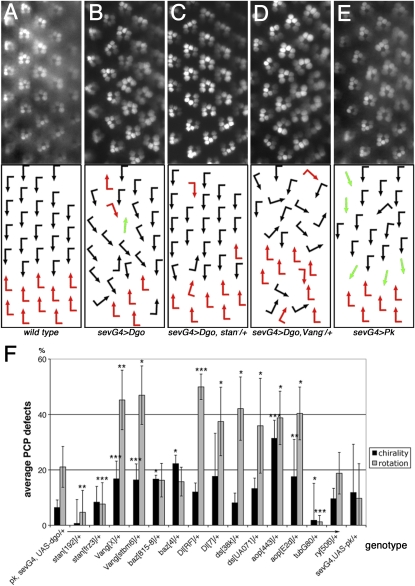
Figure 1 .
Ommatidial PCP orientation defects induced by Dgo or Pk overexpression and dominant modifications in the pilot screen. (A–E) Rhodopsin1-GFP (Rh1-GFP) pictures of rhabdomere patterns are shown on top and schematic arrows representing ommatidial orientation at bottom; dorsal is up and anterior to the left in these and all subsequent figures. The following genotypes are shown: wild type (A), pk/+, UAS-dgo/+, Rh1-GFP/+; sev-GAL4/+ (referred to as sev-GAL4, UAS-dgo) (B), stanfrz3/+, pk/+, UAS-dgo/+, Rh1-GFP/+; sev-GAL4/+ (C), Vangstbm-X/+, pk/+, UAS-dgo/+, Rh1-GFP/+; sev-GAL4/+ (D), and UAS-Pk/+, sev-GAL4/+, Rh1-GFP/+ (referred to as sev-GAL4, UAS-pk) (E). Quantifications of interactions are shown in F. Black and red arrows indicate the two chiral ommatidial forms, and green arrows represent nonchiral, symmetrical ommatidia. Note that stanfrz3/+ suppressed sev-GAL4, UAS-dgo rotation defects (C and F) and Vangstbm-X/+ enhanced sev-GAL4, UAS-dgo (D and F) chirality and rotation defects. Quantifications of additional candidate genes tested in pilot screen, showing significant modification: bazooka (baz)/D-Par3, Delta (Dl), dachsous (ds), and aop/yan (F) (*P < 0.06, **P < 0.01, and ***P < 0.005; three to four eyes and 63–173 ommatidia were scored for each genotype). Chirality and rotation defects were counted independently; negative controls did not modify sev-GAL4, UAS-dgo and GAL80ts abolished the phenotype (F).