The 1918–1919 pandemic of H1N1 virus influenza was the greatest acute plague of the 20th century. Incurring over 20 million human fatalities, however, was not a good strategy for sustaining the evolutionary fitness of the virus, because it is no longer extant; whereas, say, measles and chickenpox remain with us with no evidence of remarkable genetic change, although this may become more evident if they were to face total or near eradication through vaccination programs. The folly of flu virulence remains our chagrin, because the threat always looms over us that this family of viruses, endemic in birds, again may generate human-lethal gene reassortments. We had valid scares about that contingency with the appearance of H5N1 variant flu in Hong Kong just 3 years ago. Influenza can be regarded as a zoonosis prevalent in birds, many of them world travelers, with occasional outbreaks in humans and other animals mainly rooted in nature's own experiments in genetic engineering. Special importance is attached to reassortments between bird- and human-adapted strains most likely to occur in habitats with close contact between birds, e.g., ducks, humans, and swine (as a mixing reservoir; ref. 1). For these reasons, high urgency attaches to efforts to resurrect genetic information about the singularities of H1N1–1918. The intact virus is nowhere to be found, but genomic fragments can still be detected sensitively and diagnosed. Exemplifying the latest technical advances in the use of DNA amplification, reverse-transcriptase–PCR (RT-PCR), Jeffery Taubenberger and his associates at the Armed Forces Institute of Pathology initiated the tour de force of recovering sequences of flu from paraffin-embedded pathological specimens preserved since 1918 in the AFIP collections (2). These sources then were augmented by samples from frozen remains of an Inuit woman who succumbed to the flu in 1918 and was buried in permafrost at Brevig Mission on the Seward Peninsula of Alaska's western coast, not far from the Bering Strait. This nameless woman has left an indelible mark on world medical history (3). Now, as reported in this issue, the AFIP team has joined forces with teams from the U.S. Department of Agriculture and the Peter Palese/Adolfo García-Sastre groups at Mt. Sinai Medical School in a further quest for the RNA sequences of H1N1–1918 that might account for its historic human virulence (4).
The flu genome comprises about 13,500 bases of single-stranded RNA, disposed in eight segments varying from approximately 900 to 2,341 each. This genome is only a few millionths of the complexity of the human genome, but it is organized with great efficiency, lacks “junk R/DNA,” and encodes for a short dozen of identified gene products (Fig. 1). Many strains of flu have been sequenced fully; this feat will be achieved for H1N1–1918 with arduous labor, because the RNA, although frozen, is fragmented into snippets no larger than approximately 120 bases each. The practical way now available is to devise probes by using segments from extant flu strains, guessing at possible homologous strings, or synthesizing probes with calculated degeneracy. Until a complete genomic sequence is achieved, and it is hard to see how that will be authenticated, it is possible even that H1N1–1918 contains extraneous inserted sequences quite foreign to the canonical flu strains. Very reasonably, initial efforts focus on flu genes already identified in viruses recovered from recent outbreaks in humans, birds, swine, and other animals.
Figure 1.
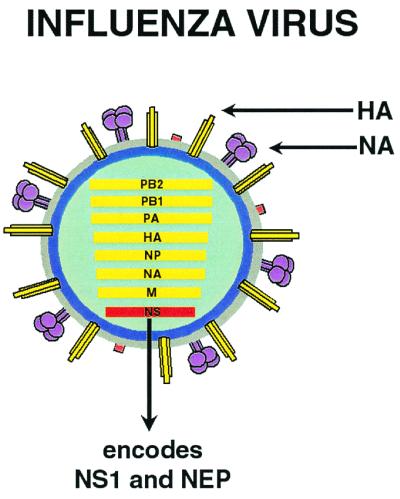
Diagram of an influenza-virus particle. The surface of each influenza virion consists of a lipid envelope in which two major viral surface antigens, the hemagglutinin (HA) and the neuraminidase (NA), are found. Within the particle are the eight negative-sense viral RNA segments encoding the viral proteins. The smallest viral segment, the NS segment, encodes two proteins: the NS1, an antagonist of the cellular type I interferon system, and the nuclear export protein (NEP), which functions in viral assembly.
Previous work has focused on two well studied gene products: hemagglutinin (HA) and neuraminidase (NA), which dominate the surface specificities of the virus and underlie most of its taxonomy (e.g., H1N1 refers to type 1 hemagglutinin, type 1 neuraminidase). These gene products are also the chief determinants of specificity in vaccine prophylaxis for flu strains circulating at any given time. HA variation can account for fluctuations of virulence and host specificity of extant flu viruses. However, nothing remarkable was seen in the HA or the NA of H1N1–1918. The next gene to be scrutinized now is NS1 (nonstructural protein 1), which the Palese/García-Sastre groups have fingered recently as an interferon antagonist and as gene essential for flu virulence in a mouse model. A reasonable conjecture was that the hypervirulence of H1N1–1918 might be lodged in its NS1, and this might be revealed in reinsertions of the 1918-NS1 segment into mouse-adapted flu strains. This challenging construct was generated in the laboratory—one hastens to footnote, under BL-3+ conditions, and under the USDA's stern regulatory scrutiny—and tested in mice. The unexpected and perhaps disappointing result was the mitigation not enhancement of virulence in this species. The incapacitation of the NS1-virulence function in the mouse was ascribed to interaction with its host factors; the other variable would be other elements of the genome of the mouse-adapted flu strain. NS1 singularity for the human virulence of H1N1–1918 is neither falsified nor corroborated by these findings.
There still remain a handful of gene candidates, including the polymerases essential for the replication of the virus. This label does not preclude any of them from also functioning in networks and pathways that are expressed as virulence. It should caution us about the nominalist fallacy to recall that the δ crystallin of the bird's lens does double duty as argininosuccinate lyase, an enzyme in the urea cycle.
In principle, the NS1 hypothesis (and its alternatives) might be tested by using similar gene constructs based on flu viruses adapted to other animal species, including primates, and challenging the corresponding hosts. Negative results would be as inconclusive as those with the mouse. Positive results, namely the association of hypervirulence with a gene sequence borrowed from N1H1–1918, would be a great advance in medical science and would offer constructive models for the development of prophylactic and therapeutic measures. They would also induce great alarm about the potential hazards to human health, if humans were also susceptible, and the virus might escape. Any such experiments should be done with strains for which current vaccines are disseminated widely and have proven effectiveness.
To conduct such experiments with human-adapted strains and challenge to human subjects as the probative step, is well nigh unthinkable. But nature is under no such restraint! The current results are a caution to look closely at the involvement of NS1 (as well as HA and NA) variation in natural outbreaks in many species and to look out for their reassortment into human strains. In addition, it might be well to undertake a special search for close homologues to 1918-NS1 in viruses circulating in avian and other species, in which they may appear to be benign in their current hosts (as in the present mouse experiments). That would be nature's inverse of the current report.
The publication by Basler et al. (4) will attract great admiration for its technical finesse and will serve as an example of the fruits from convergence of natural history, field exploration, clinical insight, and sophisticated molecular wizardry. It also will awaken anxieties about the obvious opportunities for abuse. The really fateful step was taken with the very first cultivation of pathogenic bacteria and viruses a century ago—perhaps most importantly with the discovery of the concepts of germs and communicable diseases. The notion of using ever more sophisticated technology for intentionally constructing or reconstructing ever more pathogenic variants lends further weight to that anxiety. The great debate of the mid-1970s led to sensible measures for the regulation of recombinant DNA research. There has been increasing understanding that some of nature's pathogens deserve equal or greater respect. We should be sure that we continue to devote as much reasoned ingenuity to the design of safeguards and to informed and transparent third-party scrutiny of potential hazards as we do generally to the authentication of scientific claims. We cannot afford to forego the deepest research into the plagues that beset humankind. Nor can we afford to blunder into mistakes that will do primary injury to bystanders and incur incommensurate social sanctions.
My deepest anxieties pertain to the smoldering technology and arms race that attends the power struggles in the Middle East and the economic instabilities of the former Soviet Union. Although the 1975 Biological Weapons Convention (BWC) has demilitarized the main drivers of bioweaponry technical advance, in the U.S. and in the overt activities of other formidable powers, the BWC has not been enforced successfully against Iraq and is more or less openly flouted in a handful of other countries. The United Nations (UN) Security Council is too splintered on other issues to take a firm stand on the defiance by Iraq of the UN-mandated inspections. It would not be child's play for defiant small countries to adopt advanced biotechnology into their weapons programs. But we have seen that the climactic high-science successes in one decade become fodder for high-school projects in the next. Influenza is an unlikely candidate for rational weapons development, because new strains promptly embrace the world. But that logic is insufficient reason to neglect the contingency. More likely similar principles would be applied to more governable bioagents, but any bioagents in warfare are an affront and a threat to the entire human species. Informed professionals throughout the world should be leading campaigns to insist on universal compliance with the BWC as a major bulwark of human health and associating that with the most positive measures to apply advanced biotechnology in a constructive way for dealing with nature's continued scourges.
Footnotes
See companion article on page 2746.
References
- 1.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taubenberger J K, Reid A H, Krafft A E, Bijwaard K E, Fanning T G. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 3.Reid A H, Fanning T G, Hultin J V, Taubenberger J K. Proc Natl Acad Sci USA. 1999;96:1651–1656. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler C F, Reid A H, Dybing J K, Janczewski T A, Fanning T G, Zheng H, Salvatore M, Perdue M L, Swayne D E, García-Sastre A, et al. Proc Natl Acad Sci USA. 2001;98:2746–2751. doi: 10.1073/pnas.031575198. [DOI] [PMC free article] [PubMed] [Google Scholar]


