Abstract
Purpose
The present study is a discovery mode proteomics analysis of the membrane enriched fraction of post-mortem brain tissue from Alzheimer’s disease (AD) and control cases. This study aims to validate a method to identify new proteins that could be involved in the pathogenesis of AD and potentially serve as disease biomarkers.
Experimental Design
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to analyze the membrane enriched fraction of human post-mortem brain tissue from five AD and five control cases of similar age. Biochemical validation of specific targets was performed by immunoblotting.
Results
1709 proteins were identified from the membrane enriched fraction of frontal cortex. Label free quantification by spectral counting and G-test analysis identified 13 proteins that were significantly changed in disease. In addition to Tau (MAPT), two additional proteins found to be enriched in AD, Ubiquitin carboxy-terminal hydrolase 1 (UCHL1), and syntaxin binding protein 1 (Munc-18), were validated through immunoblotting.
Discussion and clinical relevance
Proteomic analysis of the membrane enriched fraction of post-mortem brain tissue identifies proteins biochemically altered in AD. Further analysis of this sub-proteome may help elucidate mechanisms behind AD pathogenesis and provide new sources of biomarkers.
Keywords: Alzheimer’s disease, membrane enrichment, proteomics, neurodegeneration
1. Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder characterized by short-term memory loss, language impairments, deficits in visuospatial skills, and personality changes [5]. The pathological hallmarks of AD include the accumulation of extracellular beta-amyloid (Aβ) plaques, intracellular neurofibrillary tangles composed of abnormal, hyperphosphorylated tau, gliosis and neuronal cell death [6-8]. Since the proposal of the amyloid cascade hypothesis [9], the role of Aβ in disease has been advanced; however, it does not account for all of the changes seen in AD [10]. Oxidative stress, defective proteolysis, protein aggregation, altered cell signaling and chronic inflammation have all been implicated in disease pathogenesis [10]. Furthermore, an increasing number of studies have shown evidence of abnormal protein traffic within neurons early in disease [11]. Many of these abnormal cellular functions involve membrane associated proteins. Therefore, studying a membrane enriched sample may provide insights to potential biomarkers of disease.
Analysis of the proteome by liquid chromatography coupled with tandem-mass spectrometry (LC-MS/MS) has emerged as a leading technique for unbiased identification of proteins that are changed in disease [12]. LC-MS/MS is a powerful tool for analyzing pathways involved in disease pathogenesis and identifying potential biomarkers of disease. Quantitative proteomics in human tissue can be accomplished using either labeled or label-free methods. Labeled approaches can include chemical derivatization strategies using isobaric stable isotope tagging reagents, such as iTRAQ reagents [13]. Alternatively, use of stable isotope labeling with amino acids in cell culture (SILAC) has been increasingly utilized in relative quantification of tissue proteins [14]. In this approach, a small amount of heavy isotope labeled cell lysate (from one or more SILAC labeled cell lines) is added as a standard to the sample being analyzed [15, 16]. This provides an internal reference against which changes in protein abundance can be measured between proteomes. This approach has the advantage of being extremely sensitive for detecting even small changes in the proteome. However, the major drawback of this method is that adding an external reference proteome increases the complexity of an already complex mixture. As each peptide is sequenced twice (one heavy and one light), there is less coverage of the total proteome. Label-free approaches, such as spectral counting or peptide ion intensity measurements avoid this caveat, but are biased towards the most abundant proteins in a sample, which limits the detection of less abundant proteins that may be changing in disease [17].
Reducing the complexity of the sample before LC-MS/MS analysis is one way to circumvent this problem. The analysis of various subproteomes is becoming an increasingly popular strategy for identifying disease biomarkers [18]. This study presents a comparative proteomics analysis of a membrane-enriched sample of post-mortem brain tissue from AD cases and healthy controls. Label-free quantification by G-test statistical analysis of spectral counts identified significant changes in 13 proteins in AD cases compared to controls. Three of these targets, including tau, were independently validated through immunoblotting. This study highlights the utility of analyzing the membrane proteome as a method for identifying proteins that may be involved in the pathogenesis of Alzheimer’s disease.
2. Materials and Methods
2.1 Case selection
Post-mortem frontal cortex tissue from five healthy control cases and five pathologically confirmed AD cases were selected for comparison from the Emory Alzheimer’s Disease Research Center (ADRC) brain bank (Table 1). All AD cases were determined to be Braak stage V or VI and definite AD as classified by CERAD [19, 20]. Cases were matched for as closely as possible for age of death, gender and post-mortem interval (PMI). All control cases were pathologically clean except case five, which had secondary and tertiary neuropathological diagnoses (NP dx) of hemorrhagic infarcts and diffuse plaques.
Table 1.
Demographic data on cases pooled for proteomics analysis.
| Sample | Case # | primary NP dx1 | secondary NP dx | Tertiary NP dx | PMI2 | Age death | APOE3 | BRAAK | CERAD | race/gender4 |
|---|---|---|---|---|---|---|---|---|---|---|
| Control 1 | E06-41 | CTL | 10 | 57 | E3/3 | 2 | Normal | wm | ||
| Control 2 | OS00-06 | CTL | 8 | 60 | E3/4 | 0 | Normal | bf | ||
| Control 3 | OS01-112 | CTL | 6 | 65 | E3/3 | 1 | Normal | wf | ||
| Control 4 | A87-50 | CTL | 10 | 66 | E3/3 | 2 | Normal | wm | ||
| Control 5 | OS03-299 | CTL | hemorrhagic infarcts | diffuse plaques | 6 | 69 | E3/3 | 2 | Normal | wm |
| AD 1 | E07-69 | AD | 6 | 58 | E3/4 | 6 | Definite AD | wf | ||
| AD 2 | E05-67 | AD | 11.5 | 62 | E3/4 | 5 | Definite AD | wm | ||
| AD 3 | E04-17 | AD | 9 | 64 | E3/4 | 5/6 | Definite AD | wm | ||
| AD 4 | E05-04 | AD | 4.5 | 64 | E3/4 | 6 | Definite AD | wf | ||
| AD 5 | E05-56 | AD | 17 | 71 | E3/4 | 6 | Definite AD | wm | ||
NP dx = neuropathological diagnosis.
PMI = postmortem interval.
Apolipoprotein E (APOE) genotype.
w = white; b= black; m=male; f=female.
2.2 Membrane enrichment strategy
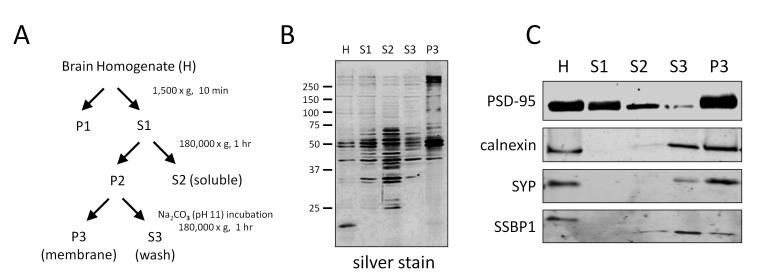
The membrane enrichment strategy employed was modified from previously published methods [21]. Homogenized frontal cortex (1.5 ml) in a low salt, buffered sucrose solution (0.24 M sucrose, 25 mM NaCl, 50 mM HEPES, pH 7.0, 20 mM iodoacetomide, 1 mM EDTA) with protease and phosphatase inhibitors was used as the starting material for each case. Briefly, the samples were thawed on ice and the homogenate (H) was sonicated (sonic dismembrator, Fisher Scientific) three times for five seconds at 20% amplitude (maximum intensity) to disrupt cell membranes, large cytoskeletal fragments and nucleic acids. Following sonication, the homogenate was centrifuged at 1500 x g for 10 minutes (Eppendorf 5417C) to sediment unhomogenized tissue and large cellular debris. The pellet (P1) was discarded. The supernatant (S1) was transferred to a polycarbonate ultracentrifuge tube and centrifuged at 180,000 x g for one hour at 4 °C (Beckman Optima TLX ultracentrifuge, TLA 100.4 rotor). After ultracentrifugation, the supernatant (S2) containing the soluble protein fraction was removed and saved. The resulting pellet (P2) was resuspended in 1 ml of 0.1 M sodium carbonate, pH 11 with protease and phosphatase inhibitors and incubated on ice for 15 minutes to strip proteins only loosely associated with the membrane. The samples were re-centrifuged at 180,000 x g for one hour at 4 °C (Beckman Optima TLX ultracentrifuge, TLA 100.4 rotor). The supernatant (S3) was removed and saved and the resulting membrane-enriched, insoluble pellet (P3) was dissolved in 100 μl 8M urea. Protein concentrations from each of the five fractions (H, S1, S2, S3, P3) were determined by the bicinchoninic acid (BCA) method (Pierce, Rockford, IL). The fractions obtained from the enrichment protocol were analyzed by silver stain. Briefly, one microgram of protein was loaded from each fraction into a 10% acrylamide gel and separated by gel electrophoresis. The gel was fixed in a solution containing 50% methanol and 5% acetic acid for 10 minutes and washed with de-ionized water. After rinsing in 0.02% sodium thiosulfate for 1 minute, the gel was stained with 0.1% silver nitrate for 10 minutes and developed with 3% sodium carbonate, 0.05% formaldehyde solution until the bands were sufficiently stained.
2.3 Proteomic Analysis
Protein (50 μg/case) from the membrane enriched fraction (P3 dissolved in 8M urea) was pooled for proteomic analysis. Peptides were obtained from an in-solution digest. Briefly, each pool was normalized to 250 μg of total protein, treated with 5 mM dithiothreitol (DTT) for 30 min at 37°C followed by 20 mM iodoacetamide (IAA) for 30 min at 37°C in the dark, and digested with 1:100 (w/w) endopeptidase LysC (Wako Chemicals, Japan) for 4 hours at 37°C. Samples were diluted with 100 mM sodium bicarbonate to a final concentration of 1.2 M urea and digested overnight with 1:50 (w/w) trypsin at 37°C. The next day, samples were acidified with 5% formic acid, 0.2% trifluoroacetic acid (TFA) and desalted by a C18 column (Sep-Pak® Cartridges, Waters, Milford, MA).
2.4 Peptide analysis by LC-MS/MS
Purified peptides were analyzed by reverse-phase liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) and each sample was analyzed in technical replicate [22]. Briefly, peptide mixtures were loaded onto a C18 column (100 μm i.d., 20 cm long, 2.7 μm HALO resin from Michrom Bioresources, Inc., Auburn, CA) and eluted over a 5-30% gradient (Buffer A: 0.1% formic acid, 0.005% heptafluorobutyric acid, and 5% AcN; Buffer B: 0.1% formic acid, 0.005% heptafluorobutyric acid, and 95% AcN). Eluates were monitored in a MS survey scan followed by ten data-dependent MS/MS scans on an LTQ-Orbitrap mass spectrometer (Thermo Finnigan, San Jose, CA). The LTQ was used to acquire MS/MS spectra (3 m/z isolation width, 35% collision energy, 5,000 AGC target, 200 ms maximum ion accumulation time). The Orbitrap was used to collect MS scans (300-1600 m/z, 1,000,000 AGC target, 1,000 ms maximum ion accumulation time, resolution 60,000). Peptide retention time was measured between replicate LC-MS/MS runs using in-house software as described previously [23].
2.5 Peptide filtering and protein identification
All data were converted from .raw files to the .dta format using ExtractMS version 2.0 (ThermoElectron) and searched against human reference database downloaded from the National Center for Biotechnology Information (November 19, 2008) using the SEQUEST Sorcerer algorithm (version 3.11, SAGE-N). Searching parameters included mass tolerance of precursor ions (±50 ppm) and product ion (±0.5 m/z), partial tryptic restriction, with a dynamic mass shift for oxidized Met (+15.9949), 2 maximal modification sites and a maximum of two missed cleavages. Only b and y ions were considered during the database match. To evaluate false discovery rate (FDR), all original protein sequences were reversed to generate a decoy database that was concatenated to the original database (77,764 entries) [24]. The FDR was estimated by the number of decoy matches (nd) and total number of assigned matches (nt). FDR = 2*nd/nt, assuming mismatches in the original database were the same as in the decoy database. To remove false positive matches, assigned peptides were grouped by a combination of trypticity (fully, partial and non-tryptic) and precursor ion-charge state. Each group was first filtered by mass accuracy (10 ppm for high-resolution MS), and by dynamically increasing correlation coefficient and ΔCn values to reduce protein FDR to less than 1 percent. All MS/MS spectra for proteins identified by a single peptide were manually inspected as described previously [25]. If peptides were shared by multiple members of a protein family, the matched members were clustered into a single group. On the basis of the principle of parsimony, the group was represented by the protein with greatest number of assigned peptides. All identified proteins (grouped and ungrouped) and peptide sequences are provided in Supplemental Table 1 and 2.
2.6 Label Free Quantification
Quantification was performed as previously described [21, 26]. Spectral counting was used to determine differences between the control and AD membrane enriched proteomes. Several of the proteins identified in this study were found exclusively in AD or control samples. For those proteins only identified in one sample (AD or control), a spectral count of 1 was applied to the sample group without detectable protein. The spectral counts were normalized to ensure that average spectral count ratio per protein was the same in the two datasets [27]. G-test was used to judge statistical significance of protein abundance difference as previously described [17, 21, 28, 29]. The G-value of each protein was calculated as shown in equation 1, where fAD and fCTL are the detected spectral counts of a given protein in the AD and control membrane fractions, respectively, and “ln” is the natural logarithm.
| (1) |
The p-value of each protein was calculated as the probability of observing a random variable larger than G from the Chi-square distribution (one degree of freedom). The frequency histogram of the p-values was created and 0.01 was set as the cutoff to detect significant changes.
2.7 Antibodies
Primary antibodies used in these studies were as follows (dilutions in parentheses): PSD95 (1:1000, mouse monocloncal; Chemicon International, Temecula, CA); Tau (1:1000, mouse monoclonal; Transduction labs); Synaptophysin (1:1000, mouse monoclonal; Boehringer); Calnexin (1:1000, rabbit polyclonal; Assay Designs, Ann Arbor, MI); SSBP1 (1:500, rabbit polyclonal; Abcam, Cambridge, MA); PGP9.5 (UCHL1) (1:1000, rabbit polyclonal; Chemicon International, Temecula CA); Munc18 (1:1000, rabbit polyclonal; Abcam, Cambridge, MA); alpha-2-macroglobulin (A2M) (1:250, mouse monoclonal; Abcam, Cambridge, MA).The antibody dilutions noted above reflect prior dilution of each antibody (1:1) with glycerol.
2.8 Immunoblotting
Equal concentrations of protein from each sample were loaded into an acrylamide gel (10% or 12%) and separated by SDS-PAGE. Proteins were transferred onto PVDF Immobilon-P membranes (Millipore, Billercia, MA) overnight at 4 °C. Immunoblots were blocked for 2 hours at room temperature with TBS/Tween and blocking buffer (USB, 5x blocking buffer ultrapure) and probed for the protein of interest with a primary antibody overnight at 4 °C. The following day, blots were incubated with fluorophore-conjugated secondary antibodies (1:20,000) for one hour in the dark. All blots were scanned and quantified using the Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE). Statistical analysis was performed using a two-tailed student’s t-test.
3. Results
3.1 Membrane enrichment strategy and LC-MS/MS
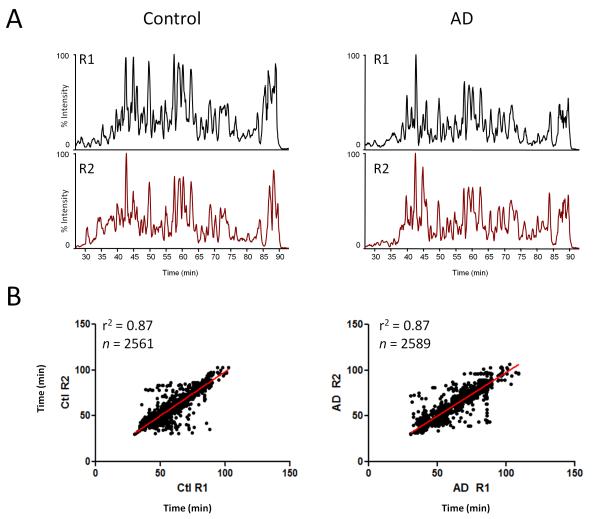
Traditional membrane purification protocols involve multiple centrifugation steps using a sucrose gradient to separate out individual membrane fractions (microsomes, endosomes, mitochondria, pre- and post-synaptic vesicles, etc.) [30]. This study sought to identify changes in the total membrane proteome and therefore, more traditional protocols were modified to obtain a sample that contained a broader membrane fraction as described in the methods and in Figure 1A. Different banding patterns in each fraction were evident by silver staining (Fig. 1B). Immunoblots demonstrate that the final membrane sample contained multiple membrane fractions. More specifically, PSD-95 (post-synaptic density), synaptophysin (synaptic vesicles), calnexin (microsomes), and SSBP1 (mitochondria) were all observed in the membrane fraction (Fig. 1C). Quantification of the PSD-95 blot demonstrated approximately a 2-fold enrichment of the protein in the membrane fraction compared to total cell lysate, indicating that this method was a reasonable strategy for enriching membrane proteins. As this was a discovery-mode proteomics experiment, cases were pooled to decrease inter-individual variability and enhance the likelihood that any changes detected would be universal to disease [31]. Peptides were extracted from the pooled samples following an in-solution digest and analyzed in technical replicate using LC-MS/MS. The extracted base peak chromatograms for peptide intensity demonstrate consistency between replicate 1 (R1) and replicate 2 (R2) within Control and AD samples. The slight variation observed between control and AD samples suggests a difference in distinct peptide abundance (Fig. 2A). Retention time for identified peptides (eluting from 30 to 100 minutes) between R1 and R2 was strongly correlated (R2=0.87), indicating chromatographic consistency between LC-MS runs (Fig. 2B). After database searching, a total of 1709 proteins in 729 homology groups were identified (Supplementary Tables 1, 2). The TMHMM 2.0 web server [32], was used to predict the number of transmembrane domains (TMD) for each protein. Overall, approximately 25% of grouped proteins (184/729) identified in the sample were predicted to have at least one TMD (Supplementary Table 1)
Figure 1. Membrane enrichment strategy.
(A) Flowchart of experimental design to enrich for membrane proteins from human brain tissue. (B) Silver stain showing differential protein banding patterns in each fraction. Immunoblot of PSD-95, a transmembrane domain-containing protein specific to the post synaptic density, demonstrates approximately 2 fold enrichment in the membrane fraction compared with total homogenate. (C) Immunoblots for calnexin, synaptophysin (SYP) and SSBP1 demonstrates that multiple membrane proteins are present in the final membrane fraction.
Figure 2. Human brain membrane factions analyzed by LC-MS/MS.
(A) Representative base peak peptide elution profiles of control and AD sample demonstrate the reproducibility between technical replicate 1 (R1) and replicate 2 (R2). (B) Peptide retention time correlation between R1 and R2 from control and AD samples is provided.
3.3 Relative quantification of proteins assessed using a label-free strategy
In this study, analysis of spectral counts by G-test was used to determine which membrane proteins were altered in AD. G-test is a statistical approach that can be used when quantifying spectral counts to determine which proteins are significantly changing between samples (e.g. control and AD). It has been proven to be a robust and reliable method for identifying changes in protein levels in label-free proteomic experiments [33-35]. To increase the stringency of the analysis, a p value cutoff of less than or equal to 0.01 was chosen to represent proteins significantly changed in disease. Using these parameters, 13 proteins altered in disease were identified (Table 2), including known markers of AD such as tau (p = 5.8E-15). From this list, ubiquitin carboxy-terminal hydrolase L1 (UCHL1) (p = 0.0015), Munc18 (p = 0.0079), and alpha-2 macroglobulin (A2M) (p = 0.0001) were also chosen to be further validated by immunoblotting given their potential association with AD [36-38].
Table 2.
Proteins significantly changing in Alzheimer's disease (AD)1
| Spectral Counts |
||||||||
|---|---|---|---|---|---|---|---|---|
| NCBI Reference | Gene ID | Description | MW | TP | % Cov. | CTL | AD | p -value |
| NP_001116539.1 | MAPT | microtubule-associated protein tau | 43 | 12 | 26 | 2 | 55 | 5.88E-15 |
| NP_001157570.1 | VCAN | versican core protein | 182 | 13 | 8 | 95 | 43 | 6.98E-07 |
| NP_000436.2 | PLEC | plectin-1 | 518 | 60 | 18 | 38 | 85 | 6.62E-05 |
| NP_000005.2 | A2M | alpha-2-macroglobulin | 163 | 25 | 26 | 47 | 19 | 0.0001 |
| NP_006588.1 | HSPA8 | heat shock cognate 71 kDa protein | 71 | 19 | 40 | 51 | 97 | 0.0005 |
| NP_001814.2 | CKB | creatine kinase B-type | 43 | 28 | 68 | 123 | 193 | 0.0007 |
| NP_004172.2 | UCHL1 | ubiquitin carboxyl-terminal hydrolase isozyme L1 | 25 | 5 | 32 | 8 | 27 | 0.0015 |
| NP_002645.3 | PKM2 | pyruvate kinase isozymes M1/M2 | 58 | 22 | 53 | 76 | 47 | 0.0022 |
| NP_031381.2 | HSP90AB1 | heat shock protein HSP 90-beta | 83 | 17 | 32 | 25 | 53 | 0.0031 |
| NP_002071.2 | GOT2 | aspartate aminotransferase, mitochondrial | 47 | 8 | 22 | 13 | 3 | 0.0056 |
| NP_003276.3 | TNR | tenascin-R | 149 | 20 | 21 | 56 | 34 | 0.0069 |
| NP_001027392.1 | STXBP1 | syntaxin-binding protein 1 (Munc-18) | 68 | 28 | 55 | 60 | 98 | 0.0080 |
| NP_002777.1 | PSMA1 | proteasome subunit alpha type-1 | 30 | 7 | 40 | 10 | 2 | 0.0104 |
MW= molecular weight (kDa), TP = Total peptides identified in control (CTL) and AD; % Cov. = % protein sequence coverage
3.4 Validation of specific targets
Immunoblotting was performed to verify the results from the label-free quantitative analysis of spectral counts. Notably, validation was carried out on individual cases rather than on the pools used for proteomics. Independently validating proteomics data is important, particularly when cases are pooled. While pooling diminishes inter-individual variability, it also opens up the possibility that one individual case could be driving the signal measured by proteomic analysis.
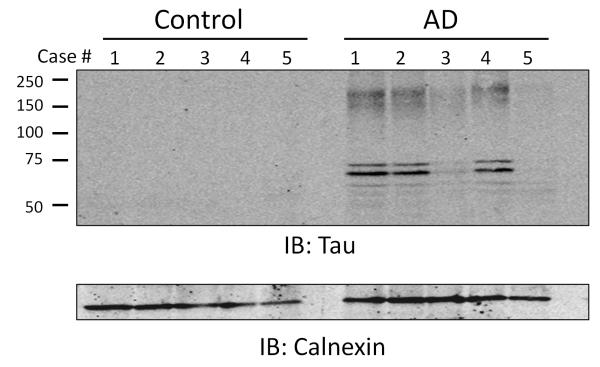
Intraneuronal inclusions of hyperphosphorylated tau are one of the pathological hallmarks of AD [19]. In this study, tau was the protein most significantly altered between AD and control cases (p = 5.88E-15). This was confirmed by immunoblotting the membrane fraction of individual cases for tau. While there was some variability in tau levels amongst AD samples with cases 1, 2, and 4 having higher levels than cases 3 and 5, none of the controls had detectable membrane associated tau (Fig. 3). Furthermore, tau measurements by immunoblotting immunoreactivity in AD cases was not correlated with Braak stage (stages listed by case in Table 1).
Figure 3. Tau enrichment in the membrane fraction of AD cases.
An immunoblot (IB) for tau in membrane fraction of individual control and AD cases is shown. Calnexin was used as a loading control. Markers on blots represent molecular weight in kDa.
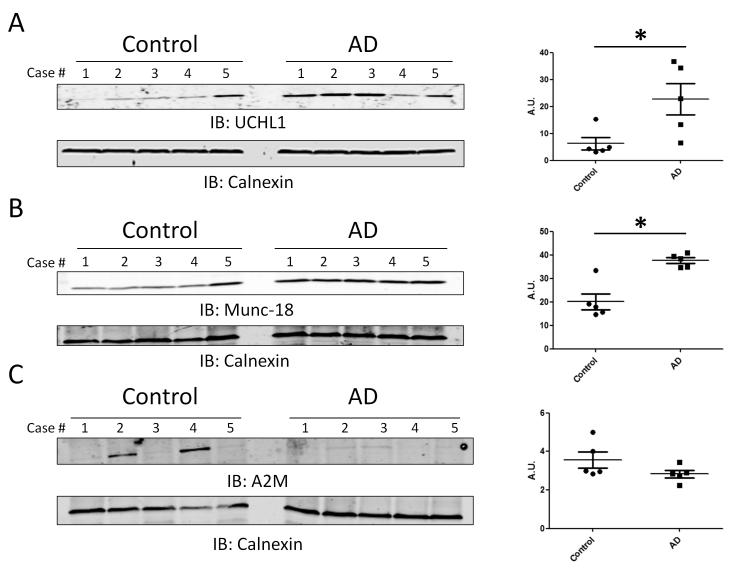
UCHL1 levels were also significantly elevated in AD by spectral counts. Blotting for UCHL1 confirmed the proteomics results with 4 of 5 AD cases showing elevated protein levels compared to only 1 control case (Student’s t-test; p = 0.0299) (Fig. 4A). While cognitively normal, this particular case (control 5) was found to have hemorrhagic infarcts and diffuse plaques on pathological examination. Increased UCHL1 is seen in other neurological conditions [39], which could account for the elevated UCHL1 levels in this case as compared to other controls. Munc-18, also enriched in AD, is a protein involved in vesicular trafficking and docking [40, 41]. This result was confirmed in individual cases by immunoblotting membrane fractions for Munc-18 (p = 0.0012) (Fig. 4B). Similar to what was seen after blotting for UCHL1, control case 5 had levels of munc18 more similar to disease cases than to other controls. A2M levels were significantly depleted in AD by spectral counts (p = 0.0001). However, immunoblotting individual cases revealed that this finding was not consistent across cases (Fig. 4C). Only 2 of 5 control cases had elevated levels of A2M, and the difference between AD and control cases was not significant by immunoblot signal determined by densitometry (p = 0.15). While this finding validates the proteomics, it demonstrates that the significant difference seen by spectral counts in the pooled samples was primarily the result extensive elevated levels in only these two cases.
Figure 4. Confirmation of proteomic changes for UCHL1, Munc-18 and A2M.
(A and B) Immunoblot (IB) analyses of the membrane fraction show that UCHL1 (p = 0.0299) and Munc-18 (p = 0.0012) levels were significantly higher in AD cases. (C) Immunoblot analysis of the membrane fraction reveals A2M levels are elevated in 2 of 5 control cases and no AD cases. This difference is not statistically significant (p = 0.15). All immunoblots are representative of three independent experiments. Calnexin was used as a loading control. Human sample information listed in Table 1. Signal intensity was determined by densitometry and statistical analysis was performed using a two-tailed student’s t-test. An asterisk (*) represents significant difference (p < 0.05)
4. Discussion
To date, there have been few reports of discovery-mode LC-MS/MS analysis of the membrane subproteome in neurodegenerative disease. This study reports changes in the membrane enriched proteome in AD. After statistical analysis of the spectral count data obtained by LC-MS/MS, 13 proteins were found significantly altered in AD. Of these proteins, tau, UCHL1, Munc-18, and A2M were independently validated by immunoblot. Confirming targets that have previously been shown to have an association with AD demonstrates that the membrane enrichment method used in this study indeed does enable sensitive discovery of proteins involved in disease pathogenesis that may be relevant as biomarkers in this or other contexts.
While the membrane enrichment strategy utilized did not produce a pure membrane sample, it did successfully enrich for membrane proteins. The percentage of proteins containing a TMD in this study was somewhat lower than the result reported in a previously published study using a similar method for membrane enrichment from cells in culture. In that study, approximately 50% of the proteins identified by LC-MS/MS had at least one TMD [42]. The lower percentage observed in brain tissue in this study is likely due in part to the relatively high amount of extracellular proteins such as myelin, versican, tenascin as well as structural proteins, that are associated with tissue rather than cells in culture.
The goal of this study was to examine a comprehensive membrane fraction of human brain tissue. For that reason, a crude membrane enrichment strategy was employed to fractionate samples to ensure that as many membranes as possible (i.e., mitochondrial membranes, plasma membranes, and vesicular membranes) were included in the final membrane enriched fraction. By doing this, it is possible that some of the purity of the sample was sacrificed. Non-transmembrane domain containing proteins, such as tubulin and actin, were still highly abundant in the membrane fraction and there are known points of contact between cytoskeleton and membranes. Nonetheless, a more stringent fractionation procedure would likely reduce the number of cytoskeletal and other non-membrane bound proteins in the membrane sample.
Tau is a microtubule associated protein important for stabilizing the neuronal cytoskeleton. In addition to binding microtubules, it also interacts with the plasma membrane [43]. This interaction appears to be mediated by phosphorylation; it was recently demonstrated that hyperphosphorylated tau loses its ability to bind the plasma membrane [44]. This may account for the intracellular accumulation of hyperphospohrylated tau observed in AD. Moreover, mislocalized tau has been shown to mediate Aβ toxicity through increased excitotoxicty [45]. Despite these findings, this study found that tau was enriched in the membrane fraction of AD cases. However, this study focused on total tau rather than hyperphosphorylated tau which may account for the difference. All of the AD cases in this study were Braak stage V or VI and therefore had high levels of pathological tau. It is possible that neurofibrillary tangles were co-purified with the membrane fraction of our sample, accounting for the increased signal seen with immunoblotting. While tau binds the plasma membrane, it does not contain a transmembrane domain. Therefore it is possible that any membrane-associated tau in the control cases was removed in the high pH wash step. There may be other modifications to tau not contained in neurofibrillary tangles in Alzheimer’s disease that make it more tightly associated with the membrane.
UCHL1 is a deubiquinating enzyme that is found primarily in neurons and neuroendocrine cells. It composes 1-2% of the soluble protein in neurons [46]. While it plays an important role in ubiquitin recycling, it has also been shown to have ubiquitin ligase activity [47]. Recently, it was reported that UCHL1 is an important regulator of survival of motor neuron (SMN) expression via ubiquitination in fibroblasts from patients with spinal muscular atrophy [48]. Increased association of UCHL1 with the membrane fraction due to farnesylation has also been reported in Parkinson’s disease and was shown to mediate alpha-synuclein toxicity in cells [49]. While certain polymorphisms in UCHL1 are reported to be protective in Parkinson’s disease, this protective effect is not seen in AD [50]. Given that UCHL1 appears to play a role in many neurodegenerative diseases, it is likely not specific to AD; however, that does not negate the fact that it may also play an important role in AD pathogenesis.
Munc-18 is involved in vesicle docking and exocytosis. It is found exclusively in presynaptic nerve terminals and binds to syntaxin-1, preventing it from interacting with docking fusion proteins [51]. Phosphorylation of Munc-18 by cyclin dependent kinase (Cdk) 5 causes Munc-18 and syntaxin-1 to dissociate and allows vesicular docking and neurotransmitter release to proceed. Phosphorylation of Munc-18 also enables Munc interacting (Mint) proteins 1 and 2 to bind Munc-18 [51]. These complexes have been postulated to increase neurotransmission and may play a role in APP processing and Aβ production. Up-regulation of this pathway has been reported in the cortex of patients with Alzheimer’s disease [37].
A2M has been implicated in AD pathogenesis due to its role in mediating Aβ clearance [52]. Several studies have also linked polymorphisms in the A2M gene to an increased risk of developing late-onset AD [38, 53, 54]. Lower levels of A2M could lead to in an inability to clear Aβ efficiently, thereby contributing to Aβ deposition in plaques. A2M was significantly depleted in AD by spectral counts (p = 0.0001); however, after immunoblotting individual cases, it became apparent that this finding was being driven by two control cases with high levels of A2M. This result illustrates a pitfall of pooling samples for proteomic experiments. Although pooling decreases inter-individual variability, it becomes difficult to ascertain if differences between pooled samples are the result of a consistent change across cases or whether they are driven by one or two outliers in the pool. This underscores the importance of independently validating findings from these studies across individual cases and that there is a large amount of inherent variability when working with human samples.
While not independently validated by immunoblotting, levels of creatine kinase B (CKB) were also significantly higher in AD cases compared to controls by proteomics analysis. Creatine kinase is a protein found in the intermembrane-space in mitochondria. It utilizes ATP to generate phosphocreatine which can then be used as a source of energy in the cell [55]. Lynn et al. recently published a study comparing the mitochondrial proteome in AD and control subjects [56]. Along with other proteins involved in ATP utilization pathways, creatine kinase was found to be enriched in AD. The results from this study support the findings by Lynn et al. and may provide further evidence that ATP utilization is abnormal in patients with AD.
One limitation to this study is that despite improved detection sensitivity, analyzing complex mixtures following LC-MS/MS by spectral counting restricts peptide identification to only the most abundant peptides. The more complex a mixture, the less likely it is that low abundance peptides will be sequenced [57] at levels high enough to reach significance. However, it is possible that changes in these proteins could have the largest biological impact. Spectral counting is not sensitive enough to detect these changes without using other methods, such as analysis of extracted peptide ion intensity. Spectral counting and G-test analysis do not take peptide signal intensity or protein coverage into consideration when determining which proteins are significantly changing. While this study successfully identified proteins with altered levels in AD, future studies using either a labeled quantitative approach or augmenting spectral count analysis with other label-free quantification methods could be used to identify less abundant proteins changing in disease. Finally, it can be difficult when analyzing post-mortem brain tissue from patients with end stage neurodegenerative diseases to dissect which changes are due to the disease process and which are a non-specific effect of neurodegeneration. In future studies, it would be important to compare these results to those from a disease control to determine whether these findings are specific to AD or represent proteins that are ubiquitously altered in neurodegeneration. Furthermore, the AD cases used in this study were all diagnosed with AD before age 65. While none of the cases had a known familial mutation causing their disease (i.e. APP (amyloid precursor protein) or presenilin 1 or 2 mutations), it is possible that these cases differ from the more common late-onset AD (diagnosed after age 65). Therefore, it would be important to verify these findings in more representative AD samples over a broader age range.
In conclusion, studying a membrane enriched fraction has several advantages for identifying proteins altered in AD. While the accumulation of Aβ certainly plays a large role in AD pathogenesis, there are other mechanisms, including abnormal vesicular trafficking and synaptic dysfunction that may also contribute to neurodegeneration [10, 11]. The small sample size in this study limits the generalization of these results and requires that the findings be confirmed with much larger samples. However, it provides support for using a membrane enriched proteome to identify biomarkers in AD. Future analysis of the membrane proteome from post-mortem brain tissue in any number of neurodegenerative diseases may offer mechanistic insights or potential new biomarkers that will aid in the diagnosis of these debilitating diseases.
Supplementary Material
Clinical Significance.
Dementia is a broad term for a variety of disorders that lead to the loss of cognitive abilities across multiple domains and impair a person’s ability to function independently. The burden of disease is dramatic: Currently over 35 million people are estimated to suffer from dementia worldwide [1]. Alzheimer’s disease (AD) is the most common cause of dementia [2] and the number of people in the United States over the age of 65 with AD is expected to triple by 2050 [3]. While acetylcholinesterase inhibitors and other drugs are used for symptomatic purposes, their effect is modest and temporary; AD is ultimately fatal. Moreover, it is becoming increasingly evident that the pharmacotherapy currently available may be most useful when started early in the disease course [4]. This underscores the need to better understand early changes that occur in AD with the hope that this will ultimately facilitate the discovery of biomarkers that can be used for diagnosis in a presymptomatic stage.
Acknowledgements
We would like to thank members of the Lah and Levey lab for constructive discussion and feedback regarding this manuscript. We also thank Dr. Jeremy H. Herskowitz for critical reading of the manuscript. Funding was provided through the National Institutes of Health through the Emory NINDS T32 Training in translational neurology grant (T32NS007480), Emory NINDS Neuroscience Core Facilities (NS055077), Emory Alzheimer’s Disease Research Center (AG025688), and National Institute of Aging (P01AG1449).
Footnotes
The authors have no conflicts of interest to report.
References
- [1].International, A. s. D. World Alzheimer Report 2009. 2009;92 [Google Scholar]
- [2].Goedert M, Spillantini MG. A century of Alzheimer’s disease. Science. 2006;314:777–781. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- [3].Association, A. s. Alzheimer’s Disease Facts and Figures. Alzheimer’s & Dementia. 2011;Volume 7 doi: 10.1016/j.jalz.2011.02.004. 2011. [DOI] [PubMed] [Google Scholar]
- [4].Daviglus ML, Bell CC, Berrettini W, Bowen PE, et al. National Institutes of Health State-of-the-Science Conference Statement: Preventing Alzheimer Disease and Cognitive Decline. Annals of Internal Medicine. 2010;153:176–181. doi: 10.7326/0003-4819-153-3-201008030-00260. [DOI] [PubMed] [Google Scholar]
- [5].Blennow K, de Leon MJ, Zetterberg H. Alzheimer’s disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- [6].Masters CL, Simms G, Weinman NA, Multhaup G, et al. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci U S A. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lee V, Balin B, Otvos L, Trojanowski J. A68: a major subunit of paired helical filaments and derivatized forms of normal Tau. Science. 1991;251:675–678. doi: 10.1126/science.1899488. [DOI] [PubMed] [Google Scholar]
- [8].Glenner GG, Wong CW. Alzheimer’s disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochemical and Biophysical Research Communications. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- [9].Hardy J. a. H., GA Alzheimer’s disase: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- [10].Pimplikar SW, Nixon RA, Robakis NK, Shen J, Tsai L-H. Amyloid-Independent Mechanisms in Alzheimer’s Disease Pathogenesis. The Journal of Neuroscience. 2010;30:14946–14954. doi: 10.1523/JNEUROSCI.4305-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Herskowitz JH, Seyfried NT, Gearing M, Kahn RA, et al. Rho Kinase II Phosphorylation of the Lipoprotein Receptor LR11/SORLA Alters Amyloid-β Production. Journal of Biological Chemistry. 2011;286:6117–6127. doi: 10.1074/jbc.M110.167239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- [13].Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Analytical and Bioanalytical Chemistry. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- [14].Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- [15].Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat Methods. 7:383–385. doi: 10.1038/nmeth.1446. [DOI] [PubMed] [Google Scholar]
- [16].Ishihama Y, Sato T, Tabata T, Miyamoto N, et al. Quantitative mouse brain proteomics using culture-derived isotope tags as internal standards. Nat Biotech. 2005;23:617–621. doi: 10.1038/nbt1086. [DOI] [PubMed] [Google Scholar]
- [17].Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, et al. Comparison of Label-free Methods for Quantifying Human Proteins by Shotgun Proteomics. Molecular & Cellular Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- [18].Matt P, Fu Z, Fu Q, Van Eyk JE. Biomarker discovery: proteome fractionation and separation in biological samples. Physiological Genomics. 2008;33:12–17. doi: 10.1152/physiolgenomics.00282.2007. [DOI] [PubMed] [Google Scholar]
- [19].Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- [20].Mirra SS, Heyman A, McKeel D, Sumi SM, et al. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- [21].Seyfried NT, Huysentruyt LC, Atwood JA, 3rd, Xia Q, et al. Up-regulation of NG2 proteoglycan and interferon-induced transmembrane proteins 1 and 3 in mouse astrocytoma: a membrane proteomics approach. Cancer Lett. 2008;263:243–252. doi: 10.1016/j.canlet.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Xu P, Duong DM, Peng J. Systematical optimization of reverse-phase chromatography for shotgun proteomics. J Proteome Res. 2009;8:3944–3950. doi: 10.1021/pr900251d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gozal YM, Duong DM, Gearing M, Cheng D, et al. Proteomics Analysis Reveals Novel Components in the Detergent-Insoluble Subproteome in Alzheimer’s Disease. Journal of Proteome Research. 2009;8:5069–5079. doi: 10.1021/pr900474t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- [25].Seyfried NT, Xu P, Duong DM, Cheng D, et al. Systematic approach for validating the ubiquitinated proteome. Anal Chem. 2008;80:4161–4169. doi: 10.1021/ac702516a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Herskowitz JH, Seyfried NT, Duong DM, Xia Q, et al. Phosphoproteomic Analysis Reveals Site-Specific Changes in GFAP and NDRG2 Phosphorylation in Frontotemporal Lobar Degeneration. Journal of Proteome Research. 2010;9:6368–6379. doi: 10.1021/pr100666c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kislinger T, Cox B, Kannan A, Chung C, et al. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- [28].Xia Q, Cheng D, Duong DM, Gearing M, et al. Phosphoproteomic analysis of human brain by calcium phosphate precipitation and mass spectrometry. J Proteome Res. 2008;7:2845–2851. doi: 10.1021/pr8000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xia Q, Liao L, Cheng D, Duong DM, et al. Proteomic identification of novel proteins associated with Lewy bodies. Front Biosci. 2008;13:3850–3856. doi: 10.2741/2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Whittiker VP. Handbook of Neurochemistry. 1969:327–364. [Google Scholar]
- [31].Kim SI, Voshol H, van Oostrum J, Hastings TG, et al. Neuroproteomics: expression profiling of the brain’s proteomes in health and disease. Neurochem Res. 2004;29:1317–1331. doi: 10.1023/b:nere.0000023618.35579.7c. [DOI] [PubMed] [Google Scholar]
- [32].Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- [33].Xia Q, Hendrickson EL, Zhang Y, Wang T, et al. Quantitative Proteomics of the Archaeon Methanococcus maripaludis Validated by Microarray Analysis and Real Time PCR. Molecular & Cellular Proteomics. 2006;5:868–881. doi: 10.1074/mcp.M500369-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Xia Q, Wang T, Park Y, Lamont RJ, Hackett M. Differential quantitative proteomics of Porphyromonas gingivalis by linear ion trap mass spectrometry: Non-label methods comparison, q-values and LOWESS curve fitting. International Journal of Mass Spectrometry. 2007;259:105–116. doi: 10.1016/j.ijms.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xia Q, Wang T, Taub F, Park Y, et al. Quantitative proteomics of intracellular Porphyromonas gingivalis. Proteomics. 2007;7:4323–4337. doi: 10.1002/pmic.200700543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Choi J, Levey AI, Weintraub ST, Rees HD, et al. Oxidative Modifications and Down-regulation of Ubiquitin Carboxyl-terminal Hydrolase L1 Associated with Idiopathic Parkinson’s and Alzheimer’s Diseases. Journal of Biological Chemistry. 2004;279:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- [37].Jacobs EH, Williams RJ, Francis PT. Cyclin-dependent kinase 5, Munc18a and Munc18-interacting protein 1/X11α protein up-regulation in Alzheimer’s disease. Neuroscience. 2006;138:511–522. doi: 10.1016/j.neuroscience.2005.11.017. [DOI] [PubMed] [Google Scholar]
- [38].Blacker D, Wilcox MA, Laird NM, Rodes L, et al. Alpha-2 macroglobulin is genetically associated with Alzheimer disease. Nat Genet. 1998;19:357–360. doi: 10.1038/1243. [DOI] [PubMed] [Google Scholar]
- [39].Lewis SB, Wolper R, Chi Y-Y, Miralia L, et al. Identification and preliminary characterization of ubiquitin C terminal hydrolase 1 (UCHL1) as a biomarker of neuronal loss in aneurysmal subarachnoid hemorrhage. Journal of neuroscience research. 2010;88:1475–1484. doi: 10.1002/jnr.22323. [DOI] [PubMed] [Google Scholar]
- [40].Tadokoro S, Kurimoto T, Nakanishi M, Hirashima N. Munc18-2 regulates exocytotic membrane fusion positively interacting with syntaxin-3 in RBL-2H3 cells. Molecular Immunology. 2007;44:3427–3433. doi: 10.1016/j.molimm.2007.02.013. [DOI] [PubMed] [Google Scholar]
- [41].Smyth AM, Rickman C, Duncan RR. Vesicle Fusion Probability Is Determined by the Specific Interactions of Munc18. Journal of Biological Chemistry. 2010;285:38141–38148. doi: 10.1074/jbc.M110.164038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Baker DL, Seyfried NT, Li H, Orlando R, et al. Determination of protein-RNA interaction sites in the Cbf5-H/ACA guide RNA complex by mass spectrometric protein footprinting. Biochemistry. 2008;47:1500–1510. doi: 10.1021/bi701606m. [DOI] [PubMed] [Google Scholar]
- [43].Brandt R, Léger J, Lee G. Interaction of tau with the neural plasma membrane mediated by tau’s amino-terminal projection domain. J Cell Biol. 1995;131:1327–1340. doi: 10.1083/jcb.131.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Maas T, Eidenmüller J, Brandt R. Interaction of Tau with the Neural Membrane Cortex Is Regulated by Phosphorylation at Sites That Are Modified in Paired Helical Filaments. Journal of Biological Chemistry. 2000;275:15733–15740. doi: 10.1074/jbc.M000389200. [DOI] [PubMed] [Google Scholar]
- [45].Ittner LM, Ke YD, Delerue F, Bi M, et al. Dendritic Function of Tau Mediates Amyloid-β Toxicity in Alzheimer’s Disease Mouse Models. Cell. 2010;142:387–397. doi: 10.1016/j.cell.2010.06.036. [DOI] [PubMed] [Google Scholar]
- [46].Day INM, Thompson RJ. UCHL1 (PGP 9.5): Neuronal biomarker and ubiquitin system protein. Progress in Neurobiology. 2010;90:327–362. doi: 10.1016/j.pneurobio.2009.10.020. [DOI] [PubMed] [Google Scholar]
- [47].Healy DG, Abou-Sleiman PM, Wood NW. Genetic causes of Parkinson’s disease: <i>UCHL-1</i>. Cell and Tissue Research. 2004;318:189–194. doi: 10.1007/s00441-004-0917-3. [DOI] [PubMed] [Google Scholar]
- [48].Hsu S-H, Lai M-C, Er T-K, Yang S-N, et al. Ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) regulates the level of SMN expression through ubiquitination in primary spinal muscular atrophy fibroblasts. Clinica Chimica Acta. 2010;411:1920–1928. doi: 10.1016/j.cca.2010.07.035. [DOI] [PubMed] [Google Scholar]
- [49].Liu Z, Meray RK, Grammatopoulos TN, Fredenburg RA, et al. Membrane-associated farnesylated UCH-L1 promotes α-synuclein neurotoxicity and is a therapeutic target for Parkinson’s disease. Proceedings of the National Academy of Sciences. 2009;106:4635–4640. doi: 10.1073/pnas.0806474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zetterberg M, Sjolander A, von Otter M, Palmer M, et al. Ubiquitin carboxy-terminal hydrolase L1 (UCHL1) S18Y polymorphism in Alzheimer’s disease. Molecular neurodegeneration. 2010;5:11. doi: 10.1186/1750-1326-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ho A, Morishita W, Hammer RE, Malenka RC, Südhof TC. A role for Mints in transmitter release: Mint 1 knockout mice exhibit impaired GABAergic synaptic transmission. Proceedings of the National Academy of Sciences. 2003;100:1409–1414. doi: 10.1073/pnas.252774899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Narita M, Holtzman DM, Schwartz AL, Bu G. α2-Macroglobulin Complexes with and Mediates the Endocytosis of β-Amyloid Peptide via Cell Surface Low-Density Lipoprotein Receptor-Related Protein. Journal of Neurochemistry. 1997;69:1904–1911. doi: 10.1046/j.1471-4159.1997.69051904.x. [DOI] [PubMed] [Google Scholar]
- [53].Liao A, Nitsch RM, Greenberg SM, Finckh U, et al. Genetic Association of an (α2-Macroglobulin (Val1000lle) Polymorphism and Alzheimer’s Disease. Human molecular genetics. 1998;7:1953–1956. doi: 10.1093/hmg/7.12.1953. [DOI] [PubMed] [Google Scholar]
- [54].Saunders AJ, Bertram L, Mullin K, Sampson AJ, et al. Genetic association of Alzheimer’s disease with multiple polymorphisms in alpha-2-macroglobulin. Human molecular genetics. 2003;12:2765–2776. doi: 10.1093/hmg/ddg310. [DOI] [PubMed] [Google Scholar]
- [55].Schlattner U, Tokarska-Schlattner M, Wallimann T. Mitochondrial creatine kinase in human health and disease. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2006;1762:164–180. doi: 10.1016/j.bbadis.2005.09.004. [DOI] [PubMed] [Google Scholar]
- [56].Lynn BC, Wang J, Markesbery WR, Lovell MA. Quantitative Changes in the Mitochondrial Proteome from Subjects with Mild Cognitive Impairment, Early Stage, and Late Stage Alzheimer’s Disease. Journal of Alzheimer’s Disease. 2010;19:325–339. doi: 10.3233/JAD-2010-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Peng J, Gygi SP. Proteomics: the move to mixtures. J. Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.