Highlights
► Mechanism of protein transfer by plasmid R1 conjugative T4 system requires relaxosome. ► Protein translocation depends on concomitant DNA transfer. ► Required functions of T4 coupling protein correlate for protein and DNA transfer.
Keywords: Horizontal gene transfer, Bacterial conjugation, Bacterial type IV secretion, Coupling protein, Plasmid R1, Relaxosome
Abstract
Bacterial conjugation disseminates genes among bacteria via a process requiring direct cell contact. The cell envelope spanning secretion apparatus involved belongs to the type IV family of bacterial secretion systems, which transport protein as well as nucleoprotein substrates. This study aims to understand mechanisms leading to the initiation of type IV secretion using conjugative plasmid paradigm R1. We analyze the general requirements for plasmid encoded conjugation proteins and DNA sequence within the origin of transfer (oriT) for protein secretion activity using a Cre recombinase reporter system. We find that similar to conjugative plasmid DNA strand transfer, activation of the R1 system for protein secretion depends on binding interactions between the multimeric, ATP-binding coupling protein and the R1 relaxosome including an intact oriT. Evidence for DNA independent protein secretion was not found.
1. Introduction
Processes of horizontal gene transfer (HGT) including transformation, transposition, transduction and conjugation contribute to genetic diversity and evolution in bacteria (Frost et al., 2005). Conjugation systems of Gram-negative and Gram-positive bacteria are the largest and most widely distributed subtype of bacterial type IV secretion systems (T4SSs) (Smillie et al., 2010). The antibiotic resistance factor R1 belongs to the F-like family of conjugative plasmids. As we see in this special issue of Plasmid, decades of research based on the R1 paradigm have contributed substantially to understanding several key aspects of the biology of plasmids and their bacterial hosts. Together with the closely related plasmids F (IncF) and R100 (IncFII) plasmid R1 has been extensively studied as a model of bacterial conjugation and its regulation.
Conjugation systems, like most T4SSs, comprise three functional substructures: cell surface pili that mediate contact between cells, a transport channel that conducts substrates across the bacterial cell envelope, and a type IV coupling protein (T4CP) that acts as substrate receptor at the cytoplasmic entrance of the secretion channel. T4CPs mediate multiple protein–protein interactions with cytoplasmic and inner membrane components of the secretion system. ATPase activity is associated with the release and unfolding of complexes between substrates and specific binding partners and is required to energize the secretion process (Alvarez-Martinez and Christie, 2009; Zechner et al., in press).
Studies of the Gram-negative conjugation paradigms including R1 have established the general mechanisms of plasmid transfer (de la Cruz et al., 2010). First, multiple proteins assemble on the plasmid origin of transfer (oriT) to form the relaxosome. This complex prepares the single-strand of plasmid DNA destined for transfer (T-strand) via the nicking activity of a relaxase enzyme. Initiation of transfer requires phosphodiester bond cleavage at nic, within oriT. The reaction is mediated by a tyrosine residue of the relaxase, so that a covalent protein-DNA adduct is formed. The T4CP specifically recognizes this nucleoprotein conjugate. In response to contact dependent initiation signals relaxase linked T-DNA is probably actively pumped through the transport apparatus. In the recipient following termination of transfer, the nicking reaction reverses, yielding the original circular plasmid molecule and freeing the relaxase. Finally stabilization of the original plasmid DNA strands by conjugative replication occurs in both donor and recipient cells.
Control of the initiation process is maintained by complex circuits that regulate transcription of conjugation genes, assembly of conjugative pili and the secretion channel connecting donor and recipient cells, and finally the enzymatic processing of plasmid DNA in preparation for secretion. Transfer is often stimulated by donor cell perception of signals in the environment (Dunny and Johnson, 2011; White and Winans, 2007; Wozniak and Waldor, 2010). Studies of the F-like transfer systems in Escherichia coli hosts have been instrumental for understanding how conjugative systems are controlled by environmental and physiological conditions as well as cellular stress (Frost and Koraimann, 2010). Conjugation systems are also activated in response to signals conveyed from recipient cells upon establishment of the donor – target cell contact (Lu and Frost, 2005). Defining the nature of these signals, their transmission to the donor cell cytoplasm, and their subsequent conversion into a secretion initiation mechanism has remained elusive in over 50 years of conjugation research.
In our work with plasmid R1 we recently postulated that bacteriophage might mimic potential recipient cells and initiate a signaling pathway that activates mechanisms typically involved in gene transfer. “Male specific” filamentous and RNA phages exploit the presence of F-like conjugative pili and the underlying envelope spanning transport machinery to gain entry to bacterial cells. The T4CP TraD of F-like plasmids is not involved in pilus biogenesis but is essential for host sensitivity to the group I RNA phages R17, f2 and MS2 (Schoulaker and Engelberg-Kulka, 1978; Valentine et al., 1969). Based on what we now know about the decisive role T4CPs play in connecting the secretion channel with the cytoplasm and in recruiting and initiating (nucleo)protein secretion, further investigation of the T4CP-dependent phage infection process seemed warranted. We analyzed the requirements for R1 conjugation proteins and found that host cells are vulnerable to infecting phage only through T4 machinery that is also competent for conjugative DNA transfer (Lang et al., 2011). Penetration of the host cell by the R17 ssRNA genome, which is covalently linked at the 3′ end to a phage protein (Krahn et al., 1972; Wong and Paranchych, 1976), required docking interactions between the plasmid R1 T4CP and catalytically active relaxase TraI bound at the plasmid origin of transfer oriT. The ATP binding activity of the T4CP was also necessary. The data support a model where the T4CP cumulatively senses an intracellular signal (substrate docking) and an extracellular signal (pilus bound by phage or a recipient cell) to coordinate a late stage assembly or gating reaction that enables bidirectional transmission of nucleoprotein substrates through the T4SS (Berry and Christie, 2011; Lang et al., 2011).
Type IV systems are remarkably versatile in that they mobilize a broad range of substrates including single proteins, protein complexes, DNA, and nucleoprotein complexes. In the few DNA transporting T4SSs where this has been analyzed, secretion of specific proteins also occurs independently of DNA transfer (Draper et al., 2005; Parker and Meyer, 2007; Vergunst et al., 2005). TraI protein can be transferred to recipient cells when it is not bound to DNA (Lang et al., 2010). However our observation that plasmid DNA and TraI interactions with its specific oriT binding sites were indispensable for R17 phage uptake led us to propose that activation of the T4 secretion channel of the R1 system requires relaxosome assembly and perception of processed ssDNA substrate regardless of the actual secretion substrate. If this is true we reasoned that a functional analysis of the requirements for protein secretion by the R1 system should reveal close correlation with those of conjugative DNA transfer. Consistent with this hypothesis, protein transfer by the R1 system was measured only under conditions supporting concomitant plasmid strand transfer.
2. Materials and methods
2.1. Strains and plasmids
All E. coli K12 strains and plasmids used in this study are described in Table 1.
Table 1.
E. coli strains used in this study.
| Description and referencea | |
|---|---|
| Strain | |
| MS411 | ilvG rfb-50 thi (M. Schembri; DTU, Denmark) |
| MS614Cm | CmR, SmR (Lang et al., 2010) |
| CSH26Cm::LTL | TcR, CSH26 galK::cat::loxP-Tet-loxP (Lang et al., 2010) |
| 61-1 | deoB-serBΔ (Roeder and Somerville, 1979) |
| Conjugative plasmids | |
| R1-16 | KmR; IncFII, fin- (Goebel et al., 1977) |
| R1-16ΔtraD | KmR, TcR; IncFII, traD::tetRA (Lang et al., 2010) |
| R1-16ΔtraY | KmR, TcR; IncFII, traY::loxPtetRAloxP (Lang et al., 2010) |
| R1-16Δnic | KmR, TcR; IncFII, nic::loxPtetRAloxP (Lang et al., 2010) |
| R1-16ΔoriT | KmR, TcR; IncFII, oriT::loxPtetRAloxP (Lang et al., 2010) |
| R1-16ΔtraM | KmR; IncFII; R1-16 carrying traM null allele; identical to R1-16M0 (Pölzleitner et al., 1997) |
| pOX38 | KmR; IncFI, derivative of F (Chandler and Galas, 1983) |
| pOX38ΔtraI | KmR, TcR; IncFI; traI::tetRA (Lang et al., 2010) |
| pOX38traD411 | KmR; IncFI, aph inserted in traD of pOX38 (Maneewannakul et al., 1996) |
| Expression vectors | |
| CFP B Sm | SmR; pBR322 expressing Cre recombinase from phage P1 (Lang et al., 2010) |
| CreTraI(3-1756) Sm | SmR; CFP B Sm with R1 traI encoding residue 3 to 1756 (Lang et al., 2010) |
| CreTraI F Sm | SmR; CFP B Sm with wild-type F traI (Lang et al., 2010) |
| pBT200 | AmpR; pTrc99A with wild-type F traD (Haft et al., 2007) |
| pBT200DiK6 | AmpR; pTrc99A encoding F TraD with 31 residue insertions after amino acid 6 (Haft et al., 2007) |
| pBT200DiK273 | AmpR; pTrc99A encoding F TraD with 31 residue insertions after amino acid 273 (Haft et al., 2007) |
| pBT200DiN702 | AmpR; pTrc99A encoding F TraD with 31 residue insertions after amino acid 702 (Haft et al., 2007) |
| pJMTraD | AmpR; pBAD24 with wild-type F traD (Lu et al., 2008) |
| pJMTraDD576∗ | AmpR; pBAD24 with partial F traD encoding residues 1-576 (Lu et al., 2008) |
| pJMTraDE709∗ | AmpR; pBAD24 with partial F traD encoding residues 1–709 (Lu et al., 2008) |
| pJMTraDF717A | AmpR; pBAD24 with F traD mutant F717A (Lu et al., 2008) |
| pMM-M0 | AmpR; pMMB67EH with site specific traM null mutant, oriT and finP (Kupelwieser et al., 1998) |
| pMM-wt | AmpR; pMMB67EH with traM, oriT and finP (Kupelwieser et al., 1998) |
| pMM-traM | AmpR; pMMB67EH with wild-type R1 traM, this study |
| pMSTraD_wt | AmpR; pMM119EH with wild-type R1 traD (Lang et al., 2010) |
| pMSTraDA | AmpR; pMSTraD_wt with point mutation in traD leading to a K198T exchange in Walker A box (Lang et al., 2010) |
| pMSTraDAB | AmpR; pMSTraD_wt with point mutations in traD leading to a K198T exchange in Walker A box and a D425N exchange in Walker B box; this study |
| pMSTraMF | AmpR; pMS119EH with wild-type F traM (Lang et al., 2010) |
| pMSTraMF K99E | AmpR; EcoRI-HindIII fragment from pRFMK99E (Lu et al., 2008)cloned into pMS119EH; this study |
| pMSYM1 | AmpR; pMS119EH with wild-type R1 traY (Lang et al., 2010) |
Antibiotic resistance: AmpR, ampicillin; CmR, chloramphenicol; KmR, kanamycin; SmR, streptomycin; TcR, tetracycline.
2.2. Enzymes, reagents and antibiotics
Plasmid DNA was purified from E. coli cells with the QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany). Restriction endonucleases, calf intestinal phosphatase, and T4 DNA ligase were purchased from Fermentas GmbH (St. Leon-Rot, Germany). DNA fragments for cloning were amplified using Phusion High-Fidelity DNA Polymerase (Finnzymes Oy, Espoo, Finland) or the Taq-Polymerase (New England Biolabs, Beverly, MA, USA). Enzymes were used according to manufacturers’ recommendations.
Antibiotics were added at the indicated concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 10 μg ml−1; kanamycin, 40 μg ml−1; streptomycin, 25 μg ml−1; tetracycline, 8 μg ml−1.
2.3. Construction of expression plasmids
The insert for pMM-traM was amplified with primers FW_TraM (5′-GTCCCGTCGACATGGCGAAAGTGCAGGCTTATGTCA-3′) and Rev_TraM (5′-GATCCCTGCAGTTATTCCT CATCATTTTCTGGAAAG-3′) from R1-16, cut with SalI/PstI and ligated with pMMB67EH. Two-step PCR was used to generate pMSTraDAB. In the first step primer sets 1 (TraDD425N_fw; 5′-TTTCTGTAATGAGTTACCCACG-3′ + SS01fw; 5′-GCCGAATTCA TGAGTTTTAACGCAAAG-3′) and 2 (TraDD425N_rev; 5′-CGTGGGTAACTCATTACAG AAA-3′ + SS02rev; 5′-CGTGAAGCTTTCAGAAATCATCTCCCG-3′) were used to amplify two fragments from pMSTraDA, such that each carried a desired point mutation in the nucleotide binding signature sequences. In the second step these two fragments were annealed and amplified with primer set 3 (SS01fw + SS02rev). The fragments were cut with EcoRI/HindIII and religated with pMS119EH.
2.4. CRAfT (Cre recombinase assay for translocation)
The Cre fusion reporter assay was performed as described previously (Lang et al., 2010). E. coli MS411 or 61-1 donor cells carrying the plasmids of interest and recipient CSH26Cm::LTL were used. Gene expression in donor cells containing plasmids derived from the pBAD vector was induced with 0.05% arabinose 1 h prior to mating. Donors were selected on plates containing appropriate antibiotics (see Table 1) and recombinants with chloramphenicol. Protein translocation frequencies are calculated as recombinants per donor. Conjugative transfer and mobilization of the R1 oriT-containing plasmids was measured in a parallel experiment using MS614Cm as recipient (Lang et al., 2010). Transconjugants were selected on chloramphenicol and kanamycin or chloramphenicol and ampicillin. The conjugation and mobilization frequencies were calculated as transconjugants per donor. The presence of all plasmids in the donor was confirmed by PCR before and after the CRAfT was performed.
3. Results
3.1. Protein secretion mediated by R1-16 proteins requires the wild type oriT in cis
The Cre recombinase assay for translocation (CRAfT) is an established method for monitoring the secretion of specific proteins to a recipient cell (Vergunst et al., 2000, 2005). In this assay (illustrated in Fig. 1), Cre lacks features that enable it to be recognized and transported by a T4SS. Fusion of a secretion protein to Cre, however, supports its transfer by the cognate T4SS. The indicator recipient strain harbors one antibiotic resistance gene interrupted by a second resistance cassette flanked by loxP sites. Recombination catalyzed by the acquired Cre fusion at loxP restores functional expression of the disrupted resistance cassette. Protein transfer to a recipient strain is thus measured by the heritable change in antibiotic resistance phenotype. We and others have applied this analysis to conjugative relaxases (Lang et al., 2010; Parker and Meyer, 2007). For this study, a fusion of the full-length traI of R1 to the 3′ end of the cre gene was used. A normal level of recombination and, indirectly, the stability of this fusion protein were confirmed through transformation of the indicator strain with the Cre fusion-plasmid and selection for recombinant cells (Lang et al., 2010). Under standard CRAfT assay conditions, donor cells harbor the fertility derepressed plasmid R1-16 to provide all of the essential components for substrate recognition, conjugative DNA processing, and transport (including wild type TraI protein). The translocation frequencies of the R1 fusion protein were measured by scoring recombinants per donor cells (Fig. 2). Protein translocation occurs with a frequency of ∼5 × 10−3 under these conditions. R1-16 plasmid transfer occurs simultaneously and provides an internal standard for the conjugation efficiency in every experiment.
Fig. 1.
Schematic illustration of the CRAfT assay and oriT deletion derivatives of R1-16 used in this study. (A) Reporter enzyme Cre recombinase (white circle) is fused to a known or putative T4 secretion protein (star). Donor cells assemble a T4SS encoded by a conjugative plasmid including a T4CP for substrate recognition (inset). T4 secretion mobilizes the conjugative plasmid and the Cre fusion to recipient cells. Protein transfer is detected by Cre catalyzed recombination in transconjugants. (B) To test the role of the relaxosome in protein translocation deletion mutations of R1-16 removed nic and the inverted repeat (R1-16Δnic) or a larger fragment including also a binding site for IHF and TraY (R1-16ΔoriT). Numbering according to (Graus-Goldner et al., 1990).
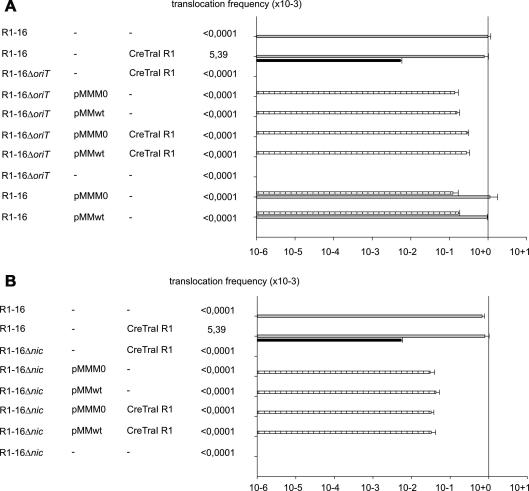
Fig. 2.
Relaxosome assembly on R1-16 is essential for Cre–TraI translocation. The protein translocation frequencies supported by R1-16 compared to R1-16ΔoriT (A), or R1-16Δnic (B) are shown (right) from donors carrying the coresident plasmids indicated (left). Values represent recombination events per donor. Black bar represents frequency of translocation (Cre–TraI R1) with statistical significance compared to the vector control (not shown). Conjugation frequencies are indicated with gray bars. Mobilization frequencies of the coresident R1 oriT plasmids (pMMwt, pMMM0) are shown with striped bars. Values represent the mean of at least three experiments. Standard deviations are shown.
We know from our previous work that truncated fragments of TraI lacking DNA binding domains are also translocated to recipient cells (Lang et al., 2010). Here we addressed whether the R1-16 oriT is necessary for protein translocation. In addition to TraI, relaxosome proteins that bind oriT with sequence specificity are the E. coli IHF and plasmid proteins TraM and TraY (Csitkovits and Zechner, 2003; Karl et al., 2001; Mihajlovic et al., 2009). Two mutant R1-16 oriT derivatives (Table 1; Fig. 2) were compared to wild type. The first construction lacks 104 bp of oriT including nic and ihfA and sby binding sites for IHF and TraY (R1-16ΔoriT). The second, (R1-16Δnic), removed 34 bp of oriT to eliminate nic, the inverted repeat and key bases for TraI recognition (Williams and Schildbach, 2006). Normal transfer (tra) gene expression from the R1-16Δnic and R1-16ΔoriT mutant plasmids was verified by measuring highly efficient conjugative mobilization of a coresident recombinant R1oriT plasmid [(Lang et al., 2011); Fig. 2]. Self-transfer of the mutant derivatives of R1-16 was not observed, as expected (Fig. 2.) Moreover, Cre-TraI protein translocation was not detected for donors carrying either mutant derivative of R1-16. Presence of the third plasmid carrying the wild type oriT failed to complement protein translocation to measurable levels. The same strains supported efficient mobilization of the recombinant oriT plasmid, however (Fig. 2). To exclude the possibility that the available TraM was depleted due to competing transfer origins (TraM binding sites are present on the R1-16 knockout derivatives as well as on the mobilizable plasmid), TraI translocation was measured in the presence (pMM-wt) or absence (pMM-M0) of additional TraM expression in trans. No difference in TraI translocation was detected. We conclude that although R1-16 conjugation proteins expressed from the oriT deficient variants are proficient for plasmid mobilization, the process of protein translocation requires that R1-16 carry an intact oriT. This finding is in complete agreement with the requirements for R1-16 mediated host cell sensitivity to R17 phage. Dependency of the T4-mediated phage uptake process on the presence of an assembled and catalytically active relaxosome on plasmid R1-16 could not be complemented in trans with a smaller oriT containing plasmid (Lang et al., 2011).
3.2. TraY is important and TraM is essential for efficient protein translocation
These observations raised a number of mechanistic questions. As a first step we sought to distinguish early stage regulation of protein transfer from other possible explanations acting at a later stage following transfer initiation. Effective nucleoprotein export (TraI-T DNA) and import (protein A-R17 RNA) by the R1 system requires productive docking interactions between relaxosome and T4CP and catalytic processing of the oriT (Lang et al., 2011). We next tested whether docking interactions involving TraM and the T4CP, which have a key role in plasmid transfer, are equally important in protein transfer. Specific interactions between the C-terminal tail of TraD and the TraM tetramerization domain link the relaxosome to the T4CP and are required for efficient conjugation (Beranek et al., 2004; Lu et al., 2008; Wong et al., 2011). Analysis of R1-16 or pOX38 traM null derivatives in a CRAfT experiment revealed no detectable DNA or protein transfer (data not shown). Supplying wild type traM in trans raised the plasmid transfer frequencies by approximately three orders of magnitude, consistent with earlier results (Pölzleitner et al., 1997). This partial complementation is typical, but remains ∼1200-fold lower than wild type gene transfer frequencies [(Pölzleitner et al., 1997); data not shown]. Expression of traM in the CRAfT assay using both F-like conjugative plasmids did not result in detectable translocation of a Cre fusion with the cognate TraI (data not shown).
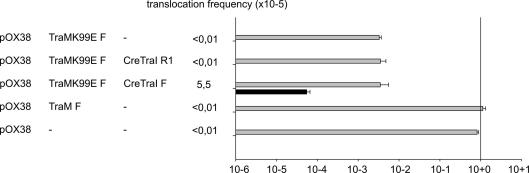
The functions of the TraMK99E mutant created by L. Frost and coworkers have been well characterized. This variant forms tetramers, binds its recognition sites in oriT and is fully functional in repressing the traM promoter; however, specific interactions with TraD are impaired in vitro and diminished conjugative DNA transfer was observed (Lu and Frost, 2005; Lu et al., 2008). Given that complementation of the traM null allele in trans is ineffective, we turned to co-expression approach for analysis of the mutant allele. Overexpression of the TraMK99E variant in trans to pOX38 in a CRAfT experiment (Fig. 3) had dominant negative effects on both DNA (∼200-fold lower) and protein transfer (∼80-fold lower). Specific TraD–TraM interactions are thus crucial to both protein and nucleoprotein export with F-like plasmids. The F secretion machinery recognizes Cre–TraIR1 poorly and transfer frequencies of 10−5 to 10−6 events per donor are typical (Lang et al., 2010). Under conditions of co-expression of wild type traM and the mutant tested here, no Cre–TraIR1 transfer was observed (Fig. 3).
Fig. 3.
Efficient protein translocation requires specific TraD–TraM interactions. The effects of expressing TraMK99E or wild type protein in trans, as indicated (left), was analyzed in a CRAfT assay. The frequencies of conjugative DNA transfer (gray bars) and translocation of the indicated Cre–TraI fusion proteins (black bars) represent are shown. Values represent the mean of at least three experiments. Standard deviations are shown.
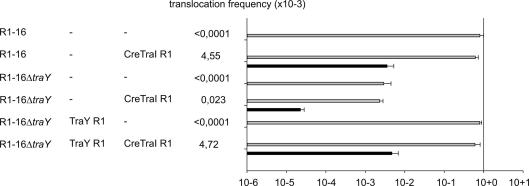
TraY is another plasmid-encoded protein imparting stability to the relaxosome. TraY proteins of different IncF plasmids specifically bind to a DNA sequence proximal to nic and stimulate TraI catalyzed nic-cleavage in vivo and in vitro (Abo and Ohtsubo, 1995; Csitkovits and Zechner, 2003; Inamoto et al., 1994; Karl et al., 2001; Nelson et al., 1995). We tested a R1-16 traY null derivative for its influence on DNA and protein transfer (Lang et al., 2011). Unlike null derivatives of traD and traM, which abolished transfer, R1-16ΔtraY lost proficiency in DNA and protein transfer by approximately two orders of magnitude compared to wild type (Fig. 4). TraY supplied in trans restored both forms of transfer to normal levels. The magnitude of reduced gene transfer is consistent with previously published data from traY null mutants of pOX38 (Maneewannakul et al., 1996). We conclude that TraY is important, but not essential, for efficient conjugative transfer of plasmid R1-16 DNA and TraI protein translocation.
Fig. 4.
TraY is important but not essential for conjugative DNA or protein transfer. The protein translocation frequencies supported by R1-16 compared to R1-16ΔtraY are shown (right) from donors carrying the coresident plasmids indicated (left). Plasmid pMSYM1 was used for complementation. Values represent recombination events per donor. Black bar represents frequency of translocation (Cre–TraIR1) with statistical significance compared to the vector control (not shown). Conjugation frequencies are indicated with gray bars. Values represent the mean of at least three experiments. Standard deviations are shown.
3.3. Mechanistic requirement for plasmid DNA during protein translocation
Taken together these data support the model that relaxosome assembly, binding to the T4CP and DNA processing within the complex is a prerequisite for protein transfer initiation even when the substrate protein cannot bind to oriT. This step seems to be crucial to early transfer regulation also in the mechanism of T4CP mediated R17 RNA import. In both whole cell assays however, the failure of a recombinant oriT plasmid to complement this requirement is unexpected. In the CRAfT assays shown in Fig. 2, the second oriT is part of a fully functional relaxosome that is also transmitted to the recipient cells. It is conceivable that the disruption of protein transfer observed with donors carrying R1-16ΔoriT or R1-16Δnic plus a coresident oriT plasmid is not due to an early stage regulatory defect but instead due to the copy number or length of the plasmid DNA being transferred.
Cre-TraI does not bind to oriT. Binding of the fusion protein to TraD may be unstable compared to the relaxosome and access of individual fusion proteins to the transporter pore may be hindered by the wild type relaxosome. It is conceivable therefore, that protein translocation happens rarely relative to a TraI-T DNA. We tested whether the presence of multiple copies of a second oriT affects protein translocation also when the R1-16 wild type plasmid is present. Consistent with a model of competitive interference or exclusion of the fusion protein, the efficiency of Cre–TraI transfer decreased substantially (∼12-fold) when either the pMM-wt or pMM-M0 plasmids were maintained in an R1-16 carrying host (not shown).
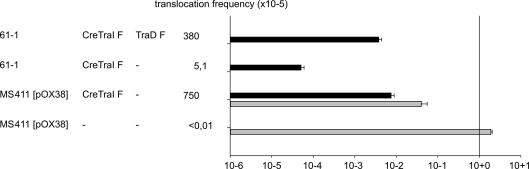
In addition to copy number differences, a second property that distinguishes R1-16 from the recombinant oriT plasmids is the ∼5-fold difference in genome length. Little is known about how macromolecules gain access to the secretion channel and whether different substrates follow the same or different pathways. As both plasmid strand transfer and protein translocation by R1 appears to require docking of a functional relaxosome to initiate transfer activity we wondered how and when unbound protein might enter the secretion channel. Since DNA strand transfer is processive, the length of the plasmid also determines the length of time the transporter is engaged in transporting the plasmid. If protein secretion is a stochastic process where free protein gains entry during plasmid transfer, then enhanced Cre recombination frequencies might be observed during conjugative transfer of longer genomes. We asked how the frequency of protein transfer mediated by an E. coli Hfr strain would compare to that mediated by the conjugative plasmid (Fig. 5). The Hfr strain 61-1 carrying a Cre–TraIF expression plasmid supported a protein transfer frequency of 5.1 × 10−5. Plasmid pOX38 in E. coli MS411 supported a 150-fold higher level of protein transfer. We know from earlier studies that TraI expressed in trans to an F plasmid has a negative effect on both DNA and protein transfer (Haft et al., 2006; Kienesberger et al., 2011; Lang et al., 2010). That negative effect on DNA transfer (∼50-fold lower) is readily seen with donor MS411 in the CRAfT assay shown in Fig. 5. To alleviate this problem, TraD can be co-expressed with the TraI fusion (Kienesberger et al., 2011; Lang et al., 2010). Coresidence of a traD expression plasmid in the Hfr strain 61-1 increased TraI translocation to a level equivalent to but not higher than that of the pOX38 donor (Fig. 5).
Fig. 5.
E. coli Hfr strain 61-1 secretes Cre–TraIF. Protein translocation frequencies (black) of TraIF fusion proteins from Hfr donor cells are compared to E. coli MS411 carrying pOX38. Gray bars indicate the observed levels of plasmid DNA transfer. Significant enhancement of protein translocation from Hfr donors due to co-overexpression of traDF and traIF is shown. All values shown were statistically compared to the vector control (data not shown). Values represent the mean of at least three experiments. Standard deviations are shown.
3.4. Similar functional domains of TraD are required for conjugative plasmid and protein transfer
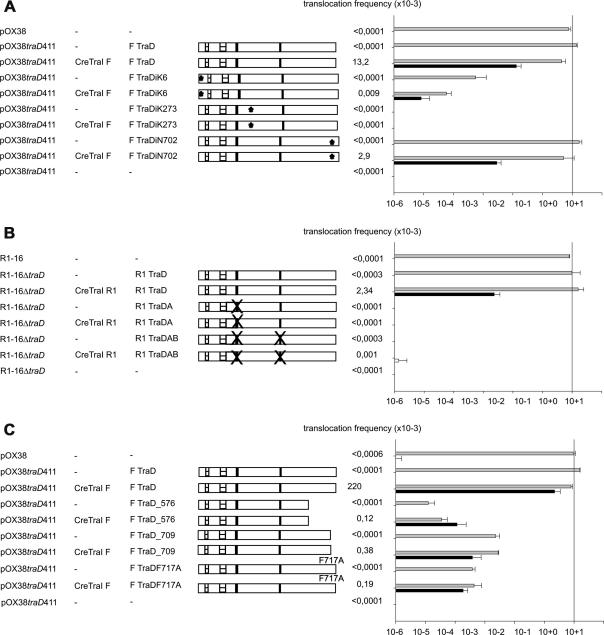
To characterize the functional contribution of TraD in protein translocation we used classes of well-defined mutations and assayed for complementation of R1-16 or pOX38 traD null derivatives. The Traxler laboratory has created a library of 31 residue insertion derivatives of TraD from plasmid F and analyzed their properties in detail (Haft et al., 2007; Lee et al., 1999). Here we confirmed that TraD oligomerization is essential for protein translocation by comparing the complementation efficiency of insertion mutants [TraDi(position of insertion)] known to disrupt association between monomers (TraDi273), or support dimer formation but not higher molecular weight complexes in vivo (TraDiK6) (Haft et al., 2007). The traD:iK6 allele supported ∼7500-fold lower frequencies of plasmid transfer and ∼1600-fold less protein transfer than wild type traD (Fig. 6A). No transfer was detected when traD:iK273 was present, consistent with the abolished conjugation phenotype observed earlier (Haft et al., 2007). TraD:i702 supports normal self associations in vivo (Haft et al., 2007) and was compared as a positive control in this experiment. Wild type plasmid transfer, but slightly lower (5-fold) protein translocation was observed with this allele. Complementation of transfer with the wild type allele in this series of assays (panel A) is lower than that observed with the constructions tested in panel C. We believe that this arises because expression from the pTrc99A vector-based constructions provided by the Traxler laboratory is lower than that of the high copy pBAD based constructions used in experiments of panel C.
Fig. 6.
Mutant forms of TraD proficient for conjugation also support TraI translocation. traD null derivatives of pOX38Km or R1-16 were complemented with TraD variants carrying either (A) 31 residue insertions (∗), (B) point mutations (×) abolishing NTP binding and hydrolysis, or (C) C-terminal deletions or mutations. Mutant variants of TraD are illustrated left. Striped boxes indicate predicted transmembrane domains. Black vertical stripes represent Walker A and B motifs. The translocation frequencies of the indicated Cre–TraI fusion proteins are given as recombination event per donor (right). Black bars show frequencies of translocation with statistical significance compared to the vector control (not shown). Conjugation frequencies are indicated with gray bars. Values are the mean of at least three experiments. Standard deviations are shown.
Mutations disrupting the Walker A motif for NTP binding (TraDK198T) or a double disruption eliminating also the Walker B site (TraDK198T/D425 N) were available for the traD allele of plasmid R1 and tested here. Each allele reduced plasmid and protein transfer to the limits of detection (Fig. 6B). Earlier studies have shown that the C terminal domain of TraD is involved in several important functional interactions with protein TraM (Beranek et al., 2004; Lu et al., 2008; Sastre et al., 1998; Wong et al., 2011). The loss of some amino acids from the C-terminus allows the F system to transfer a broader range of mobilizable plasmids at the expense of a decreased efficiency in interacting with the F plasmid relaxosome (Sastre et al., 1998). As shown in Fig. 6C, complementation of pOX38traD411 plasmid transfer with the TraD576 truncation variant was effective at a very low frequency (∼10−5). Interestingly, this mutation supports 3-fold more efficient protein translocation than plasmid transfer. A truncation lacking the last nine amino acids of TraD supported a modest 3-fold increase in protein transfer compared to TraD576 and a ∼80-fold increased frequency of plasmid transfer. The single residue exchange TraDF717A resulted in even lower plasmid- (6-fold) and protein- (2-fold) transfer compared with TraD709. In summary these results show that efficient TraI translocation requires TraD proficient in oligomer formation (Fig. 6A), NTP binding (Fig. 6B), and C-terminal mediated contact with the relaxosome (Fig. 6C), including specific interactions with TraM (Fig. 3).
4. Discussion
This study aimed to shed light on the activation mechanisms initiating (nucleo)protein transfer by the R1 T4SS. Recognition of the TraI protein as a secretion substrate requires a translocation signal (TS) presented by approximately 300 amino acids at one of two independently functional positions on the protein (Lang et al., 2010). Productive interactions with TraD that lead to secretion appear to occur even though these protein fragments are not likely to be integrated in the relaxosome. The results of this study indicate that protein translocation by the R1-16 system nevertheless depends on the formation of a relaxosome that is proficient in the same functions needed by a relaxosome involved in plasmid DNA transfer. Our previous analysis of the mechanistic requirements for the TraD dependent process of R17 phage uptake revealed that plasmid DNA and TraI interactions with its specific oriT binding sites were indispensable for nucleoprotein import via this machinery (Lang et al., 2011). The similarity of data from the different studies is compelling and argues that activation of the T4 secretion channel of the R1 system requires relaxosome assembly and perception of processed ssDNA substrate regardless of the actual secretion substrate.
In contrast to the few other DNA transporting systems where this has been investigated (Draper et al., 2005; Parker and Meyer, 2007; Vergunst et al., 2005) we found no evidence for DNA independent protein translocation by the R1 system. The mechanistic requirement is apparently manifest at the early docking and activation stage. We further hoped to gain insight to whether single proteins gain entry to the secretion channel during plasmid transfer (rather than preceding plasmid uptake or following termination) by comparing protein translocation driven by a plasmid or Hfr donor. The Cre-recombinase reaction monitored in recipient cells requires four monomers of Cre protein (Hoess and Abremski, 1984) and the low levels typically detected in our CRAfT assays were below a maximum level of detection. Therefore we tested whether engagement of the secretion apparatus with processive transfer of the E. coli 61-1 chromosome would raise the level of observed Cre recombination in the recipient population. The frequencies observed were equivalent to but not higher than that of the pOX38 donor. Addressing these mechanistic questions will obviously require higher resolution approaches.
As key regulators of secretion, T4CPs control not only initiation by selective binding of secretion substrates, their adaptor proteins and chaperones, they also interact with extracytoplasmic components of the secretion system. T4CPs act as molecular motors, harnessing the energy of ATP hydrolysis to invoke structural changes and through this activity are postulated to have several downstream functions during substrate passage across the cell envelope. Current models based on evidence from DNA mobilizing systems project that ATP hydrolysis by T4CPs energizes substrate movement (Cascales and Christie, 2004b; Llosa et al., 2002). Moreover T4 ATPases are proposed to control conformational change in system components that might regulate checkpoints in the secretion process (Cascales and Christie, 2004a, 2004b). The multisubunit T4 core complex forms a cylindrical structure with a central channel of about 110 Å diameter (Chandran et al., 2009; Fronzes et al., 2009). The N-termini of 14 monomers of VirB10 are anchored in the inner membrane. These protomers extend to the outer membrane where C-terminal domains associate to form a pore structure. VirB10 undergoes a structural transition when it senses modulation of the ATP binding or hydrolyzing activity of the inner membrane ATPases VirB11 or the T4CP (Cascales and Christie, 2004a; Jakubowski et al., 2009). Accordingly VirB10 is proposed to transduce energy from the inner membrane ATPases into conformational changes in the channel during active secretion. Recent phenotypic evidence monitoring release of the 69 kDa secretion protein VirE2 of the Agrobacterium tumefaciens T-DNA delivery system supports the model that the C-terminus of VirB10 is important to regulating passage of substrates across the outer membrane (Banta et al., 2011). It is not yet clear whether the pathway of substrate entry to the secretion channel is generally conserved and whether the same pathway is taken by all substrates of a given system. To address these challenging questions it will be important to use genetic approaches with a number of whole cell assays dedicated to transfer of a range of substrates. The capacity of male specific phage to exploit T4SSs competent for substrate translocation to gain entry to the host cell interior provides a fantastic opportunity to investigate the mechanistic basis underlying activation of a T4 secretion channel upon perception of extracellular and intracellular signals. The R1 system also has excellent potential to provide important insights in that three distinct activities, nucleoprotein export, protein export and nucleoprotein import are amenable to high through put phenotypic screening. The current study included a small number of known functional classes of mutations, nonetheless mutant variants of TraD which mediate distinct gain and loss of function phenotypes for different processes of substrate transfer were identified. The truncated traD576 allele is unique in that higher levels of protein translocation than plasmid transfer were observed (Fig. 6C). Also the observation that overexpression of TraMK99E in trans to a wild type pOX38 plasmid has a more pronounced inhibitory affect on DNA transfer compared to protein transfer is intriguing. The higher impact of this TraM variant on DNA transfer lowered the relative ratio of DNA/protein transfer by ∼6-fold. We conclude that a broad mutagenesis strategy combined with a phenotypic screen for several secretion substrates in parallel will be a promising approach to gain new insights to the mechanisms of conjugation systems and T4SS generally.
Plasmid R1 has been a highly successful model system for understanding the biology of plasmids and their bacterial hosts. We are confident that studies based on this model will advance our understanding for years to come.
Acknowledgments
We gratefully acknowledge L. Frost and B. Traxler for providing plasmids and S. Mihajlovic for constructing pMSTraDAB. This research was supported by the Austrian Science Fund (FWF) P-24016 and P-18607.
Communicated by Dr. F. de la Cruz
References
- Abo T., Ohtsubo E. Characterization of the functional sites in the oriT region involved in DNA transfer promoted by sex factor plasmid R100. J. Bacteriol. 1995;177:4350–4355. doi: 10.1128/jb.177.15.4350-4355.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Martinez C.E., Christie P.J. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta L.M. An Agrobacterium VirB10 mutation conferring a type IV secretion system gating defect. J. Bacteriol. 2011;193:2566–2574. doi: 10.1128/JB.00038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek A. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J. Bacteriol. 2004;186:6999–7006. doi: 10.1128/JB.186.20.6999-7006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry T.M., Christie P.J. Caught in the act: the dialogue between bacteriophage R17 and the type IV secretion machine of plasmid R1. Mol. Microbiol. 2011;82:1039–1043. doi: 10.1111/j.1365-2958.2011.07870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E., Christie P.J. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl. Acad. Sci. USA. 2004;101:17228–17233. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E., Christie P.J. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., Galas D.J. Cointegrate formation mediated by Tn9. II. Activity of IS1 is modulated by external DNA sequences. J. Mol. Biol. 1983;170:61–91. doi: 10.1016/s0022-2836(83)80227-7. [DOI] [PubMed] [Google Scholar]
- Chandran V. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csitkovits V.C., Zechner E.L. Extent of single-stranded DNA required for efficient TraI helicase activity in vitro. J. Biol. Chem. 2003;278:48696–48703. doi: 10.1074/jbc.M310025200. [DOI] [PubMed] [Google Scholar]
- de la Cruz F. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 2010;34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- Draper O. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc. Natl. Acad. Sci. USA. 2005;102:16385–16390. doi: 10.1073/pnas.0506081102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G.M., Johnson C.M. Regulatory circuits controlling enterococcal conjugation: lessons for functional genomics. Curr. Opin. Microbiol. 2011;14:174–180. doi: 10.1016/j.mib.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronzes R. Structure of a type IV secretion system core complex. Science. 2009;323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L.S., Koraimann G. Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol. 2010;5:1057–1071. doi: 10.2217/fmb.10.70. [DOI] [PubMed] [Google Scholar]
- Frost L.S. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- Goebel W. Dissociation and recombination of fragments with defined functions of the antibiotic resistance factor R1. In: Drews J., Högenauer G., editors. vol. 2. Springer Verlag; NewYork: 1977. (Topics in infectious diseases). [Google Scholar]
- Graus-Goldner A. The sequences of genes bordering oriT in the enterotoxin plasmid P307: comparison with the sequences of plasmids F and R1. Plasmid. 1990;24:119–131. doi: 10.1016/0147-619x(90)90014-4. [DOI] [PubMed] [Google Scholar]
- Haft R.J. In vivo oligomerization of the F conjugative coupling protein TraD. J. Bacteriol. 2007;189:6626–6634. doi: 10.1128/JB.00513-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft R.J. General mutagenesis of F plasmid TraI reveals its role in conjugative regulation. J. Bacteriol. 2006;188:6346–6353. doi: 10.1128/JB.00462-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess R.H., Abremski K. Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc. Natl. Acad. Sci. USA. 1984;81:1026–1029. doi: 10.1073/pnas.81.4.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamoto S. Site- and strand-specific nicking at oriT of plasmid R100 in a purified system: enhancement of the nicking activity of TraI (helicase I) with TraY and IHF. J. Biochem. 1994;116:838–844. doi: 10.1093/oxfordjournals.jbchem.a124604. [DOI] [PubMed] [Google Scholar]
- Jakubowski S.J. Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol. Microbiol. 2009;71:779–794. doi: 10.1111/j.1365-2958.2008.06565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl W. Transfer protein TraY of plasmid R1 stimulates TraI-catalyzed oriT cleavage in vivo. J. Bacteriol. 2001;183:909–914. doi: 10.1128/JB.183.3.909-914.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienesberger S. Interbacterial macromolecular transfer by the Campylobacter fetus subsp. venerealis type IV secretion system. J. Bacteriol. 2011;193:744–758. doi: 10.1128/JB.00798-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn P.M. Stages in phage R17 infection. VI. Injection of A protein and RNA into the host cell. Virology. 1972;47:628–637. doi: 10.1016/0042-6822(72)90552-1. [DOI] [PubMed] [Google Scholar]
- Kupelwieser G. Transfer protein TraM stimulates TraI-catalyzed cleavage of the transfer origin of plasmid R1 in vivo. J. Mol. Biol. 1998;275:81–94. doi: 10.1006/jmbi.1997.1436. [DOI] [PubMed] [Google Scholar]
- Lang S. Molecular recognition determinants for type IV secretion of diverse families of conjugative relaxases. Mol. Microbiol. 2010;78:1539–1555. doi: 10.1111/j.1365-2958.2010.07423.x. [DOI] [PubMed] [Google Scholar]
- Lang S. An activation domain of plasmid R1 TraI protein delineates stages of gene transfer initiation. Mol. Microbiol. 2011;82:1071–1085. doi: 10.1111/j.1365-2958.2011.07872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.H. Analysis of F factor TraD membrane topology by use of gene fusions and trypsin-sensitive insertions. J. Bacteriol. 1999;181:6108–6113. doi: 10.1128/jb.181.19.6108-6113.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa M. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- Lu J., Frost L.S. Mutations in the C-terminal region of TraM provide evidence for in vivo TraM–TraD interactions during F-plasmid conjugation. J. Bacteriol. 2005;187:4767–4773. doi: 10.1128/JB.187.14.4767-4773.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J., et al., 2008. Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol. Microbiol. [DOI] [PubMed]
- Maneewannakul K. Construction of derivatives of the F plasmid pOX-tra715: characterization of traY and traD mutants that can be complemented in trans. Mol. Microbiol. 1996;22:197–205. doi: 10.1046/j.1365-2958.1996.00087.x. [DOI] [PubMed] [Google Scholar]
- Mihajlovic S. Plasmid R1 conjugative DNA processing is regulated at the coupling protein interface. J. Bacteriol. 2009;191:6877–6887. doi: 10.1128/JB.00918-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W.C. The traY gene product and integration host factor stimulate Escherichia coli DNA helicase I-catalyzed nicking at the F plasmid oriT. J. Biol. Chem. 1995;270:28374–28380. [PubMed] [Google Scholar]
- Parker C., Meyer R.J. The R1162 relaxase/primase contains two, type IV transport signals that require the small plasmid protein MobB. Mol. Microbiol. 2007;66:252–261. doi: 10.1111/j.1365-2958.2007.05925.x. [DOI] [PubMed] [Google Scholar]
- Pölzleitner E. TraM of plasmid R1 controls transfer gene expression as an integrated control element in a complex regulatory network. Mol. Microbiol. 1997;25:495–507. doi: 10.1046/j.1365-2958.1997.4831853.x. [DOI] [PubMed] [Google Scholar]
- Roeder W., Somerville R.L. Cloning the trpR gene. Mol. Gen. Genet. 1979;176:361–368. doi: 10.1007/BF00333098. [DOI] [PubMed] [Google Scholar]
- Sastre J.I. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J. Bacteriol. 1998;180:6039–6042. doi: 10.1128/jb.180.22.6039-6042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoulaker R., Engelberg-Kulka H. Escherichia coli mutant temperature sensitive for group I RNA bacteriophages. J. Virol. 1978;25:433–435. doi: 10.1128/jvi.25.1.433-435.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie C. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine R.C. The F-pilus of E. coli. Adv. Microb. Physiol. 1969;3:1–52. [Google Scholar]
- Vergunst A.C. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- Vergunst A.C. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. USA. 2005;102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C.E., Winans S.C. Cell-cell communication in the plant pathogen Agrobacterium tumefaciens. Philos. Trans. R Soc. Lond. B Biol. Sci. 2007;362:1135–1148. doi: 10.1098/rstb.2007.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.L., Schildbach J.F. Examination of an inverted repeat within the F factor origin of transfer: context dependence of F TraI relaxase DNA specificity. Nucleic Acids Res. 2006;34:426–435. doi: 10.1093/nar/gkj444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, J. J., et al., 2011. Structural basis of cooperative DNA recognition by the plasmid conjugation factor, TraM. Nucleic Acids Res. [DOI] [PMC free article] [PubMed]
- Wong K., Paranchych W. The preservation of the secondary structure of R17 RNA during penetration into host bacteria. Virology. 1976;73:476–488. doi: 10.1016/0042-6822(76)90409-8. [DOI] [PubMed] [Google Scholar]
- Wozniak R.A., Waldor M.K. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- Zechner, E. L., et al., in press. Assembly and mechanisms of bacterial type IV secretion machines. Phil. Trans. R. Soc. B. [DOI] [PMC free article] [PubMed]