Summary
Pluripotency is a central, well-studied feature of embryonic development, but the role of pluripotent cell regulation in somatic tissue regeneration remains poorly understood. In planarians, regeneration of entire animals from tissue fragments is promoted by the activity of adult pluripotent stem cells (cNeoblasts). We utilized transcriptional profiling to identify planarian genes expressed in adult proliferating, regenerative cells (neoblasts). We also developed quantitative clonal analysis methods for expansion and differentiation of cNeoblast descendants that, together with RNAi, revealed gene roles in stem cell biology. Genes encoding two zinc finger proteins, Vasa, a LIM domain protein, Sox and Jun-like transcription factors, two candidate RNA-binding proteins, a Setd8-like protein, and PRC2 (Polycomb) were required for proliferative expansion and/or differentiation of cNeoblast-derived clones. These findings suggest that planarian stem cells utilize molecular mechanisms found in germ cells and other pluripotent cell types, and identify novel genetic regulators of the planarian stem cell system.
Introduction
Pluripotent cells, such as embryonic stem (ES) cells and induced pluripotent (iPS) cells provide attractive opportunities for regenerative medicine. These cells can be propagated long term in cell culture, yet retain the ability to differentiate into all specialized cell types of the body. The possibility of using pluripotent cells to regenerate adult somatic tissues in vivo, however, remains a daunting challenge: successful regeneration must balance precise regulation of cell proliferation, lineage commitment, migration, and differentiation, all while avoiding tumor formation.
Model organisms will likely play a crucial role in uncovering important general principles and gene networks that can regulate pluripotency for tissue regeneration. Planarians, an emerging model system for molecular studies of regeneration, are free-living freshwater flatworms that can regenerate any body part – including the head – in about a week (Reddien and Sánchez Alvarado, 2004). Planarians can be reared in large numbers in the laboratory and a recently sequenced genome (Robb et al., 2007), robust histological methods (Pearson et al., 2009), molecular tools (Newmark and Sánchez Alvarado, 2000), and RNAi (Reddien et al., 2005a; Sánchez Alvarado and Newmark, 1999) currently facilitate the identification of genes controlling regeneration. Planarians are triploblastic (possessing derivatives of all three germ layers) and contain a population of adult proliferative somatic cells (neoblasts) that have similar morphology (Reddien and Sánchez Alvarado, 2004), can be identified by common expression of PIWI-encoding genes (Reddien et al., 2005b), and have RNA-rich subcellular bodies known as chromatoid bodies (Higuchi et al., 2007). Recently, the neoblast population was demonstrated to contain clonogenic cells (cNeoblasts) that give rise to descendants spanning multiple germ layers, can restore tissue turnover and regenerative ability to irradiated animals, and can even generate entire adult bodies in single-cell transplantation experiments (Wagner et al., 2011). Genetic investigation of planarian stem cell biology, therefore, provides an opportunity to discover gene networks promoting pluripotency, maintenance of pluripotency within adult tissues, and deployment of stem cells for the regeneration of missing cell types.
Here, we took a genome-level approach to profile and functionally characterize a large set of intrinsic regulators of planarian stem cell biology. Motivated by the clonal repopulation of proliferative cells observed in planarians following treatment with low irradiation doses (Wagner et al., 2011), we developed quantitative functional assays for stem cell-initiated clonal expansion and differentiation that permit rapid identification of RNAi phenotypes. Numerous candidate regulatory genes identified by expression analyses were thus assessed and functionally classified by RNAi and clonal analysis. Together, these studies establish a simple yet powerful framework for investigating planarian stem cell regulation, and reveal a large panel of genetic factors utilized by this highly regenerative stem cell system.
Results
Transcriptional Profiling the Proliferative Cell Compartment of Adult Planarians
To identify candidate genetic regulators of planarian stem cells, we performed genome-level microarray studies comparing the expression profiles of untreated animals to animals exposed to a lethal dose (6,000 rads) of γ-irradiation. In planarians, proliferative cells (Dubois, 1949; Reddien et al., 2005b) and their associated transcripts (Eisenhoffer et al., 2008; Rossi et al., 2007) are rapidly and specifically eliminated following lethal irradiation. We generated a comprehensive oligonucleotide array (representing approximately 37,936 sequences) from EST data and predicted genes present in the Schmidtea mediterranea genome (Robb et al., 2007; Sánchez Alvarado et al., 2002; Zayas et al., 2005). To specifically profile proliferating cells (neoblasts), rather than their post-mitotic progeny, we identified the first transcripts depleted within 6, 12, 24, and 48 hours after exposure to lethal (6,000 rad) irradiation. As expected, transcripts for smedwi-1, PCNA, mcm2, and RRM2-1, which are specifically expressed in proliferative cells (Eisenhoffer et al., 2008; Guo et al., 2006; Hayashi et al., 2010; Wagner et al., 2011), were rapidly depleted within 24 hours of irradiation (Fig. 1A). Transcripts marking post-mitotic cell types, NB.21.11E, NB.32.1G, AGAT-1, MCP-1, and ODC-1 (Eisenhoffer et al., 2008), were not eliminated until 48 hours or later, and transcripts expressed in multiple differentiated cell types remained unaffected (Fig. 1A and Supplemental Fig. 1). We confirmed these trends by whole-mount triple fluorescent in situ hybridization (FISH) experiments with animals fixed 24 hours after irradiation (Fig. 1B).
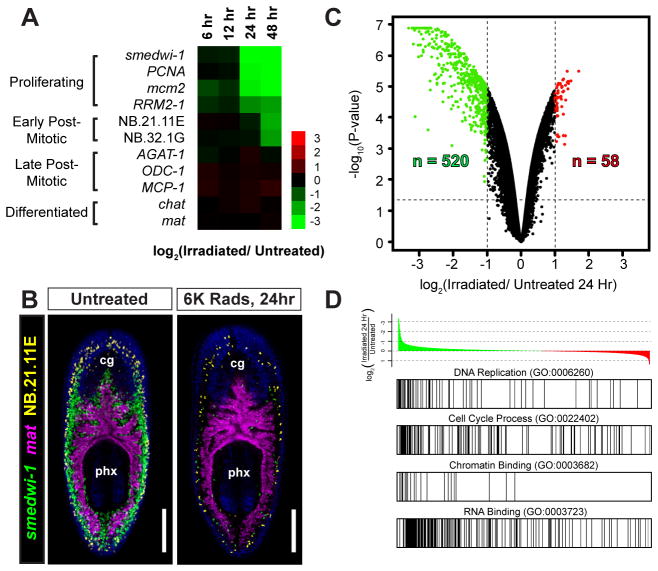
Figure 1. Identification of Irradiation-Sensitive Transcripts in Adult Planarians by Microarrays.
(A) Heat map illustrating mRNA depletion kinetics following 6,000 Rads γirradiation. Markers for proliferating cells (smedwi-1, PCNA, mcm2, RRM2-1), post-mitotic cells (NB.21.11E, NB.32.1G, AGAT-1, ODC-1, and MCP-1), neurons (chat), and intestine (mat) are shown. (B) Whole-mount triple-fluorescence in situ hybridization (FISH) shows the anatomical distribution of proliferative cells (neoblasts) with untreated and 24-hour irradiated animals. Shown are projections through z-stacks of multiple confocal planes in the interior of entire animals. Pharynx (px) and cephalic ganglia (cg) are indicated. Ventral views, anterior up. Scale bars, 200 μm. (C) Volcano plot showing transcripts depleted (green) or upregulated (red) 24 hours after irradiation. See Supplemental Table 1. (D) Gene set enrichment analysis (GSEA) with annotated gene list pre-ranked by log2 ratio (24-hour irradiated/untreated). Example gene sets enriched among irradiation-depleted transcripts are shown. See Supplemental Table 2.
The vast majority of transcripts examined (98.7%) did not show differential expression between untreated and 24 hour-irradiated animals. Of the 578 transcripts that showed differential expression, 90% were downregulated and 67% displayed sequence similarity (BLASTx, E value < 1×10−10) to human genes (Fig. 1C and Supplemental Table 1). Gene Set Enrichment Analysis (Subramanian et al., 2005) revealed that Gene Ontology (GO) terms associated with cell proliferation, e.g., DNA_Replication (GO:0006260) and Cell_Cycle_Process (GO:022402), were enriched within the 24-hour dataset (Fig. 1D; adj. p value < 0.001, and p = 0.0107, respectively). Chromatin_Binding (GO:0003682), RNA-Binding (GO:0003723) and DNA-Binding (GO:0003677) terms were also enriched (Fig. 1D and Supplemental Table 2), suggesting that genes regulating pluripotency and stem cell maintenance might also be contained within this dataset.
Expression of Candidate Regulatory Genes in Proliferative Cells
We determined the patterns and irradiation sensitivity of expression for identified candidate regulatory genes by whole-mount in situ hybridization. We prioritized for analysis genes predicted to encode DNA-, RNA-, or protein-binding domains (see Supplemental Table 3). Proliferative cells (e.g., smedwi-1+ cells) are irradiation-sensitive, absent from head tips, and distributed throughout the body mesenchyme (Reddien et al., 2005b). We identified 28 genes expressed in irradiation-sensitive patterns similar to the distribution of smedwi-1+ cells (Fig. 2A). Several of these genes are predicted to encode chromatin-modifying protein domains, including Smed-mrg-1 (MORF-related), Smed-nsd-1 (NSD1-like SET domain), Smed-rbbp4-1 (Retinoblastoma binding protein 4), and Smed-setd8-1 (SET domain containing 8) (Supplemental Table 3). In addition, three members of Polycomb repressive complex 2 (PRC2) were also identified: Smed-ezh (Enhancer of Zeste), Smed-sz12-1 (Suppressor of Zeste), and Smed-eed-1 (Embryonic Ectoderm Development), all well-established regulators of cell fate and stem cell activity (Boyer et al., 2006; Ezhkova et al., 2011; Lee et al., 2006; Ringrose and Paro, 2004).
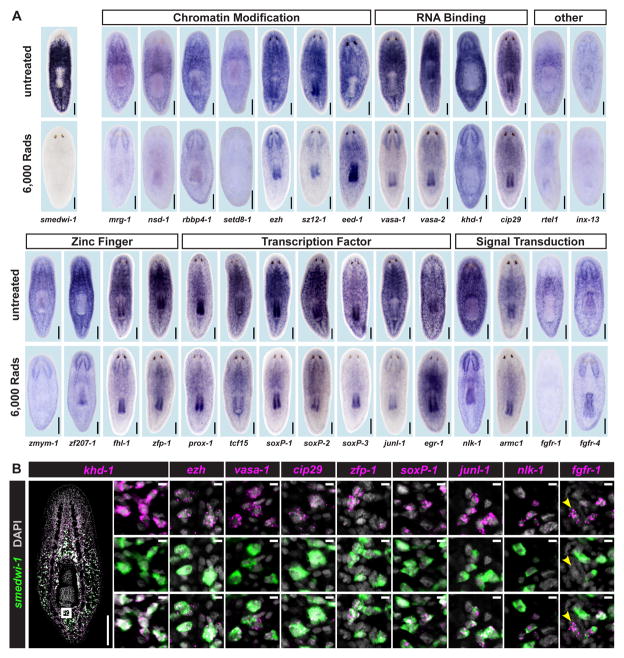
Figure 2. Irradiation-Sensitive Transcripts are Expressed in smedwi-1+ Proliferating Cells.
(A) Whole-mount in situ hybridization (ISH) in untreated animals and animals fixed 5 days after 6,000 Rads γ-irradiation. Genes were annotated by BLASTx and PFAM (See also Supplemental Table 3). (B) Expression of genes identified by microarray, analyzed by double FISH with a smedwi-1 RNA probe (proliferative cell marker). Zoomed images are single confocal planes from tail regions. Most cells detected by FISH co-expressed smedwi-1; cells with little/no smedwi-1 gene expression are labeled by arrowheads. Some transcripts (e.g., ezh and cip29) are expressed at low levels with background signal (scattered magenta dots) also visible. See also Supplemental Figure 2. Shown are representative ventral views, anterior up. Scale bars 200 μm, 10 μm (zoomed images).
Consistent with the well-known expression of germ cell-associated factors in planarian mitotic cells (Guo et al., 2006; Reddien et al., 2005b; Salvetti et al., 2005; Shibata et al., 2010; Shibata et al., 1999; Solana et al., 2009) we also identified four genes with sequence similarity to RNA-binding proteins. Two of these genes, Smed-vasa-1 and Smed-vasa-2, encode DEAD-Box 4 helicase domains and are similar to the germline-associated helicase, Vasa. Neoblast expression of Vasa-like PL10 genes (DjvlgA and DjvlgB) and DjVas-1, a vasa ortholog, have been described in a related planarian species, Dugesia japonica (Mochizuki et al., 2001; Rouhana et al., 2010; Shibata et al., 1999). DjVas-1 is required for regeneration in D. japonica, but the cellular basis for this phenotype is unclear (Rouhana et al., 2010). We also identified Smed-khd-1 (a gene encoding a KH-domain protein) and Smed-cip29 (similar to Cytokine-Induced Protein 29 kDa) genes, which both encode proteins that might function in RNA metabolism (Sugiura et al., 2007; Valverde et al., 2008; Wang et al., 2002; Yamazaki et al., 2010). Similar expression patterns for a cip29-like gene have also been reported for Dugesia japonica (Rossi et al., 2007).
To date, the potential role of transcription factors and signaling proteins in planarian stem cell regulation has only received minor attention. We identified seven genes encoding predicted transcription factors expressed in proliferative cells: Smed-prox-1 (Prospero Homeobox 1), Smed-tcf15 (Transcription Factor 15), Smed-soxP-1, Smed-soxP-2, and Smed-soxP-3 (three genes encoding SRY [sex-determining region Y] box containing proteins), Smed-junl-1 (c-jun-like), and Smed-egr-1 (Early Growth Response protein), as well as four genes predicted to encode zinc finger domain-containing proteins, Smed-zfmym-1, Smed-zf207-1, Smed-fhl-1 (a Four and a Half LIM Domain protein), and Smed-zfp-1. Finally, four neoblast-expressed genes encode proteins with similarity to signal transduction proteins: Smed-nlk-1 (Nemo-like kinase), Smed-armc1 (Armadillo repeat-containing 1), as well as Smed-fgfr-1 and Smed-fgfr-4 (Fibroblast growth factor receptors). Expression of FGFR-like genes in proliferative cells has been reported in Dugesia japonica (Ogawa et al., 2002), but no functional roles for these genes in planarian stem cells are described.
Several genes identified by the microarray studies described above were expressed in smedwi-1-like, irradiation-sensitive patterns, but appeared enriched in specific subpopulations of cells. Some genes (e.g. prox-1, tcf15, armc1, fgfr-1) were expressed at high levels in irradiation-sensitive cells near the midline of the animal, whereas others (e.g. egr-1, soxP-3) were found in patterns that extended to the periphery of the body and/or regions slightly anterior to the photoreceptors, similar to previously described post-mitotic cell populations (Fig. 2A) (Eisenhoffer et al., 2008). In addition, expression of many genes was also detected in the pharynx, and/or in the central nervous system (Fig. 2A). Of the 22 transcripts detectable by FISH, all displayed overlapping expression with smedwi-1 (Fig. 2B and Supplemental Fig. 2), confirming expression within dividing cells. Together, these data significantly extend the list of genes with identified expression in the neoblasts of planarians.
Identification of Genes Required for Recovery from Sublethal Irradiation
To investigate the functions of identified candidate regulatory genes, we first developed a sensitized assay capable of screening for weak (or strong) phenotypic defects in stem cell activity. Animals exposed to a high irradiation dose (6,000 rads) experience complete failure of tissue turnover (Bardeen and Baetjer, 1904; Dubois, 1949) and display stereotypical signs of stem cell loss, including tissue regression, ventral curling, and ultimate lysis (Reddien et al., 2005b). By contrast, delivery of sublethal irradiation doses (e.g. 1,000 – 1,250 rads) initially reduces dividing cell numbers, but such animals can ultimately restore normal proliferative cell levels (Salvetti et al., 2009) and regenerative ability (Wagner et al., 2011). For example, smedwi-1+ cell numbers were dramatically reduced seven days after exposure to 1,250 rads, but were restored to approximately normal levels over the next 14 days (Fig. 3A). Accordingly, under mock RNAi conditions, most animals exposed to 1,250 rads survived and remained healthy for over two months of observation (Fig. 3B–D). We reasoned that 1,250-rad irradiated animals would be highly sensitized to defects in stem cell function. In such a background, even subtle RNAi phenotypes might render animals unable to restore tissue turnover and manifest as a lethal (easily scored) phenotype.
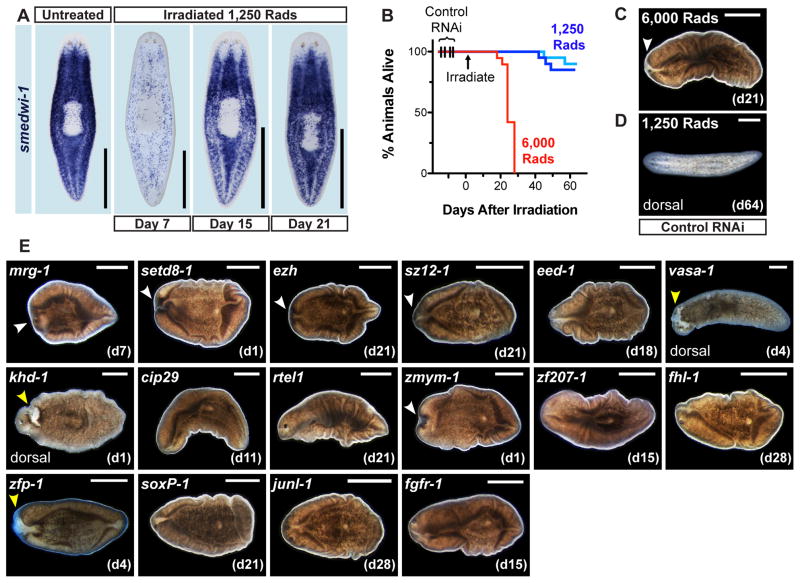
Figure 3. Identification of RNAi Phenotypes after Sublethal Irradiation.
(A) Proliferative cell repopulation detected by smedwi-1 ISH in animals fixed 7, 15, and 21 days after 1,250 Rads γ-irradiation. Ventral views, anterior up. (B) Survival curves of control RNAi animals exposed to 1,250 Rads (two independent experiments) and 6,000 Rads (n = 19–20 animals per sample). (C–E) Representative views of irradiated RNAi animals, anterior left. Images are ventral views unless otherwise noted. (C) 6,000 Rad-irradiated animals experienced head regression (white arrowheads) and ventral curling by day 21. (D) Most sublethally irradiated (1,250 Rad) animals, by contrast, were visibly normal on day 64. (E) Representative live images of animals with RNAi phenotypes after 1,250 rad exposure. Images are from the first day post-irradiation (noted in parenthesis) when tissue failure appeared; only cases where >50% of animals displayed defects are shown. White arrowheads: tissue regression. yellow arrowhead: epidermal lesions. Scale bar, 500 μm. See also Supplemental Figure 3 and Supplemental Table 4.
Animals were fed dsRNA for the 28 identified gene candidates, exposed to 1,250 rads irradiation, and assessed for the ability to survive. RNAi of 16 genes yielded robust RNAi phenotypes, similar to the effects of lethal irradiation (i.e., head regression, curling, and lysis; Fig. 3E, Supplemental Fig. 3, Supplemental Table 4). fgfr-1(RNAi) animals exhibited head regression and curling, then recovered (head regeneration) (Fig. 3E and Supplemental Table 4), possibly explained if RNAi effects wore off. With three phenotypes (khd-1, zfp-1, vasa-1), lesions developed on the dorsal epidermis, suggesting a mode of tissue failure at least in part distinct from that of lethal irradiation (Fig. 3E and Supplemental Fig. 3A). Sublethal irradiation, therefore, provides a sensitive, readily scalable screening strategy for planarians that can facilitate identification of novel stem cell RNAi phenotypes.
Assessment of Stem Cell Regulation by Single-Colony Analysis
Failed recovery RNAi phenotypes observed following 1,250 rads irradiation (described above) could occur because of defects in a wide range of processes including stem cell self-renewal, lineage commitment, cell proliferation, DNA repair, migration, and/or differentiation. In order investigate identified RNAi phenotypes in greater cellular detail and to distinguish between these possibilities, we developed methods for quantitative analysis of individual planarian stem cell clones. High doses of irradiation (e.g., 6,000 rads) completely eliminate proliferating cells from planarian tissues. Much lower doses (e.g., 1,500–1,750 rads) similarly deplete the vast majority (>99%) of proliferating cells, but permit the survival of rare, isolated neoblasts. These remaining clonogenic neoblasts (“cNeoblasts”) divide to produce, from a single-cell starting point, clonally-derived clusters of cellular descendants (i.e., “colonies”) (Wagner et al., 2011). Between days 7 and 14 post-irradiation colonies exhibited robust (approximately 10-fold) and significant (p < 0.01, Student’s t-test) increase of smedwi-1+ cell numbers (Fig. 4A–C). In addition to smedwi-1+ neoblasts, expanding colonies also produce post-mitotic differentiating cell types that can be labeled with RNA probes to the NB.21.11E and AGAT-1 genes (Wagner et al., 2011). NB.21.11E+ and AGAT-1+ cells are mesenchymal cell types that are located subepidermally, adjacent to the body-wall musculature (Eisenhoffer et al., 2008). Because these cells have a short (2–4 day) lifespan, they are initially depleted in irradiated animals and are only later produced within expanding smedwi-1+ cell colonies. Notably, colonies exhibited a linear relationship between numbers of dividing and differentiating cells (Fig. 4B–C). Single colony analysis can therefore be used to simultaneously and quantitatively assess multiple aspects of stem cell function. First, colony cell number can be readily quantified as a readout of neoblast “expansion” (i.e., self-renewal). Second, the ratio of proliferating to post-mitotic cell types can be used to assess the rate of differentiation within a single colony. Each of these processes (expansion and differentiation) can be measured independently within individual colonies under RNAi conditions, providing a powerful framework for deciphering phenotypes of candidate stem cell regulatory genes.
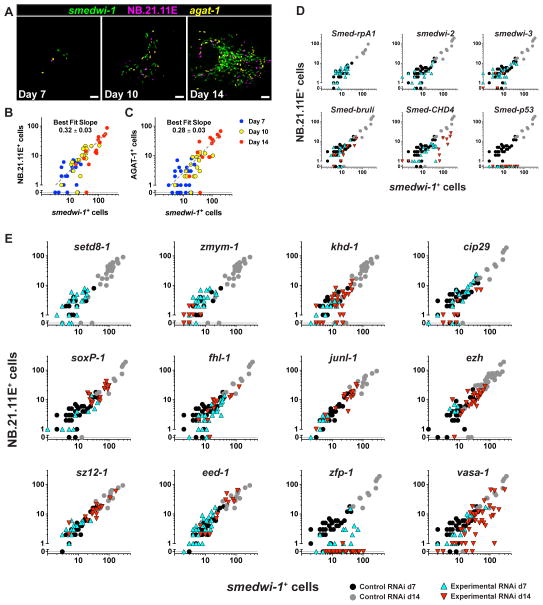
Figure 4. Genes Required for Proliferative Cell Expansion and Differentiation Identified by RNAi and Clonal Analysis.
(A) Representative colonies from animals exposed to 1,750 Rads analyzed by triple FISH. Ventral views, adjacent to the pharynx, anterior left. Proliferating cells (smedwi-1+), and two post-mitotic cell types (NB.21.11E+ and AGAT-1+) are labeled. Scale bars, 50 μm. (B–C) Log-scale plots of raw cell count data. Each dot represents an individual colony. smedwi-1+ cell numbers per colony are plotted against numbers of NB.21.11E+ (B) and AGAT-1+ cells (C). (D–E) Animals irradiated 1,500 – 1,750 Rads fed one RNAi food dose displayed reduced numbers of smedwi-1+ and/or NB.21.11E+ cells per colony. RNAi was administered by feeding 4 days after irradiation except for junl-1, ezh, sz12-1, and eed-1, which were fed seven days prior to irradiation. See also Supplemental Figure 4. For statistical analysis of colony phenotypes, see Supplemental Table 5.
We first tested the effectiveness of colony analysis by studying genes with previously reported RNAi phenotypes. Animals were fed a single dose of dsRNA 4 days after exposure to 1,500 rads irradiation, and fixed on days 7 and 14. With this protocol, colonies initiate prior to dsRNA administration, and consequences of RNAi on these colonies are thereafter assessed. We devised criteria for several colony phenotype classes. “Colony loss” phenotypes, for example, were designated when RNAi caused complete disappearance of colony cells between days 7 and day 14 post-irradiation. “Failed expansion” phenotypes were classified as those in which both smedwi-1+ and NB.21.11E+ day-14 colony cell counts were significantly decreased (t-test, p < 0.05) relative to control RNAi colonies. “Failed differentiation” phenotypes were designated when post-mitotic to proliferative cell count ratios deviated significantly (ANCOVA, p < 0.01) from those of control RNAi colonies (see Materials and Methods). We first analyzed, as a control, the RNAi phenotype of Smed-rpa-1, a gene encoding a protein similar to human Replication Protein A1 (RPA1). RPA1 is required for DNA replication, and Smed-rpa1 RNAi leads to decreased mitotic activity and tissue failure in adult planarians (Reddien et al., 2005a). Animals fed dsRNA for Smed-rpa1 experienced complete disappearance of colony cells (i.e., “colony loss”) between days 7 and 14 post-irradiation (Fig. 4D). Comparable results were obtained with additional cell cycle genes Smed-rplp0 (similar to human Ribosomal Protein, large, P0), Smed-cdc23 (similar to human CDC23), and Smed-cyclinL1 (similar to human Cyclin L1) (Supplemental Fig. 4A–C), indicating that RNAi of genes required for cell division disrupts continued presence of colony cells, as expected. Furthermore, these results demonstrate that a single RNAi dose can be highly effective for triggering colony phenotypes.
We next analyzed the RNAi phenotype of Smed-bruli, a gene similar to the germline-associated RNA-binding protein, Bruno (Guo et al., 2006). bruli is expressed in dividing cells in adult planarians, and bruli RNAi leads to gradual neoblast loss, possibly reflecting a stem cell self-renewal defect (Guo et al., 2006). Such a hypothesis predicts that following bruli RNAi, smedwi-1+ colonies should continue to produce differentiating cells, but fail to expand. Indeed, bruli RNAi resulted in colonies with a “failed expansion” colony phenotype in which both smedwi-1+ cell (t-test, p = 0.016) and NB.21.11E+ cell (p = 0.0065) numbers were significantly reduced by day 14, relative to controls, but possessed normal ratios of smedwi-1+ to NB.21.11E+ cells (Fig 4D and Supplemental Table 5). These results are consistent with a role for bruli in stem cell self-renewal.
smedwi-2 and smedwi-3 encode two Piwi homologs that, like bruli, are expressed in dividing cells of adult planarians (Palakodeti et al., 2008; Reddien et al., 2005b). Previous studies demonstrated that RNAi of smedwi-2 or smedwi-3 leads to failure of regeneration and tissue homeostasis. Furthermore, RNAi of smedwi-2 (and to a lesser extent smedwi-3) ultimately depletes proliferative cells (Palakodeti et al., 2008; Reddien et al., 2005b). We observed a “colony loss” phenotype following smedwi-2 RNAi (Fig. 4D). smedwi-3 RNAi produced a similar effect, although some colonies, which had failed to expand, were still present on day 14 (Fig. 4D). Colony assays, therefore, demonstrate clear intrinsic defects in stem cell division/expansion for both smedwi-2 and smedwi-3(RNAi) animals.
To assess stem cell differentiation, we examined Smed-CHD4 and Smed-p53, two genes with previously described roles in the production of post-mitotic cell types (Pearson and Sánchez Alvarado, 2010; Scimone et al., 2010). As predicted, RNAi of CHD4 and p53 each resulted in “failed differentiation” colony phenotypes, with significant reductions (p ≤ 0.0107) in NB.21.11E+ and AGAT-1+ cell numbers on day 14, and significant deviations in ratios of smedwi-1+ to NB.21.11E+ (or AGAT-1+) cells (Fig. 4D, Supplemental Fig. 4D–E, Supplemental Table 5). In addition to differentiation defects, CHD4(RNAi) and p53(RNAi) planarians have been reported to ultimately experience exhaustion of proliferative cell activity. p53 RNAi colonies also displayed significantly lower numbers of smedwi-1+ cells (p = 0.0018), confirming a combined defect in both colony expansion and differentiation (Fig. 4D, Supplemental Table 5). By contrast, CHD4(RNAi) colonies displayed approximately normal numbers of smedwi-1+ cells on day 14, indicating an initial persistence of colony expansion activity even while differentiation was diminished (Fig. 4D and Supplemental Table 5). Taken together, these RNAi experiments demonstrate the power of colony analysis for examination of stem cell phenotypes. In particular, bruli and CHD4 RNAi phenotypes also indicate that expansion and differentiation of colony cells are distinct biological processes that can be independently assessed and phenotypically decoupled.
In analyses of any stem cell system, distinguishing primary defects associated with gene loss of function from indirect changes in stem cell behavior (e.g., as a consequence of tissues becoming defective) is a major challenge. Prior strategies for studying stem cell function in planarians often involve slow-unfolding phenotypes that can confound interpretation. The study of RNAi phenotypes by colony analysis in planarians resolves many of these issues because stem cell defects can be assessed before substantial tissue degeneration has occurred. This approach using planarians and cNeoblast colonies is sensitive, direct, and allows rapid determination of defects immediately following initial gene inhibition.
RNAi and Clonal Analysis Identify Regulatory Roles for Neoblast Genes
We next assessed the functions of genes associated with “failed recovery” RNAi phenotypes following sublethal irradiation (described above) using colony analysis. One gene, setd8-1, exhibited a “colony loss” phenotype (Fig. 4E), indicating that this gene is required for the persistence of cell division in growing colonies. Accordingly, human SETD8/PR-SET7 (an H4K20me1 lysine methyltransferase) is known to promote a silent chromatin state and is required for cell cycle progression (Nishioka et al., 2002; Oda et al., 2009).
Nine genes had “failed colony expansion” RNAi phenotypes (Fig. 4E and Supplemental Table 5). Included in this class were zmym-1, khd-1, cip29, soxP-1, fhl-1, and junl-1. Interestingly, RNAi of all three genes encoding components of the Polycomb PRC2 complex, ezh, sz12-1, and eed-1, also resulted in failed colony expansion phenotypes, indicating that Polycomb proteins might also play a key role in the regulation of planarian stem cell self-renewal.
We uncovered roles for two genes in the process of differentiation. Following RNAi of Smed-zfp-1 or Smed-vasa-1, day 14 colonies displayed significantly distorted ratios (p < 0.0001) of proliferative to post-mitotic cells (Fig. 4E, Supplemental Fig. 4, and Supplemental Table 5). The zfp-1 RNAi phenotype was particularly strong, with day 14 colonies completely devoid of both NB.21.11E+ and AGAT-1+ cells (Fig. 4E, Supplemental Fig. 4F). In addition to defects in post-mitotic cell numbers, RNAi of zfp-1 or vasa-1 also led to reduced numbers of smedwi-1+ cells in day 14 colonies (Fig. 4E, Supplemental Table 5). We utilized zfp-1 to test colony formation with an independent method; following transplantation of zfp-1 RNAi cells into irradiated non-RNAi hosts, similar colony abnormalities were observed (Supplemental Fig. 4H). zfp-1 and vasa-1 are similar to p53 in that RNAi phenotypes include aspects of both failed colony expansion and failed differentiation. However, unlike p53, which is expressed predominantly in post-mitotic cells (Pearson and Sánchez Alvarado, 2010), both vasa-1 and zfp-1 are primarily expressed within the proliferative smedwi-1+ population (Fig. 2A-B). Together, these data suggest that vasa-1 and zfp-1 function within proliferative cells, prior to significant expression of p53, to promote the process of differentiation. Furthermore, these data demonstrate that vasa-1, a gene similar to the germ cell-promoting VASA helicase, is integral to the process of somatic cell differentiation in planarian adults.
Genes with Colony RNAi Phenotypes are Required for Tissue Homeostasis and Regeneration
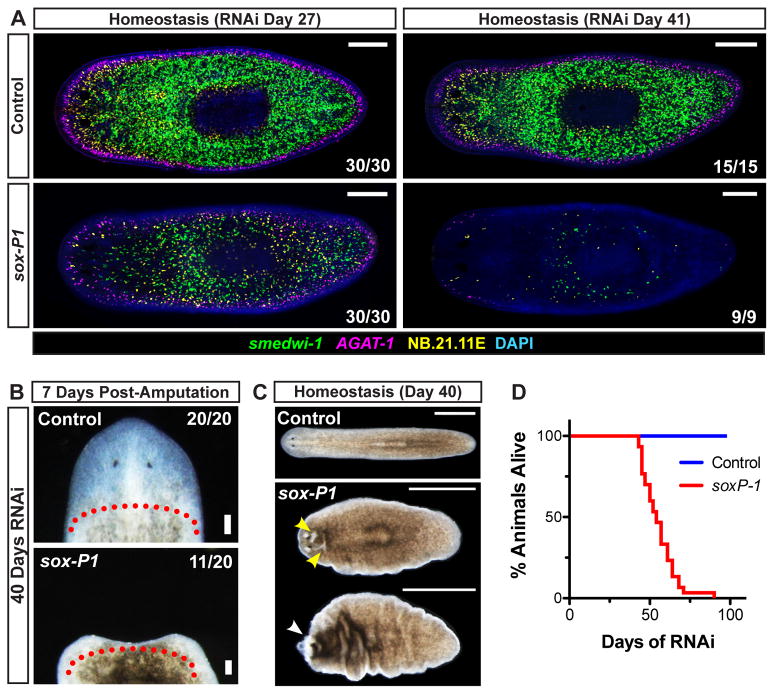
Because homeostasis and regeneration RNAi data can be predictors of colony phenotypes (Fig. 4D), we tested whether RNAi phenotypes initially identified by colony analyses would manifest in adult planarians in the absence of irradiation. Smed-soxP-1 encodes SRY-box-containing (Sox) transcription factor (Supplemental Fig. 5) and displayed a failed expansion RNAi phenotype in colony assays. Long-term experiments in unirradiated animals revealed that soxP-1(RNAi) animals experience progressive loss of smedwi-1+ cells, resulting in near complete depletion after 41 days of RNAi (Fig. 5A). Animals amputated after 40 days of soxP-1 RNAi either lysed (9/20) or failed to regenerate (11/20) (Fig. 5B), and animals exposed to continuous soxP-1 RNAi in the absence of injury developed epidermal lesions, experienced regression of head tissue (Fig. 5C), and all ultimately died (n > 30 animals/sample) (Fig. 5D). Together these data indicate soxP-1 is necessary for long-term maintenance of adult stem cell activity in the absence of irradiation. Notably, SOXP-1 represents the first known transcription factor expressed broadly in the neoblast population and required for its maintenance.
Figure 5. Smed-soxP-1 is Required for Maintenance of smedwi-1+ Cells, Tissue Homeostasis, and Regeneration.
(A) Animals were assessed for proliferative (smedwi-1+) and post-mitotic (AGAT-1+ and NB.21.11E+) cell types after several weeks of RNAi. Shown are representative confocal planes, anterior left. (B–C) Regenerative ability and tissue homeostasis were assessed after 40 days of RNAi. (B) Representative live images of head regions 7 days post-amputation are shown, anterior up. Approximate amputation plane is indicated by dotted line. (C–D) Tissue maintenance and animal survival after continuous RNAi feedings every 3–4 days (n ≥ 30 animals/sample). (C) Representative live images of whole animals undergoing tissue failure, anterior left. Epidermal lesions and head regression are indicated by yellow and white arrowheads, respectively. (D) Survival curves. Scale bars 200 μm (A), 100μm (B), 1mm (C).
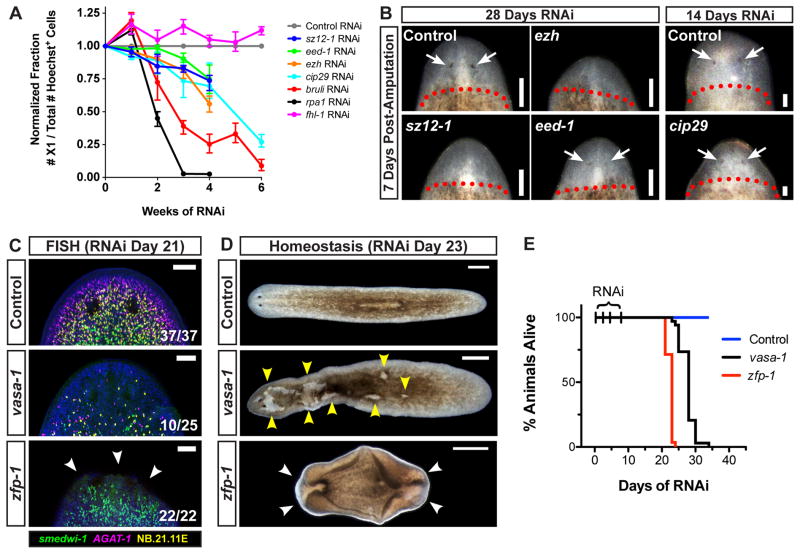
We investigated additional genes displaying expansion RNAi phenotypes by long term RNAi timecourse experiments. Flow cytometry-based quantification of the X1 cell population (~90% of which express smedwi-1) is an established, reliable method for assessing the presence of proliferating cells (Hayashi et al., 2006; Palakodeti et al., 2008; Reddien et al., 2005b; Scimone et al., 2010). We predicted that similar to soxP-1 and bruli (Guo et al., 2006), RNAi of ezh, eed-1, sz12-1, fhl-1, or cip29 would lead to a loss of proliferating cells, but at a rate much slower than that of a gene required for cell division (e.g., rpa1). Indeed, the proportion of X1 cells present in planarian tissues gradually decreased over a 4–6 week RNAi timecourse for ezh, eed-1, sz12-1, and cip29, similar to the decline measured in bruli RNAi animals (Fig. 6A). Proliferative cells were depleted much more rapidly following rpa1 RNAi (Fig. 6A). RNAi of fhl-1 did not result in decline of cell proliferation (Fig. 6A), indicating that not all genes required for colony expansion will necessarily display defects during normal homeostasis. Loss of proliferative cell activity in ezh, eed-1, sz12-1, or cip29 RNAi animals was correlated with reduced size of regenerative blastemas, and faint or absent photoreceptors when compared to control RNAi animals (Fig. 6B). These data indicate impairment, although not a complete elimination, of regenerative ability. These results confirm that ezh, eed-1, sz12-1, and cip29 do indeed promote the long-term maintenance of proliferative cell activity and regenerative ability in adult planarians, even under normal homeostasis conditions.
Figure 6. Genes Necessary for Colony Expansion and Differentiation are Required for Tissue Maintenance.
(A) Animals fed RNAi food for several weeks were assessed for proliferative cell presence by flow cytometry. Numbers of proliferating X1 cells, represented as a fraction of total Hoechst+ cells, were normalized to internal RNAi controls. Three biological replicates were used per time point. Shown are means; error bars denote data range. (B) Regenerative ability after 14 or 28 days of continuous RNAi feeding. Shown are head regions, anterior up, seven days after decapitation. Dotted line, approximate amputation planes. Arrows, photoreceptors. (C–E) RNAi animals were assessed for tissue maintenance defects. (C) Confocal projections from animals after 21 days of RNAi. Dorsal views of head regions, anterior up. Arrows, tissue regression sites. (D–E) Tissue maintenance and animal survival after RNAi (n = 29–34 worms/sample). (D) Images after 23 days of RNAi. Anterior, left. Yellow arrows, lesions. White arrows, tissue regression and ventral curling. (E) Survival curves. Scale bars 100 μm (B–C), 500 μm (D).
We next tested whether zfp-1 and vasa-1, two genes for which RNAi caused failed colony expansion and differentiation, might similarly be required for tissue maintenance in non-irradiated animals. For both zfp-1 and vasa-1(RNAi) animals, successful head regeneration was observed 7 days after amputation, although regenerated tissues subsequently regressed (Supplemental Fig. 6). 21 days of continuous RNAi treatment resulted in dramatic reductions in the numbers of smedwi-1+, NB.21.11E+, and AGAT-1+ cells in head regions of some vasa-1(RNAi) animals, as well as dramatic reductions in NB.21.11E+ and AGAT-1+ cell numbers in all zfp-1(RNAi) animals examined (Fig. 6C). Similar to phenotypes observed following 1,250 rads irradiation, lesions appeared on the dorsal surface of vasa-1(RNAi) animals, and zfp-1(RNAi) animals experienced tissue regression and ventral curling (Fig. 6D). These signs of tissue failure were accompanied by complete lethality within 30 days (Fig. 6E). Together, these data indicate that failures of colony expansion and differentiation observed following zfp-1 or vasa-1 RNAi reflect a general requirement of these genes in stem cell-mediated maintenance of adult tissues.
Discussion
Planarians Provide an Ideal in vivo System for Gene-Function Studies of Somatic Pluripotency
Investigations into the molecular basis of pluripotency have long focused on germ cell regulation and embryonic cells in culture. Genetic screens in D. melanogaster and C. elegans, for example, have identified regulatory factors important for germline development and maintenance, and early embryonic fate decisions (Rongo and Lehmann, 1996; Seydoux and Braun, 2006). Recent efforts investigating regulation of embryonic stem (ES) cells and induced pluripotent (iPS) cells additionally provide a window into the molecular underpinnings of early events in mammalian development (Young, 2011).
Increasingly, it has become evident that several genes associated with germ cell biology are also expressed in multipotent, adult, somatic cell types across disparate animal phyla including cnidarians, sponges, lophotrochozoans, and echinoderms (Juliano et al., 2010). These organisms provide a novel physiological context – adult tissue maintenance and regeneration – and useful evolutionary positions for studying the pluripotent state. Planarians, with the availability of robust culturing methods, a sequenced genome, molecular tools, and RNAi, are well suited for this task. Planarians possess remarkable regenerative abilities and are also now known to contain pluripotent stem cells in the adult animal (Wagner et al., 2011). The neoblast population therefore presents an attractive, experimentally accessible system for gene function studies of somatic pluripotency.
Sublethal Irradiation and Clonal Analysis Enable Rapid Elucidation of Stem Cell Phenotypes
Limitations of currently available methods prompted us to develop novel functional assays for studying stem cell regulation in planarians. One of the most striking properties of stem cells is the ability to reconstitute and restore tissues of an irradiated host animal (Osawa et al., 1996; Spangrude et al., 1988; Till and McCulloch, 1961). Such a capacity for cNeoblasts has been recently shown by single-cell transplantation into lethally irradiated hosts (Wagner et al., 2011), and by sublethal irradiation experiments (Salvetti et al., 2009; Wagner et al., 2011). Here we demonstrated the feasibility of using sublethal irradiation to screen for “failed recovery” phenotypes as a first step in identifying genes regulating planarian stem cell activity. Additionally, we determined that individual stem cell colonies undergo a stereotyped process of expansion and post-mitotic cell production. Single colony analysis can therefore be used to quantitatively assess these features under RNAi conditions.
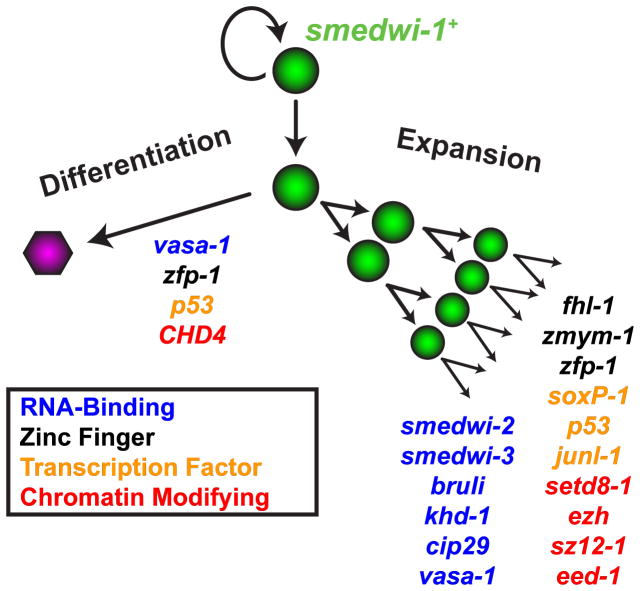
Phenotypes described by colony assays can mirror those elucidated during homeostasis and regeneration (and vice versa). For example, we assessed the requirements of planarian CHD4 and p53 homologs in colonies. RNAi of either CHD4 or p53 was previously described to affect the numbers of differentiating neoblast-descendent cells (Pearson and Sánchez Alvarado, 2010; Scimone et al., 2010), and RNAi of either gene caused a robust defect in colony differentiation. cNeoblast colonies provide several key advantages over previously available functional assays in planarians: they are quantitative in nature, they tend to generate rapid phenotypes (prior to significant somatic tissue damage which can confound phenotype interpretation), they involve a synchronized starting point, and they assess gene function under conditions of rapid symmetric stem cell expansion, a process which might only occur intermittently during normal homeostasis. Together, these methods represent a significant step forward in the ability to study stem cell phenotypes in planarians. Using clonal analysis and RNAi, we categorized several known and novel genetic factors with “failed differentiation” or “failed expansion” phenotypes in planarians (Fig. 7).
Figure 7. RNA-Binding, Transcription, and Chromatin Modifying Factors Regulate Clonogenic Expansion and Differentiation of Planarian Stem Cells.
Genes expressed within the proliferative cell compartment are required for expansion and differentiation activity associated with clonogenic cells (cNeoblasts). Several of these genes (e.g. p53, zfp-1, vasa-1) are involved in both proliferative cell expansion and differentiation.
Regulation of Neoblasts Involves Molecular Features Shared with other Stem Cell Types
We identified three planarian genes Smed-ezh, Smed-sz12-1, and Smed-eed-1 (encoding predicted proteins similar to Polycomb Repressive Complex 2 (PRC2) components) that are expressed in proliferative cells and required for colony expansion and long-term maintenance of proliferative cell activity. In C. elegans, Mes proteins including MES-2 (a homologue of Drosophila PRC2 protein, Enhancer of Zeste) are required for X-chromosome dosage compensation and are essential for germ cell development (Fong et al., 2002; Garvin et al., 1998). Polycomb-mediated regulation of pluripotency, furthermore, is found in mammalian embryonic stem cells in which PRC2 complex components bind and deposit repressive H3K27me3 histone marks at promoters of developmental regulators (Boyer et al., 2006; Lee et al., 2006). In mouse ES cells, PRC2 inhibition leads to upregulation of genes normally expressed during differentiation, though these ES cells can still propagate in vitro and remain pluripotent (Boyer et al., 2006; Chamberlain et al., 2008). Similarly, loss of PRC2 in adult cells such as hair follicles of the epidermis and skeletal muscle satellite cells leads to transcriptional derepression and stem cell dysfunction (Ezhkova et al., 2011; Juan et al., 2011). In these in vivo contexts, however, stem cell failure is associated with loss of proliferative activity (Ezhkova et al., 2011; Juan et al., 2011). In planarians, loss of proliferative cells following RNAi of PRC2 genes therefore resembles in vivo phenotypes observed for adult stem cells. Future studies of PRC2 function in planarians will therefore provide an important in vivo context for further investigating the role of this complex in pluripotent cells.
We also identified a requirement for Smed-soxP-1, a gene encoding a Sox transcription factor, in the process of colony expansion, however we could not classify the Sox subfamily to which SOXP-1 belongs. The discovery of soxP-1 is significant because this encodes the first known transcription factor expressed broadly in neoblasts and required for their maintenance. Sox transcription factors regulate cell fate in a wide range of contexts from sex determination (Sinclair et al., 1990) to pluripotency (Avilion et al., 2003; Boyer et al., 2005). It will be interesting to compare the functions of SOXP-1 and to those of Sox proteins in other organisms, such as Sox2 in ES cells.
Polycomb and Sox proteins are not exclusively utilized in pluripotent cells, and are routinely deployed in other physiological contexts (e.g., adult stem cell regulation (Ezhkova et al., 2011; Juan et al., 2011) and tissue patterning (Denell, 1978)). By contrast, expression and function of conserved RNA-binding germline-associated genes (e.g., vasa, bruno, piwi) might be more restricted to instances of pluripotent cell activity (Juliano et al., 2010). Cancer cells, which share many common features with pluripotent cells, do in some cases express germline-associated genes and/or require them for tumorigenesis (Janic et al., 2010; Simpson et al., 2005). In planarian adults, homologs of germline regulators (e.g., bruli, smedwi-2, smedwi-3, DjPum, and Spoltud-1) are expressed within proliferating cells and required for their maintenance (Guo et al., 2006; Palakodeti et al., 2008; Reddien et al., 2005b; Salvetti et al., 2005; Solana et al., 2009). Here, we determined that smedwi-2, smedwi-3, and bruli are all required for proliferation and/or expansion of cNeoblast descendants. We also defined a functional role for Smed-vasa-1, a gene highly similar to Drosophila and human Vasa proteins, in planarian stem cell regulation. Drosophila Vasa contains a DEAD-box RNA helicase domain (Lasko and Ashburner, 1988), and is required for accumulation and proper translational regulation of germ cell mRNAs, oocyte patterning, and differentiation (Styhler et al., 1998; Tomancak et al., 1998). Similarly, we demonstrated that Smed-vasa-1 is required for proper differentiation and expansion of somatic stem cell colonies, and for maintenance of tissue integrity in unirradiated animals. Interestingly, the mode of tissue failure in Smed-vasa-1 RNAi animals (lesions) is reminiscent of other phenotypes related to loss of differentiation activity (e.g., p53 or zfp-1 RNAi animals). These findings, in particular those for Smed-vasa-1, support the idea that pluripotent stem cells and germ cells can share common molecular features.
Novel Genetic Regulators of Pluripotent Stem Cell Activity
In addition to the genes we characterized with previously studied functions in other organisms, we also identified phenotypes for a number of genes with understudied roles in stem cell biology (Fig 7). For example, we identified novel functional roles in stem cell regulation for three genes encoding zinc finger proteins (zmym-1, fhl-1, and zfp-1), one additional transcription factor (junl-1), and two genes encoding putative RNA-binding proteins (khd-1 and cip29) (Sugiura et al., 2007; Valverde et al., 2008). Given the extensive role of RNA-binding proteins in germ cell and planarian stem cell biology, further study of khd-1 and cip29 might reveal additional molecular characteristics of pluripotency and the undifferentiated state. Our expression analyses also identified additional candidate regulatory genes expressed in neoblasts, including those encoding additional transcription factors (soxP-2, soxP-3, prox-1, tcf15, and egr-1), which were not associated with detected RNAi phenotypes. Furthermore, RNAi of some genes (rtel1, fgfr-4, znf207-1, and mrg-1) caused tissue maintenance defects after sublethal irradiation but did not cause detected defects in colony assays. Future in depth studies of identified phenotypes and exploration of potential redundancy with those genes without phenotypes are likely to reveal additional important regulatory aspects of the planarian stem cell system. An additional interesting future direction will involve exploring the expression of genes identified here and any candidate heterogeneity that might exist in the neoblasts. Given that a sequenced genome and now suitable functional assays provide an avenue for large-scale unbiased screens, Schmidtea mediterranea presents a clear, tractable in vivo model system for future molecular dissection of stem cell and regenerative biology.
Experimental Procedures
Planarian Culture and Irradiation
Strain ClW4 was maintained as described (Newmark and Sánchez Alvarado, 2000), including Gentamycin. Irradiation experiments involved ≥seven days starved, size-matched animals and dual Gammacell-40 137 Cesium sources delivering 79–82 rads/min. Irradiated animals were washed every 3–4 days.
RNAi and Molecular Biology
cDNAs (Sánchez Alvarado et al., 2002) and libraries from regenerating animals were used. Genes were cloned into pGEM (Promega). pPR244 usage and bacterial growth for RNAi were as described (Reddien et al., 2005a). Bacteria were resuspended in 66% homogenized beef liver (in water) at 1/300th the initial bacterial culture volume. Negative RNAi controls used unc-22, a C. elegans gene lacking significant similarity to the S. mediterranea genome. Flow cytometry with RNAi phenotypes was performed as described (Scimone et al., 2010). Microarray and Gene Set Enrichment Analysis (GSEA) methods can be found in Supplemental Material.
Histology
RNA probes were in vitro transcribed by T7 polymerase (Promega) using DIG-, FITC- (Roche), or DNP- (Perkin Elmer) modified ribonucleotides, purified by ethanol precipitation with 7.5M ammonium acetate, and resuspended in deionized formamide. Tyramide-conjugated fluorophores were generated from AMCA, Fluorescein, Rhodamine (Pierce), and Cy5 (GE Healthcare) N-Hydroxysuccinimide (NHS) esters. in situ hybridizations were performed as described (Pearson et al., 2009). For double/triple-color fluorescence in situ hybridization (FISH), HRP-inactivation was performed between labelings in 4% formaldehyde, 45 min.
Single-Colony Analysis
Doses causing 30–50% of irradiated animals to lack any colony were optimized, such that single or sparse colonies were examined in the remaining animals. 1,750 and 1,500 rads were used for 2–4mm and < 2mm animals, respectively. Colonies that merged or were in close proximity (<50–100μm) were not analyzed. Proliferative to post-mitotic cell ratios were assessed by linear regression (best-fit slope and 95% confidence intervals). Student’s t-test (two-tailed, unequal variance) was used to determine significant changes in cell numbers. Analysis of covariance (ANCOVA) was used to test for significant changes in cell-type ratios (relative to control RNAi ratios) by comparing slopes of linear regression lines fit to day 14 colony data plots (see Supplemental Table 5).
Supplementary Material
Highlights.
Expression profiling identifies regulatory genes expressed in neoblasts
Clonal stem cell assay and RNAi identify stem cell expansion/differentiation genes
Planarian stem cells share molecular features with embryonic and germ cells
Acknowledgments
We thank G. Bell for microarray assistance; J. van Wolfswinkel for cDNA sequence assembly, D. Wenemoser and K. Li for fgfr cloning; and P. Hsu, R. Young, D. Kim, S. Lapan, and all Reddien Lab members for comments and discussion. P.W.R. is an early career scientist of the Howard Hughes Medical Institute and an associate member of the Broad Institute of Harvard and MIT. We acknowledge support from the NIH (R01GM080639) and the Keck Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Gene Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeen C, Baetjer F. The inhibitive action of the Roentgen rays on regeneration in planarians. J Exp Zool. 1904;1:191–195. [Google Scholar]
- Boyer L, Plath K, Zeitlinger J, Brambrink T, Medeiros L, Lee T, Levine S, Wernig M, Tajonar A, Ray M, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denell RE. Homoeosis in Drosophila. II. a Genetic Analysis of Polycomb. Genetics. 1978;90:277–289. doi: 10.1093/genetics/90.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois F. Contribution á l ’ètude de la migration des cellules de règènèration chez les Planaires dulcicoles. Bull Biol Fr Belg. 1949;83:213–283. [Google Scholar]
- Eisenhoffer GT, Kang H, Sánchez Alvarado A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3:327–339. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Gene Dev. 2011;25:485–498. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong Y, Bender L, Wang W, Strome S. Regulation of the Different Chromatin States of Autosomes and X Chromosomes in the Germ Line of C. elegans. Science. 2002;296:2235–2238. doi: 10.1126/science.1070790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin C, Holdeman R, Strome S. The phenotype of mes-2, mes-3, mes-4 and mes-6, maternal-effect genes required for survival of the germline in Caenorhabditis elegans, is sensitive to chromosome dosage. Genetics. 1998;148:167–185. doi: 10.1093/genetics/148.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Peters AHFM, Newmark PA. A bruno-like Gene Is Required for Stem Cell Maintenance in Planarians. Dev Cell. 2006;11:159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ. 2006;48:371–380. doi: 10.1111/j.1440-169X.2006.00876.x. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Shibata N, Okumura R, Kudome T, Nishimura O, Tarui H, Agata K. Single-cell gene profiling of planarian stem cells using fluorescent activated cell sorting and its “index sorting” function for stem cell research. Dev Growth Differ. 2010;52:131–144. doi: 10.1111/j.1440-169X.2009.01157.x. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Hayashi T, Hori I, Shibata N, Sakamoto H, Agata K. Characterization and categorization of fluorescence activated cell sorted planarian stem cells by ultrastructural analysis. Dev Growth Differ. 2007;49:571–581. doi: 10.1111/j.1440-169X.2007.00947.x. [DOI] [PubMed] [Google Scholar]
- Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010;330:1824–1827. doi: 10.1126/science.1195481. [DOI] [PubMed] [Google Scholar]
- Juan AH, Derfoul A, Feng X, Ryall JG, Dell’Orso S, Pasut A, Zare H, Simone JM, Rudnicki MA, Sartorelli V. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Gene Dev. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano CE, Swartz SZ, Wessel GM. A conserved germline multipotency program. Development. 2010;137:4113–4126. doi: 10.1242/dev.047969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan SW, Reddien PW. dlx and sp6–9 Control Optic Cup Regeneration in a Prototypic Eye. PLoS Genet. 2011;7:e1002226. doi: 10.1371/journal.pgen.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335:611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono KI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T. Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev Genes Evol. 2001;211:299–308. doi: 10.1007/s004270100156. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Kobayashi C, Hayashi T, Orii H, Watanabe K, Agata K. Planarian fibroblast growth factor receptor homologs expressed in stem cells and cephalic ganglions. Dev Growth Differ. 2002;44:191–204. doi: 10.1046/j.1440-169x.2002.00634.x. [DOI] [PubMed] [Google Scholar]
- Osawa M, Hanada Ki, Hamada H, Nakauchi H. Long-Term Lymphohematopoietic Reconstitution by a Single CD34-Low/Negative Hematopoietic Stem Cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14:1174–1186. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B, Sánchez Alvarado A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development. 2010;137:213–221. doi: 10.1242/dev.044297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn. 2009;238:443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P, Bermange A, Murfitt K, Jennings J, Sánchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005a;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005b;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Robb SMC, Ross E, Sánchez Alvarado A. SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res. 2007;36:D599–D606. doi: 10.1093/nar/gkm684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongo C, Lehmann R. Regulated synthesis, transport and assembly of the Drosophila germ plasm. Trends Genet. 1996;12:102–109. doi: 10.1016/0168-9525(96)81421-1. [DOI] [PubMed] [Google Scholar]
- Rossi L, Salvetti A, Marincola FM, Lena A, Deri P, Mannini L, Batistoni R, Wang E, Gremigni V. Deciphering the molecular machinery of stem cells: a look at the neoblast gene expression profile. Genome Biol. 2007;8:R62. doi: 10.1186/gb-2007-8-4-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Shibata N, Nishimura O, Agata K. Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev Biol. 2010;341:429–443. doi: 10.1016/j.ydbio.2010.02.037. [DOI] [PubMed] [Google Scholar]
- Salvetti A, Rossi L, Bonuccelli L, Lena A, Pugliesi C, Rainaldi G, Evangelista M, Gremigni V. Adult stem cell plasticity: neoblast repopulation in non-lethally irradiated planarians. Dev Biol. 2009;328:305–314. doi: 10.1016/j.ydbio.2009.01.029. [DOI] [PubMed] [Google Scholar]
- Salvetti A, Rossi L, Lena A, Batistoni R, Deri P, Rainaldi G, Locci MT, Evangelista M, Gremigni V. DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development. 2005;132:1863–1874. doi: 10.1242/dev.01785. [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. PNAS. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA, Robb SM, Juste R. The Schmidtea mediterranea database as a molecular resource for studying Platyhelminthes, stem cells and regeneration. Development. 2002;129:5659–5665. doi: 10.1242/dev.00167. [DOI] [PubMed] [Google Scholar]
- Scimone ML, Meisel J, Reddien PW. The Mi-2-like Smed-CHD4 gene is required for stem cell differentiation in the planarian Schmidtea mediterranea. Development. 2010;137:1231–1241. doi: 10.1242/dev.042051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Srivastava M, Bell GW, Reddien PW. A regulatory program for excretory system regeneration in planarians. Development. 2011;138:4387–4398. doi: 10.1242/dev.068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Shibata N, Rouhana L, Agata K. Cellular and molecular dissection of pluripotent adult somatic stem cells in planarians. Dev Growth Differ. 2010;52:27–41. doi: 10.1111/j.1440-169X.2009.01155.x. [DOI] [PubMed] [Google Scholar]
- Shibata N, Umesono Y, Orii H, Sakurai T, Watanabe K, Agata K. Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev Biol. 1999;206:73–87. doi: 10.1006/dbio.1998.9130. [DOI] [PubMed] [Google Scholar]
- Simpson AJG, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Solana J, Lasko P, Romero R. Spoltud-1 is a chromatoid body component required for planarian long-term stem cell self-renewal. Dev Biol. 2009;328:410–421. doi: 10.1016/j.ydbio.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangrude G, Heimfeld S, Weissman I. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Swan A, Suter B. vasa is required for Gurken accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 1998;125:1569–1578. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura T, Sakurai K, Nagano Y. Intracellular characterization of DDX39, a novel growth-associated RNA helicase. Exp Cell Res. 2007;313:782–790. doi: 10.1016/j.yexcr.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Till J, McCulloch E. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat Res. 1961;14:213–222. [PubMed] [Google Scholar]
- Tomancak P, Guichet A, Zavorszky P, Ephrussi A. Oocyte polarity depends on regulation of gurken by Vasa. Development. 1998;125:1723–1732. doi: 10.1242/dev.125.9.1723. [DOI] [PubMed] [Google Scholar]
- Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xu J, Zeng L, Ye X, Wu Q, Dai J, Ji C, Gu S, Zhao C, Xie Y, et al. Cloning and characterization of a novel human STAR domain containing cDNA KHDRBS2. Mol Biol Rep. 2002;29:369–375. doi: 10.1023/a:1021246109101. [DOI] [PubMed] [Google Scholar]
- Weissman I. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Fujiwara N, Yukinaga H, Ebisuya M, Shiki T, Kurihara T, Kioka N, Kambe T, Nagao M, Nishida E, et al. The closely related RNA helicases, UAP56 and URH49, preferentially form distinct mRNA export machineries and coordinately regulate mitotic progression. Mol Biol Cell. 2010;21:2953–2965. doi: 10.1091/mbc.E09-10-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayas RM, Hernández A, Habermann B, Wang Y, Stary JM, Newmark PA. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain. PNAS. 2005;102:18491–18496. doi: 10.1073/pnas.0509507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.