | - | 1.57×10−08 | −0.18 | 3.71×10−02 | EXOG | intron | |
| 2 | 6 | rs9470361 | A/G | 0.25 | 0.87 | 0.08 | 3.00×10−27 | 14.6 | - | - | 3.00×10−27 | - | - | CDKN1A | intergenic |
| 3 | 6 | rs11153730 | C/T | 0.49 | 0.59 | 0.07 | 1.26×10−18 | 5.3 | - | - | 1.26×10−18 | - | - | C6orf204/SLC35F1/PLN/BRD7P3 | intergenic |
| 4 | 1 | rs9436640 | G/T | 0.46 | −0.59 | 0.07 | 4.57×10−18 | 51.2 | - | - | 4.57×10−18 | - | - | NFIA | intron |
| 5 | 5 | rs13165478 | A/G | 0.36 | −0.55 | 0.07 | 7.36×10−14 | 64.6 | - | - | 7.36×10−14 | - | - | HAND1/SAP30L | intergenic |
| 6 | 7 | rs1362212 | A/G | 0.18 | 0.69 | 0.09 | 1.12×10−13 | 0.0 | - | - | 1.12×10−13 | - | - | TBX20 | intergenic |
| 7 | 14 | rs11848785 | G/A | 0.27 | −0.50 | 0.08 | 1.04×10−10 | 0.0 | - | - | 1.04×10−10 | - | - | SIPA1L1 | intron |
| 8 | 12 | rs883079 | C/T | 0.29 | 0.49 | 0.08 | 1.33×10−10 | 8.3 | - | - | 1.33×10−10 | - | - | TBX5 | 3′-UTR |
| 9 | 12 | rs10850409 | A/G | 0.27 | −0.49 | 0.08 | 3.06×10−10 | 0.0 | - | - | 3.06×10−10 | - | - | TBX3 | intergenic |
| 10 | 10 | rs7342028 | T/G | 0.27 | 0.48 | 0.08 | 4.95×10−10 | 0.0 | - | - | 4.95×10−10 | - | - | VTI1A | intron |
| 11 | 18 | rs991014 | T/C | 0.42 | 0.42 | 0.07 | 6.20×10−10 | 0.0 | - | - | 6.20×10−10 | - | - | SETBP1 | intron |
| 12 | 2 | rs17020136 | C/T | 0.21 | 0.51 | 0.08 | 1.90×10−9 | 0.0 | - | - | 1.90×10−9 | - | - | HEATR5B/STRN | intron |
| 13 | 3 | rs4687718 | A/G | 0.14 | −0.63 | 0.11 | 6.25×10−9 | 0.0 | - | - | 6.25×10−9 | - | - | TKT/PRKCD/CACNA1D | intron |
| 14 | 2 | rs7562790 | G/T | 0.40 | 0.39 | 0.07 | 8.22×10−9 | 0.0 | - | - | 8.22×10−9 | - | - | CRIM1 | intron |
| 15 | 1 | rs17391905 | G/T | 0.05 | −1.35 | 0.23 | 8.72×10−9 | 4.0 | -1.17 | 0.005 | 3.26×10−10 | - | - | C1orf185/RNF11/CDKN2C/FAF1 | intergenic |
| 16 | 17 | rs9912468 | G/C | 0.43 | 0.39 | 0.07 | 1.06×10−8 | 28.2 | - | - | 1.06×10−8 | - | - | PRKCA | intron |
| 17 | 7 | rs7784776 | G/A | 0.43 | 0.39 | 0.07 | 1.42×10−8 | 0.0 | 0.36 | 0.015 | 1.28×10−9 | - | - | IGFBP3 | intergenic |
| 18 | 1 | rs4074536 | C/T | 0.29 | −0.42 | 0.07 | 2.36×10−8 | 0.5 | - | - | 2.36×10−8 | - | - | CASQ2 | missense |
| 19 | 13 | rs1886512 | A/T | 0.37 | −0.40 | 0.07 | 4.31×10−8 | 0.0 | -0.28 | 0.047 | 1.27×10−8 | - | - | KLF12 | intron |
| 20 | 3 | rs2242285 | A/G | 0.42 | 0.37 | 0.07 | 4.79×10−8 | 35.4 | 0.29 | 0.040 | 1.09×10−8 | - | - | LRIG1/SLC25A26 | intron |
| 21 | 10 | rs1733724 | A/G | 0.25 | 0.49 | 0.09 | 1.26×10−7 | 0.0 | 0.34 | 0.035 | 3.05×10−8 | - | - | DKK1 | intergenic |
| 22 | 17 | rs17608766 | C/T | 0.16 | 0.53 | 0.10 | 3.71×10−7 | 13.8 | 0.92 | 4.7×10−5 | 4.75×10−10 | - | - | GOSR2 | intron, 3′ |
In each locus at least one marker exceeds the genome-wide significance threshold of P < 5 × 10−8. At locus 1, six signals were identified (r2<0.05) that exceeded genome-wide threshold. In a multiSNP model that included all 6 SNPs, there was evidence that at least 4 of these SNPs were independently associated with QRS duration. The bolded allele is the coded allele. Beta values (β) estimate the difference in QRS interval in milliseconds per copy of the coded allele, adjusted for the covariates in the model. Chr, chromosome; AF, coded allele frequency; SE, standard error; GC, genomic control adjusted; UTR, untranslated region. AF is an average weighted by study size.
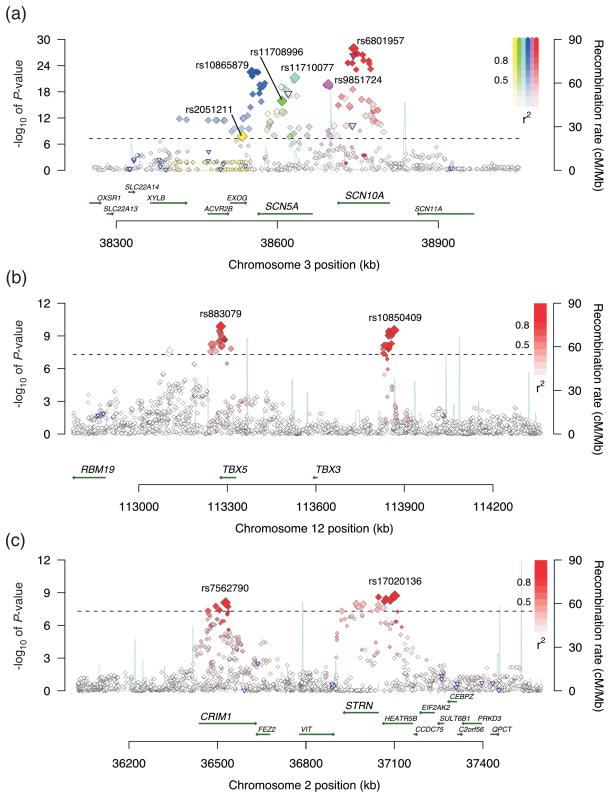
Across the genome, the most significant association for QRS interval duration (locus 1) was on chromosome 3p22 (Figure 2a), where we identified six potentially independent association signals based on the linkage disequilibrium (LD) patterns in HapMap-CEU (pairwise r2 among index SNPs < 0.05). In conditional analyses where all six SNPs were included in the same regression model, there was compelling evidence that at least four SNPs from this region were independently associated with QRS duration (Table 1). Two of these associations were in or near SCN10A, a voltage-gated sodium channel gene. Variation at this locus was recently associated with QRS duration in two GWAS. The top SNP identified in those two studies, rs6795970, was in strong LD with our top signal, rs6801975 (r2=0.93).8, 9 Two additional signals were identified in SCN5A, a sodium channel gene adjacent to SCN10A (Table 1).
Figure 2. Association plots for select loci.
Each SNP is plotted with respect to its chromosomal location (x-axis) and its P-value (y-axis on the left). The tall blue spikes indicate the recombination rate (y-axis on the right) at that region of the chromosome. The blue-outlined triangles indicate coding region SNPs. (a) Locus 1 (SCN5A/SCN10A) on chromosome 3: The six index signals are named with their rs numbers and highlighted in different colors (yellow, green, teal, blue, purple, and red). Other SNPs in linkage disequilibrium with the index SNP are denoted in the same color. Color saturation indicates the degree of correlation with the index SNP. (b) Locus 8 (TBX5) and locus 9 (TBX3) on chromosome 12. (c) Locus 12 (HEATR5B/STRN) and locus 14 (CRIM1) on chromosome 2.
The second most significant locus (locus 2) was on chromosome 6p21 near CDKN1A, a cyclin dependent kinase inhibitor. The CDKN1A locus was recently associated with QRS interval duration in an Icelandic population.9 The index SNP in the prior report, rs1321311, was in strong LD with our top signal, rs9470361 (r2=0.88). Another cyclin dependent kinase inhibitor (CDKN2C) was located in locus 15, which encompasses several other genes including C1orf185, RNF11, and FAF1.
Locus 3 on chromosome 6q22 contains the PLN/SLC35F1/C6orf204/BRD7P3 cluster of genes. PLN encodes phospholamban, a key regulator of sarcoplasmic reticulum calcium reuptake. Significant associations were found in several other regions harboring calcium-handling genes, including locus 12 (STRN/HEATR5B), locus 16 (PRKCA), and locus 18 (CASQ2).
Locus 4 mapped to an intronic SNP in NFIA, a transcription factor. Several other significant loci also mapped in or near transcription factors including locus 5 (HAND1), locus 6 (TBX20), locus 8 (TBX5), locus 9 (TBX3), and locus 19 (KLF12). Common variation in TBX5 was recently associated with QRS duration.9 The index signal in the prior report, rs3825214, was in moderate LD with our top signal, rs883079 (r2=0.67).
Additional regions identified include locus 7 (SIPA1L1), locus 10 (VTI1A), locus 11 (SETBP1), locus 13 (TKT/CACNA1D/PRKCD), locus 14 (CRIM1), locus 17 (nearest gene, IGFBP3, is 660kb away), and locus 20 (LRIG1).
Collectively, the identified index SNPs across these 20 loci explained approximately 5.7% (±2.3%) of the observed variance in QRS duration, consistent with a polygenic model in which each of the discovered variants exerts only a modest effect on QRS interval. None of these index SNPs showed a significant interaction with sex or age after Bonferroni correction (Supplementary Table 2). We observed moderate levels of heterogeneity of the effect (25 < I2 < 75) for several index SNPs (Table 1). However, only HAND1/SAP30L showed significant evidence of heterogeneity using Cochran’s Q test corrected for 23 independent genome-wide variants (Cochran’s P = 0.005).
Extension of findings in an additional 7170 individuals
Based on the discovery meta-analysis, we selected the index SNPs at four loci (loci 15, 17, 19, and 20) with P-values ranging between 5 × 10−8 and 5 × 10−9 and from all five loci with P-values ranging from 1 × 10−6 to 5 × 10−8 (Methods) for genotyping in an additional 7170 European individuals in order to boost power. In a joint analysis combining all 47,577 individuals, the significance for the four loci with P-values between 5 × 10−8 and 5 ×10−9 increased, indicating these represent true positive associations (Table 1). The joint analysis also provided further evidence for two other loci (locus 21 near DKK1, and locus 22 tagged by an intronic SNP in GOSR2) that reached genome-wide significance, bringing the total number of significant loci to 22 with 25 independently associated index SNPs (Table 1). The index SNP (rs1733724) in DKK1 was previously associated with QRS duration in an Icelandic population.9
Association with conduction defect
Based on this series of QRS associations, we sought to test the hypothesis that QRS prolonging alleles, on average, increase risk of ventricular conduction defects. To address this question, we calculated a risk score in each individual by adding up the number of QRS prolonging alleles identified in this study, weighted by the observed effect sizes (β-estimates) from the final meta-analysis. In an independent set of 522 individuals from the ARIC and RS studies with bundle branch block or nonspecific prolongation of QRS interval (QRS>120 ms) compared with those with normal conduction (N = 12,804), each additional copy of a QRS prolonging allele was associated with a 8% increase in risk of ventricular conduction defect (P = 0.004). This result was largely driven by those with non-specific intraventricular conduction defects as opposed to those with left or right bundle branch block (Supplementary Tables 3a and 3b). Similar results were observed using an unweighted genotype risk score.
Putative functional variants
Of 612 genome-wide significant SNPs, one in SCN5A (rs1805124, H558R, P = 2.4×10−18), two in SCN10A (rs12632942, L1092P, P = 5.1×1011, and rs6795970, A1073V, P = 5×10−27), one in C6orf204 near PLN (rs3734381, S137G, P=1.1×10−10), and one in CASQ2 (index SNP rs4074536, T66A, P = 2.4×10−8) were nonsynonymous (Figure 2 and Supplementary Figure 2). The PolyPhen-2 program predicts all five of these variants to be benign, which is consistent with small-effect associations: each copy of the minor allele was associated with cross-sectional differences in QRS duration of less than 1ms.
The 25 index SNPs (from Table 1) were subsequently tested for association with gene cis-expression levels in 1,240 PAXgene whole blood samples10. Four cis-eQTLs were detected after stringent Bonferroni correction (Supplementary Figure 3). The most striking eQTLs were observed for probes in exonic regions of TKT (rs4687718, P = 5.87×10−70) and CDKN1A (rs9470361, P = 1.41×10−10) and an intronic probe for C6orf204 near PLN (rs11153730, P = 1.54×10−10). We additionally assessed cis-regulation for all HapMap SNPs for these three loci (± 250kb around the SNPs). The top eSNP for TKT (rs9821134) and C6orf204 (rs11970286) were in moderate to high LD (r2 = 0.47 and 0.91, respectively) with the top QRS signals at these loci. However, the top eSNP for CDKN1A, rs735013, was only weakly correlated with the QRS index SNP rs9470361 (r2 = 0.089). In conditional analysis that included both CDKN1A locus SNPs in the regression model, both rs735013 and rs9470361 remained independently associated with expression levels (P = 1.7 × 10−9 and 2.3 × 10−5, respectively). Additionally, rs735013 itself was marginally associated with QRS duration (coded allele frequency = 0.39; β = 0.33 ms (±0.07); P = 2.4 × 10–6). Whether these associations in whole blood samples will be similar to associations in cardiac myocytes and conduction tissue deserves further investigation.
Pleiotropic effects of variants associated with QRS duration and other ECG measurements
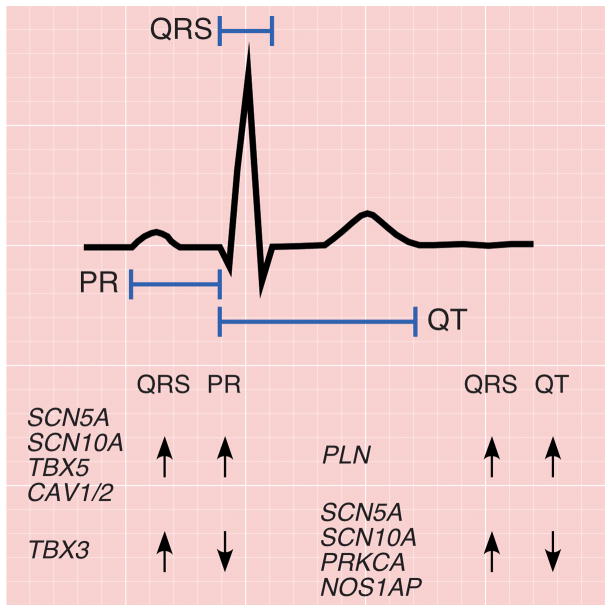
To explore the shared genetic underpinnings between atrial and ventricular depolarization and conduction (as measured by PR and QRS intervals) as well as ventricular depolarization and repolarization (QRS and QT intervals), we examined the effects of published PR and QT SNPs with respect to QRS interval. Several QRS loci were previously associated with PR or QT intervals, including PLN, TBX5/3, and SCN5A/10A, the last of which is associated with all three traits (Supplementary Table 4a). We also tested nine PR SNPs and 16 QT SNPs for their effect on QRS duration (Supplementary Table 4b).11–13 Our results suggest roles for CAV1/2 (rs3807989, P = 5.8 × 10−6) and NOS1AP (rs12143842, P = 1.3 × 10−4) in QRS duration. Indeed CAV1/2 was recently associated with QRS interval.9
QRS duration is positively correlated with both PR interval (r = 0.09) and QT interval (r = 0.44).9 To test if these relationships are also observed genetically, we compared the directionality of the association of SNPs at the published PR and QT loci with those for QRS duration. Generally, the effects of SNPs on PR interval were positively correlated with their effects on QRS duration (r = 0.53). With the exception of TBX3, the loci influencing both PR and QRS (SCN5A, SCN10A, TBX5, and CAV1/2), do so in a concordant fashion (i.e. variants that prolong PR also prolong QRS duration) (Figure 3 and Supplementary Tables 4a and 4b). By contrast, while QT and QRS are positively correlated at the population level, the effects of SNPs on QT interval were marginally negatively correlated with their effects on QRS (r = −0.08). Of the index SNPs at the four loci significantly associated with both QT and QRS (SCN5A/SCN10A, PRKCA, NOS1AP, PLN), only the PLN locus SNPs showed effects in the same direction (Figure 3 and Supplementary Tables 4a and 4b).
Figure 3. Pleiotropic associations of PR, QRS, and QT loci.
Electrocardiographic tracing delineating the PR, QRS, and QT intervals. PR and QRS intervals reflect myocardial depolarization and conduction time through the atria and down the atrioventricular node (PR) and throughout the ventricle (QRS), and are weakly positively correlated (r=0.09). The majority of loci that influence both PR and QRS (SCN5A, SCN10A, TBX5, CAV1/2), do so in a concordant fashion (i.e. variants that prolong PR also prolong QRS duration). The notable exception is a region on chromosome 12, where variants in the TBX5 locus have a concordant effect whereas those in nearby TBX3 have a discordant effect. By contrast, although QRS (ventricular depolarization) and QT (ventricular repolarization) are moderately positively correlated, the majority of loci (SCN5A, SCN10A, PRKCA, NOS1AP) that influence both phenotypes do so in a discordant fashion (i.e. variants that prolong QRS shorten the QT interval). The exception is the locus at PLN, where variants have a concordant effect.
Bioinformatic network analysis of QRS-associated loci
To examine the relationships between genetic loci associated with QRS duration, we developed an in silico relational network linking the loci based on published direct gene product interactions obtained from curated databases (Supplementary Figure 4).14 Most loci meeting genome-wide significance mapped to this network after a minimum number of “linker” nodes were incorporated to create a spanning network. This analysis provides a graphical overview of the interconnections among QRS-associated genetic loci and highlights both known and putative molecular mechanisms regulating ventricular conduction (see Discussion). Several of the “linker” nodes incorporated in the network, such as calmodulin, connexin 43 (GJA1), NEDD4, KCNMA1, and RYR2 are known modulators of cardiac electrical activity. Functional enrichment analysis of the QRS-associated network nodes (loci with P <5×10−8) using two independent software tools revealed that programs involved in heart development were highly over-represented (P-value range: 5.8×10−6 – 9.6×10−5).15, 16
Functional effects of the SCN10A locus in a mouse model of cardiac conduction
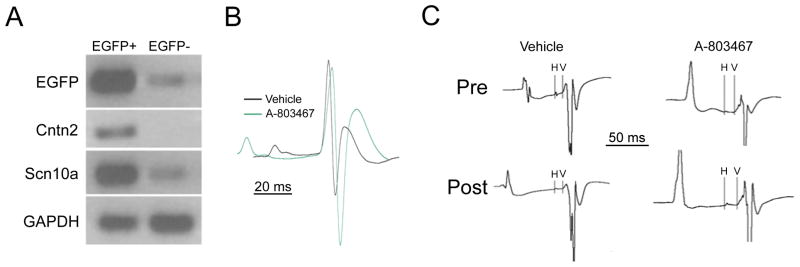
We undertook functional studies to determine whether our most significant locus was associated with ventricular conduction in mice. Transcriptional profiling suggests that Scn10a/Nav1.8 mRNA is expressed in ventricular myocardium and at higher levels in the specialized conduction system.17 These data were confirmed and extended by qPCR (Figure 4a), demonstrating a 25.7 ± 1.1 fold enrichment of Scn10a/Nav1.8 in Purkinje cells compared to working ventricular myocytes (n=3 for each cell type; p=0.002).
Figure 4. Expression and function of Scn10a in the murine heart.
Panel A. Neonatal ventricular myocytes from Cntn2-EGFP BAC transgenic mice were FACS sorted and EGFP+ and EGFP- pools were analyzed by RT-PCR. Transcripts encoding EGFP, Cntn2 and Scn10a were highly enriched in the EGFP+ fraction. Quantitative RT-PCR demonstrated 25.7 fold enrichment of Scn10a/Nav1.8. Panel B. Representative telemetric electrocardiographic recordings (lead II configuration) obtained 30 minutes after administration of vehicle alone (black tracing) or the Scn10a/Nav1.8 antagonist A-803467 (green tracing). The two tracings are aligned at the onset of the QRS wave and both PR interval and QRS interval prolongation were observed in drug-treated mice. Panel C. Representative intracardiac recordings showing HV intervals obtained prior to (Pre) and after (Post) administration of vehicle or A-803467. Significant HV prolongation was observed in drug-treated mice.
Telemetric electrocardiographic recordings (lead II position) were obtained in conscious mice treated with A-803467, a potent Scn10a/Nav1.8 antagonist, which blocks Nav1.8 100 times more potently than Nav1.5 with the doses used.18 These studies demonstrated a significant increase in QRS duration (11.6 ± 2.6 ms to 14.5 ± .54 ms; n = 7; P<0.001), whereas vehicle alone was without effect (11.4 ± .29 ms to 11.9 ± .42 ms; n = 7; P=NS). PR interval was also increased in drug-treated mice, from 31.4 ± .98 ms to 42.5 ± 3.3 ms; n=7; P< 0.01), whereas vehicle alone resulted in no significant change (32.6 ± 1.0 ms to 33.4 ± .69 ms; n=7; P=NS) (Figure 4b). To further delineate the site of ventricular conduction slowing, we performed intra-cardiac recordings from mice treated with A-803467. These studies confirmed the significant increase in QRS duration (from 12.26 ± 0.62 ms to 14.56 ± 0.58 ms; n=7; P =0.015), whereas vehicle alone was without significant effect (12.39 ± 0.52 ms to 13.65 ± 0.97 ms; n = 5, P = NS). A-803467 treatment resulted in a 35.7% ± 1.2% increase in HV interval (from 9.33 ± 0.74 ms to 12.67 ± 1.06 ms; P = .009), whereas vehicle alone was without significant effect (10.67 ± .83 ms to 11.17 ± 1.10 ms; P = NS) (Figure 4c). Taken together, these data indicate that the QRS prolongation may primarily reflect conduction slowing in the specialized ventricular conduction system.
Discussion
Our meta-analysis of 14 genome-wide association studies consisting of 40,407 individuals of European descent, with additional genotyping in 7170 Europeans, yielded genome-wide significant associations of QRS duration with common variants in 22 loci. Variations in four of these loci (locus 1, SCN5A/10A; locus 2, CDKN1A; locus 8, TBX5; and locus 21, DKK1) were previously associated with QRS duration in smaller independent studies using both candidate gene and genome-wide approaches.7–9 The 22 loci include genes in a number of interconnected pathways, including some previously known to be involved in cardiac conduction, such as sodium channels, calcium-handling proteins, and transcription factors, as well as novel processes not known to be involved in cardiac electrophysiology, such as kinase inhibitors, growth factor-related genes, and others.
The electrocardiographic QRS interval reflects ventricular depolarization and conduction time. Ventricular myocyte depolarization occurs via cardiac membrane excitatory inward currents mediated by voltage-gated sodium channels.19 The primary determinants of conduction velocity are the magnitude of excitatory inward currents flowing through these sodium channels, the extent of cell-to-cell communication via gap junction/connexin coupling, and cell and tissue architecture and morphology.19 Multiple pathways suggested in this study determine or modulate these key components of ventricular depolarization and conduction. Candidate genes in these pathways are briefly discussed in Box 1.
Box 1. Noteworthy genes within loci associated with QRS duration.
Of the 22 loci identified, common variants in four loci (SCN5A/SCN10A, CDKN1A, TBX5,and DKK1) were previously associated with QRS duration in genetic association studies. Mutations in two (SCN5Aand TBX5) lead to inherited syndromes associated with conduction disease. Animal experiments demonstrate a role for several additional loci (HAND1, TBX3,andTBX5) in cardiac ventricular conduction, as detailed below. The remainder are novel QRS loci, and their role in cardiac conduction remains to be elucidated.
-
Cardiac sodium channel genes:
SCN5A (locus 1): SCN5A encodes the cardiac Nav1.5 sodium channel and is well known to influence cardiac conduction, as well as other cardiovascular and electrophysiologic phenotypes.20, 21
SCN10A (locus 1): SCN10A encodes the Nav1.8 sodium channel, present in both ventricular myocardium and conduction fibers. Selective SCN10A blocker prolongs QRS interval.
-
Calcium handling proteins:
CASQ2 (locus 18): CASQ2 regulates opening of the ryanodine receptor (RYR2).37, 38 Cellular depolarization via Na-channels triggers calcium influx through L-type calcium channels, which in turn provokes RYR2-mediated calcium release from the sarcoplasmic reticulum. CASQ2 mutations have been associated with catecholaminergic polymorphic ventricular tachycardia.39, 40
PLN (locus 3): Calcium uptake into the sarcoplasmic reticulum by SERCA2a is regulated by phospholamban (PLN).41 The phosphorylation state of PLN is dependent on signaling pathways involving phosphatases and kinases including PRKCA.41 We previously demonstrated that this locus is associated with both cardiac electrical properties (QT interval duration, heart rate) and size (left ventricular end diastolic dimension) in GWA analyses.11, 12, 42, 43
PRKCA (locus 16): Protein kinase C alpha activity affects dephosphorylation of the sarcoplasmic reticulum Ca2+ ATPase-2 (SERCA-2) pump inhibitory protein phospholamban (PLN), and alters sarcoplasmic reticulum Ca2+ loading and the Ca2+ transient.44
STRN (locus 12): Striatin is a Ca2+/calmodulin binding protein that directly binds to caveolin scaffolding protein. Striatin has recently been implicated in a canine model of arrhythmogenic right ventricular cardiomyopathy.45, 46
-
Transcription factors:
TBX3 (locus 9) and TBX5 (locus 8): TBX3 and TBX5 encode transcription factors found in the cardiac conduction system. TBX5 (activator) competes with TBX3 (repressor) for the regulation of working myocardial genes such as GJA1.47, 48 Common variations near TBX3 and TBX5 were associated with PR and QRS durations.9, 13 Mutations in TBX3 and TBX5 have been associated with rare inherited syndromes manifested by an array of defects including ventricular structural and/or conduction defects.
TBX20 (locus 6): TBX20 demarcates the left and right ventricles49 and mutations in TBX20 have been implicated in multiple structural defects in mouse and human models.50, 51
HAND1 (locus 5): HAND1 encodes a transcription factor essential to cardiac morphogenesis,52 with a mutation identified in human hearts with septal defects.53 Over-expression of Hand1 in the adult mouse heart leads to loss of connexin43 (GJA1) expression, QRS prolongation, and predisposition to ventricular arrhythmia.54
NFIA (locus 4) and KLF12 (locus 19): Little is known about the role of Nuclear Factor One (NFIA) and Kruppel like protein 12 (KLF12) in cardiac tissue development.
-
Cyclin dependent kinase inhibitors:
CDKN1A (locus 2): CDKN1A is a negative regulator of cell cycle entry into G2/M phase, and is upregulated by ERBB2 activation. ERBB2, a member of the EGF receptor family of tyrosine kinases, is essential for proper heart development, and its ligand neuregulin-1 promotes formation of the murine cardiac conduction system.55 Furthermore, ERBB2 can modulate gap junction assembly and alter appropriate phosphorylation of connexin 43 in glial cells.56 In addition, CDKN1A is upregulated by PRKCA (locus 16).57
CDKN2C (locus 15): A member of the family of cyclin-dependent kinase inhibitors that prevent the activation of the CDK kinases, thus functioning as a cell growth regulator that controls cell cycle G1 progression.
-
Other pathways:
CRIM1 (locus 14): CRIM1 (locus 14), a cell-surface transmembrane protein that may bind to various members of the TGF-beta superfamily of ligands, is expressed in mouse and human cardiac tissues.58, 59 CRIM1 interacts with bone morphogenetic proteins, which induce the expression of CDKN1A (p21).58, 60
LRIG1 (locus 20): LRIG1 is upregulated in malignancies. It negatively regulates the proto-oncogenic, tyrosine kinase receptor family ERBB2.61
SETBP1 (locus 11): SETBP1 (locus 11) encodes a ubiquitously expressed protein that binds to the SET gene.62 The SETBP1-SET interaction has been hypothesized to be a component in tumor development.
TKT (locus 13): Transketolase (TKT) is a ubiquitous enzyme used in multiple metabolic pathways, including the pentose phosphate pathway.63
DKK1 (locus 21): DKK1, implicated in several tumors, inhibits the Wnt signaling pathway.64 Wnt signaling is an important modulator of connexin43 dependent intercellular coupling in the heart.65 In cardiac tissue it has an embryologic role with regard to axial development.66
SIPA1L1 (locus 7): SIPA1L1 appears to play a role in non-canonical Wnt signaling and contributes to development.67
Our strongest association signal (locus 1) mapped in or near two voltage-gated sodium channel genes: SCN5A and SCN10A. SCN5A encodes the cardiac Nav1.5 sodium channel and is well known for its role in cardiac conduction, and other cardiovascular and electrophysiologic phenotypes.20, 21 SCN10A encodes the Nav1.8 sodium channel. We provide novel data demonstrating that the SCN10A transcript and product is preferentially expressed in the mouse His-Purkinje system compared with the ventricular myocardium, and that Nav1.8 channel blockers result in QRS and HV interval prolongation, indicative of a slowing of impulse propagation in the specialized ventricular conduction system and delayed activation of the ventricular myocardium. Interestingly, Chambers et al. recently reported shortening of the PR interval in Scn10a knockout mice and concluded that Scn10a prolongs cardiac conduction and that rs6795970, encoding a Nav1.8 A1073V variant, is a gain-of-function allele.8 Alternatively, the more rapid conduction they observed in the knockout mice could reflect compensatory upregulation of TTX-sensitive currents, a 22 phenomenon observed in Nav1.8-deficient DRG neurons.
We, and others, demonstrated previously that, in addition to their association with QRS duration, variants in SCN5A and SCN10A are associated with atrial conduction (PR interval) and myocardial repolarization (QT interval), as well as atrial and ventricular fibrillation.8, 9, 13 These results emphasize the crucial role played by these genes in cardiac conduction and the generation of arrhythmias.
Calcium regulation is integral to impulse propagation, modulating cellular electrophysiology including sodium channel and gap junction function, as well as tissue architecture.20, 23, 24 Several of the loci associated with QRS duration contain genes directly related to calcium processes. As depicted in Supplementary Figure 4 and detailed in Box 1, these genes encode interrelated proteins that influence Ca2+ signaling (PLN in locus 3; PRKCA in locus 16; and CASQ2 in locus 18) and downstream effects (STRN in locus 12).
Transcription factors regulating embryonic electrophysiologic development are critical for the integrity of impulse conduction.25 We identified six transcription factors (TBX3 in locus 9; TBX5 in locus 8; TBX20 in locus 6; HAND1 in locus 5; NFIA in locus 4; and KLF12 in locus 19) in loci associated with QRS duration. Several of these transcription factors impact cardiac morphogenesis and may influence conduction by altering cellular and tissue architecture. Intriguingly, they may also have direct electrophysiologic consequences by modifying factors involved in impulse conduction. For example, HAND1 and T-box factors regulate connexin 40 (GJA5) and/or connexin 43 (GJA1), and TBX5 binds to the ATP2A2 (SERCA2A) promoter.26
Our study suggests a number of processes and pathways not previously known to be involved in cardiac electrophysiology, including cyclin dependent kinase inhibitors and genes related to tumorigenesis and cellular transformation. How these novel processes influence QRS duration remains to be defined.
In pleiotropic analyses, most variants influencing both PR and QRS, with the exception of TBX3, were concordant in effect direction, consistent with the known shared physiologic processes underlying the two traits: depolarization and conduction time in the sino-atrial, atria and atrioventricular node (PR) and depolarization and conduction time in the ventricles (QRS). By contrast, although QRS (ventricular depolarization) and QT (ventricular repolarization) are moderately positively correlated, most loci influencing both traits showed discordant effect directions (with the exception of the PLN locus). Investigating the physiologic foundations for these concordant and discordant PR-QRS and QT-QRS relationships could be particularly informative for elucidating the mechanisms by which these loci influence cardiac depolarization, conduction and repolarization.
Several limitations of our study should be considered. First, although we have identified 22 loci significantly associated with QRS duration, the broad nature of linkage disequilibrium among common variants generally precludes an unambiguous identification of the culprit variant or of the functional gene. For several genes (SCN5A, SCN10A, C6orf204, CASQ2), there are common coding SNPs in high LD with the index SNP, which may lend some support for a functional role for these genes. Furthermore, our expression analysis in blood revealed very strong cis-eQTL associations for TKT and CDKN1A, lending additional support to these genes as functional candidates. It would be desirable to perform similar eQTL analyses based on expression data in myocardial cells or conduction tissue. For our top signal in SCN10A, a gene which until recently was not known to be expressed in the heart, our functional work in mice confirm that SCN10A is involved in ventricular depolarization and conduction. Further fine-mapping is needed at all 22 loci to conclusively test all genetic variation (rare and common) for a role in QRS modulation.
To minimize the potential for confounding due to population substructure, we limited the analyses to individuals of European descent, for whom we could assemble the largest number of samples. At the individual study level, the GWAS showed very little evidence for gross stratification (genomic inflation factor, λGC, values ranged from 1.00 to 1.05). However, one of our QRS loci, mapping to HAND1/SAP30L, showed evidence of heterogeneity. In genetic association studies, heterogeneity can be due to sampling error, differences in phenotypic measurement, differences in LD structure between populations, technical artifacts, or genuine biological heterogeneity, but it would be difficult to conclude on the basis of our data here which is the most likely explanation.27
Our study underscores the power of a large genome-wide association study to extend prior biological understanding of cardiac ventricular conduction. Better understanding of the complex biologic pathways and molecular genetics associated with cardiac conduction and QRS duration may offer insight into the molecular basis underlying the pathogenesis of conduction abnormalities that can result in increased risk of sudden death, heart failure, and cardiac mortality.
Methods
Methods and any associated references are available in the online version of the paper.
Supplementary Material
Acknowledgments
Acknowledgements are available in the Supplementary Note.
Author contributions: Study concept and design:
N.S., A.A., D.E.A., P.I.W.d.B., E.B., H.C., A.C., C.M.v.D., M.E., S.B.F., G.I.F., A.R.F., J.F., V.G., P.v.d.H., S.R.H., A.A.H., A.H., A.I., S.K., H.K.K., C.N.-C., B.A.O., A.Pf., P.P.P., B.M.P., J.I.R., I.R., H.S., E.Z.S., B.H.C.S., A.G.U., A.V.S., U.V., H.V., T.J.W., J.F.W., A.F.W., N.J.S., Y.J.
Acquisition of data:
A.A., D.E.A., L.H.v.d.B., R.A.d.B., E.B., M.J.C., A.C., J.M.C., A.F.D., M.D., C.M.v. D., R.S.N.F., A.R.F., L.F., S.G., H.J.M.G., T.B.H., P.v.d.H., C.H., G.v.H., A.I, W.H.L.K., N.K., J.A.K., A.K., L.L., M.L., F.-Y.L., I.M.L., G.t.M., P.B.M., G.N., C.N.-C., B.A.O., R.A.O., S.Pe., A.Pf., A.Pe., O.P., B.M.P., J.Q., F.R., J.I.R., I.R., N.J.S., C.S., M.P.S.S., M.F.S., E.Z.S., B.H.C.S., A.T., A.G.U., D.J.v.V., C.B.V., R.K.W., C.W., J.F.W., J.C.M.W., D.L., T.D.S
Statistical analysis and interpretation of data:
A.A., D.E.A., T.A., P.I.W.d.B., N.S., E.B., A.C., L.A.C., M.E., K.E., G.I.F., A.R.F., L.F., J.F., C.F., S.A.G., W.H.v.G., S.G., V.G., P.v.d.H., C.H., S.R.H., A.I., T.J., W.H.L.K., X.L., K.D.M., I.M.L., M.M., I.M.N., S.Pa., A.Pf., O.P., B.M.P., K.R., H.S., A.T., A.V.S., S.H.W., A.Y.W., N.J.S.
Drafting of the manuscript:
N.S., A.A., D.E.A, F.W.A., P.I.W.d.B., M.D., C.M.v.D,. M.E., G.I.F., J.F., S.A.G., V.G., C.H., A.I., Y.J., S.K., J.W.M., I.M.N., O.P., N.J.S., H.S., C.N-C., P.v.d.H.
Critical revision of the manuscript:
A.A., D.E.A., T.A., F.W.A., J.C.B., R.A.d.B., E.B., H.C., M.J.C., A.C., J.M.C., L.A. C., A.F.D., M.D., C.M.v.D., M.E., K.E, S.B.F., G.I.F., A.R.F., J.F., W.H.v.G., V.G., T.B.H., P.v.d.H., C.H., S.R.H., G.v.H., A.A.H., A.H., A.I., Y.J., T.J., S.K., W.H.L.K., N.K., J.A.K., A.K., H.K.K., L.L., D.L., M.L., J.W.M., I.M.L., T.M., M.M, P.B.M., G.N., C.N.-C., I.M.N., C.J.O., B.A.O., S.Pa., S.Pe., A.Pf., A.Pe., O.P., B.M.P., F.R., J.I.R., I.R., M.P.S.S., M.F.S., D.S.S., H.S., B.H.C.S., E.Z.S., A.T., A.G.U., D.J.v.V., U.V., H.V., T.J.W., H.-E.W., A.V.S., S.H.W., J.F.W., J.C.M.W., A.F.W.
Obtained funding:
L.H.v.d.B., E.B., H.C., M.J.C., A.C., J.M.C., A.F.D., C.M.v.D., S.B.F., G.I.F., W.H. v.G., H.J.M.G., V.G., P.v.d.H., A.H., Y.J., S.K., H.K.K., L.L., P.B.M, G.N., C. N.-C., C.J.O., B.A.O., R.A.O., P.P.P., B.M.P., J.I.R., I.R., N.J.S., N.S, T.D.S., A.G.U., D.J.v.V., U.V., H.V., T.J.W., R.K.W., H.-E.W., C.W., J.F.W., A.F.W., D.L.
URL Section
MACH - http://www.sph.umich.edu/csg/abecasis/mach/
MANTEL - http://www.broadinstitute.org/~debakker/mantel.html
MetABEL - http://mga.bionet.nsc.ru/~yurii/ABEL/
METAL - http://www.sph.umich.edu/csg/abecasis/metal/
PLINK - http://pngu.mgh.harvard.edu/~purcell/plink/
IMPUTE - https://mathgen.stats.ox.ac.uk/impute/impute.html
SNPTEST - http://www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html
1000 Genomes project - www.1000Genomes.org
SNAP - http://www.broadinstitute.org/mpg/snap/
Ingenuity - http://www.ingenuity.com
DAVID - http://david.abcc.ncifcrf.gov
Footnotes
Note: Supplementary information is available online.
Competing interests statement: Aravinda Chakravarti is a paid member of the Scientific Advisory Board of Affymetrix, a role that is managed by the Committee on Conflict of Interest of the Johns Hopkins University School of Medicine.
Reference List
- 1.Desai AD, et al. Prognostic Significance of Quantitative QRS Duration. Am J Med. 2006;119:600–606. doi: 10.1016/j.amjmed.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 2.Elhendy A, Hammill SC, Mahoney DW, Pellikka PA. Relation of QRS duration on the surface 12-lead electrocardiogram with mortality in patients with known or suspected coronary artery disease. Am J Cardiol. 2005;96:1082–1088. doi: 10.1016/j.amjcard.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 3.Oikarinen L, et al. QRS duration and QT interval predict mortality in hypertensive patients with left ventricular hypertrophy: the Losartan Intervention for Endpoint Reduction in Hypertension Study. Hypertension. 2004;43:1029–1034. doi: 10.1161/01.HYP.0000125230.46080.c6. [DOI] [PubMed] [Google Scholar]
- 4.Dhingra R, et al. Electrocardiographic QRS duration and the risk of congestive heart failure: the Framingham Heart Study. Hypertension. 2006;47:861–867. doi: 10.1161/01.HYP.0000217141.20163.23. [DOI] [PubMed] [Google Scholar]
- 5.Busjahn A, et al. QT interval is linked to 2 long-QT syndrome loci in normal subjects. Circulation. 1999;99:3161–3164. doi: 10.1161/01.cir.99.24.3161. [DOI] [PubMed] [Google Scholar]
- 6.Hanson B, et al. Genetic factors in the electrocardiogram and heart rate of twins reared apart and together. Am J Cardiol. 1989;63:606–609. doi: 10.1016/0002-9149(89)90907-7. [DOI] [PubMed] [Google Scholar]
- 7.Bezzina CR, et al. Common sodium channel promoter haplotype in asian subjects underlies variability in cardiac conduction. Circulation. 2006;113:338–344. doi: 10.1161/CIRCULATIONAHA.105.580811. [DOI] [PubMed] [Google Scholar]
- 8.Chambers JC, et al. Genetic variation in SCN10A influences cardiac conduction. Nat Genet. 2010;42:149–152. doi: 10.1038/ng.516. [DOI] [PubMed] [Google Scholar]
- 9.Holm H, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 10.Dubois PC, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newton-Cheh C, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfeufer A, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfeufer A, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calvano SE, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 15.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 16.Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pallante BA, et al. Contactin-2 Expression in the Cardiac Purkinje Fiber Network. Circ Arrhythm Electrophysiol. 2010;3:186–194. doi: 10.1161/CIRCEP.109.928820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvis MF, et al. A-803467, a potent and selective Nav1.8 sodium channel blocker, attenuates neuropathic and inflammatory pain in the rat. Proc Natl Acad Sci U S A. 2007;104:8520–8525. doi: 10.1073/pnas.0611364104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desplantez T, Dupont E, Severs NJ, Weingart R. Gap junction channels and cardiac impulse propagation. J Membr Biol. 2007;218:13–28. doi: 10.1007/s00232-007-9046-8. [DOI] [PubMed] [Google Scholar]
- 20.Abriel H. Cardiac sodium channel Na(v)1.5 and interacting proteins: Physiology and pathophysiology. J Mol Cell Cardiol. 2010;48:2–11. doi: 10.1016/j.yjmcc.2009.08.025. [DOI] [PubMed] [Google Scholar]
- 21.Remme CA, Wilde AA, Bezzina CR. Cardiac sodium channel overlap syndromes: different faces of SCN5A mutations. Trends Cardiovasc Med. 2008;18:78–87. doi: 10.1016/j.tcm.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Akopian AN, et al. The tetrodotoxin-resistant sodium channel SNS has a specialized function in pain pathways. Nat Neurosci. 1999;2:541–548. doi: 10.1038/9195. [DOI] [PubMed] [Google Scholar]
- 23.Saimi Y, Kung C. Calmodulin as an ion channel subunit. Annu Rev Physiol. 2002;64:289–311. doi: 10.1146/annurev.physiol.64.100301.111649. [DOI] [PubMed] [Google Scholar]
- 24.Potet F, et al. Functional Interactions between Distinct Sodium Channel Cytoplasmic Domains through the Action of Calmodulin. J Biol Chem. 2009;284:8846–8854. doi: 10.1074/jbc.M806871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolf CM, Berul CI. Inherited conduction system abnormalities--one group of diseases, many genes. J Cardiovasc Electrophysiol. 2006;17:446–455. doi: 10.1111/j.1540-8167.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Y, et al. Tbx5-dependent pathway regulating diastolic function in congenital heart disease. Proc Natl Acad Sci U S A. 2008;105:5519–5524. doi: 10.1073/pnas.0801779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lebrec JJ, Stijnen T, van Houwelingen HC. Dealing with heterogeneity between cohorts in genomewide SNP association studies. Stat Appl Genet Mol Biol. 2010;9 doi: 10.2202/1544-6115.1503. Article. [DOI] [PubMed] [Google Scholar]
- 28.Hofman A, et al. The Rotterdam Study: 2010 objectives and design update. Eur J Epidemiol. 2009;24:553–572. doi: 10.1007/s10654-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor Popul Biol. 2001;60:155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- 31.de Bakker PI, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 33.Johnson AD, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sreejit P, Kumar S, Verma RS. An improved protocol for primary culture of cardiomyocyte from neonatal mice. In Vitro Cell Dev Biol Anim. 2008;44:45–50. doi: 10.1007/s11626-007-9079-4. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Lee P, et al. Conditional lineage ablation to model human diseases. Proc Natl Acad Sci U S A. 1998;95:11371–11376. doi: 10.1073/pnas.95.19.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei L, Hanna AD, Beard NA, Dulhunty AF. Unique isoform-specific properties of calsequestrin in the heart and skeletal muscle. Cell Calcium. 2009;45:474–484. doi: 10.1016/j.ceca.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 38.Terentyev D, et al. Abnormal interactions of calsequestrin with the ryanodine receptor calcium release channel complex linked to exercise-induced sudden cardiac death. Circ Res. 2006;98:1151–1158. doi: 10.1161/01.RES.0000220647.93982.08. [DOI] [PubMed] [Google Scholar]
- 39.Priori SG, et al. Clinical and molecular characterization of patients with catecholaminergic polymorphic ventricular tachycardia. Circulation. 2002;106:69–74. doi: 10.1161/01.cir.0000020013.73106.d8. [DOI] [PubMed] [Google Scholar]
- 40.Postma AV, et al. Absence of calsequestrin 2 causes severe forms of catecholaminergic polymorphic ventricular tachycardia. Circ Res. 2002;91:e21–e26. doi: 10.1161/01.res.0000038886.18992.6b. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Goldhaber JI. Return of calcium: manipulating intracellular calcium to prevent cardiac pathologies. Proc Natl Acad Sci U S A. 2004;101:5697–5698. doi: 10.1073/pnas.0401518101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasan RS, et al. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA. 2009;302:168–178. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eijgelsheim M, et al. Genome-wide association analysis identifies multiple loci related with resting heart rate. HumMolGenet. 2010 Oct;19(19):3885–94. doi: 10.1093/hmg/ddq303. Hum Mol Gen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braz JC, et al. PKC-alpha regulates cardiac contractility and propensity toward heart failure. Nat Med. 2004;10:248–254. doi: 10.1038/nm1000. [DOI] [PubMed] [Google Scholar]
- 45.Baillat G, et al. Molecular cloning and characterization of phocein, a protein found from the Golgi complex to dendritic spines. Mol Biol Cell. 2001;12:663–673. doi: 10.1091/mbc.12.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meurs KM, et al. Genome-wide association identifies a deletion in the 3’ untranslated region of Striatin in a canine model of arrhythmogenic right ventricular cardiomyopathy. Hum Genet. 2010 doi: 10.1007/s00439-010-0855-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boogerd KJ, et al. Msx1 and Msx2 are functional interacting partners of T-box factors in the regulation of Connexin43. Cardiovasc Res. 2008;78:485–493. doi: 10.1093/cvr/cvn049. [DOI] [PubMed] [Google Scholar]
- 48.Hoogaars WM, et al. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004;62:489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 49.Singh R, et al. Tbx20 interacts with smads to confine tbx2 expression to the atrioventricular canal. Circ Res. 2009;105:442–452. doi: 10.1161/CIRCRESAHA.109.196063. [DOI] [PubMed] [Google Scholar]
- 50.Posch MG, et al. A Gain-of-function TBX20 Mutation Causes Congenital Atrial Septal Defects, Patent Foramen Ovale and Cardiac Valve Defects. J Med Genet. 2009 doi: 10.1136/jmg.2009.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bakker ML, et al. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ Res. 2008;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- 52.Riley P, Anson-Cartwright L, Cross JC. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet. 1998;18:271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- 53.Reamon-Buettner SM, et al. A functional genetic study identifies HAND1 mutations in septation defects of the human heart. Hum Mol Genet. 2009;18:3567–3578. doi: 10.1093/hmg/ddp305. [DOI] [PubMed] [Google Scholar]
- 54.Breckenridge RA, et al. Overexpression of the transcription factor Hand1 causes predisposition towards arrhythmia in mice. J Mol Cell Cardiol. 2009;47:133–141. doi: 10.1016/j.yjmcc.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 55.Rentschler S, et al. Neuregulin-1 promotes formation of the murine cardiac conduction system. Proc Natl Acad Sci U S A. 2002;99:10464–10469. doi: 10.1073/pnas.162301699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hofer A, et al. C-erbB2/neu transfection induces gap junctional communication incompetence in glial cells. J Neurosci. 1996;16:4311–4321. doi: 10.1523/JNEUROSCI.16-14-04311.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Besson A, Yong VW. Involvement of p21(Waf1/Cip1) in protein kinase C alpha-induced cell cycle progression. Mol Cell Biol. 2000;20:4580–4590. doi: 10.1128/mcb.20.13.4580-4590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilkinson L, et al. CRIM1 regulates the rate of processing and delivery of bone morphogenetic proteins to the cell surface. J Biol Chem. 2003;278:34181–34188. doi: 10.1074/jbc.M301247200. [DOI] [PubMed] [Google Scholar]
- 59.Kolle G, Georgas K, Holmes GP, Little MH, Yamada T. CRIM1, a novel gene encoding a cysteine-rich repeat protein, is developmentally regulated and implicated in vertebrate CNS development and organogenesis. Mech Dev. 2000;90:181–193. doi: 10.1016/s0925-4773(99)00248-8. [DOI] [PubMed] [Google Scholar]
- 60.Pardali K, Kowanetz M, Heldin CH, Moustakas A. Smad pathway-specific transcriptional regulation of the cell cycle inhibitor p21(WAF1/Cip1) J Cell Physiol. 2005;204:260–272. doi: 10.1002/jcp.20304. [DOI] [PubMed] [Google Scholar]
- 61.Laederich MB, et al. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem. 2004;279:47050–47056. doi: 10.1074/jbc.M409703200. [DOI] [PubMed] [Google Scholar]
- 62.Minakuchi M, et al. Identification and characterization of SEB, a novel protein that binds to the acute undifferentiated leukemia-associated protein SET. Eur J Biochem. 2001;268:1340–1351. doi: 10.1046/j.1432-1327.2001.02000.x. [DOI] [PubMed] [Google Scholar]
- 63.Zhao J, Zhong CJ. A review on research progress of transketolase. Neurosci Bull. 2009;25:94–99. doi: 10.1007/s12264-009-1113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fedi P, et al. Isolation and biochemical characterization of the human Dkk-1 homologue, a novel inhibitor of mammalian Wnt signaling. J Biol Chem. 1999;274:19465–19472. doi: 10.1074/jbc.274.27.19465. [DOI] [PubMed] [Google Scholar]
- 65.Ai Z, Fischer A, Spray DC, Brown AM, Fishman GI. Wnt-1 regulation of connexin43 in cardiac myocytes. J Clin Invest. 2000;105:161–171. doi: 10.1172/JCI7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Korol O, Gupta RW, Mercola M. A novel activity of the Dickkopf-1 amino terminal domain promotes axial and heart development independently of canonical Wnt inhibition. Dev Biol. 2008;324:131–138. doi: 10.1016/j.ydbio.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsai IC, et al. A Wnt-CKIvarepsilon-Rap1 pathway regulates gastrulation by modulating SIPA1L1, a Rap GTPase activating protein. Dev Cell. 2007;12:335–347. doi: 10.1016/j.devcel.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.