Abstract
Maximum likelihood methods for the estimation of linkage disequilibrium between biallelic DNA-markers in half-sib families (half-sib method) are developed for single and multifamily situations. Monte Carlo computer simulations were carried out for a variety of scenarios regarding sire genotypes, linkage disequilibrium, recombination fraction, family size, and number of families. A double heterozygote sire was simulated with recombination fraction of 0.00, linkage disequilibrium among dams of δ = 0.10, and alleles at both markers segregating at intermediate frequencies for a family size of 500. The average estimates of δ were 0.17, 0.25, and 0.10 for Excoffier and Slatkin (1995), maternal informative haplotypes, and the half-sib method, respectively. A multifamily EM algorithm was tested at intermediate frequencies by computer simulation. The range of the absolute difference between estimated and simulated δ was between 0.000 and 0.008. A cattle half-sib family was genotyped with the Illumina 50K BeadChip. There were 314,730 SNP pairs for which the sire was a homo-heterozygote with average estimates of r2 of 0.115, 0.067, and 0.111 for half-sib, Excoffier and Slatkin (1995), and maternal informative haplotypes methods, respectively. There were 208,872 SNP pairs for which the sire was double heterozygote with average estimates of r2 across the genome of 0.100, 0.267, and 0.925 for half-sib, Excoffier and Slatkin (1995), and maternal informative haplotypes methods, respectively. Genome analyses for all possible sire genotypes with 829,042 tests showed that ignoring half-sib family structure leads to upward biased estimates of linkage disequilibrium. Published inferences on population structure and evolution of cattle should be revisited after accommodating existing half-sib family structure in the estimation of linkage disequilibrium.
TRADITIONAL methods for gene mapping are based on linkage, which requires a family structure because loci are mapped by tracing inheritance of marker alleles in progeny from at least one ancestor. The DNA markers of choice were microsatellites because they were abundant and very informative. Linkage maps of microsatellites were developed for farm animal species with a half-sib structure such as cattle (Da and Lewin, 1995; Ma et al. 1996; Kappes et al. 1997; Barendse et al. 1997; Våge et al. 2000). In recent years, a revolution has been initiated in human genetics with the large-scale DNA sequencing of the HAP MAP project (2007), which allowed the discovery of vast amounts of single nucleotide polymorphism (SNP). SNP sequences were used in arrays allowing interrogation of the human genome from thousands to over a million SNPs. The biggest interest in humans is the application of this technology for identification of variants that are associated with genetic diseases in the so-called case-control studies.
The development of SNP arrays in human genetics was followed by animal geneticists. There are commercially available arrays for over 50 or 60 thousands SNPs for the cow, sheep, and swine. The statistical treatment of SNP arrays in animal populations is carried out without consideration for the breeding structure currently present in farm animals but lacking in experiments for case-control studies in human populations. Contrary to human populations, animals at the farm are highly related because of the use of artificial insemination (AI) and intensive breeding. For example, dairy bulls with high estimated breeding values might have over a million daughters (http://www.crv4all.com/eng/halloffame/Sunny_Boy_HOFame.pdf). Analysis of linkage disequilibrium (LD) between pairs of SNPs in cattle populations has been carried out either using the expectation maximization (EM) algorithm for unrelated individuals (e.g., Sargolzaei et al. 2008) or using the most likely of phased haplotypes (e.g., McKay et al. 2007; Qanbari et al. 2010). The first method ignores that the contribution of haplotypes from sires to progeny exceeds its true counts in the population because each offspring receives one haplotype from their sire. The second method ignores that marker informativity might cause a systematic increase or decrease of informative sire haplotype counts (and consequently informative haplotype counts from dams) depending on the genetic distance between markers. Consequently, bias in the estimation of LD using half-sib data might occur.
The objective of this article is to develop maximum likelihood methods for the estimation of linkage disequilibrium between codominant DNA markers in half-sib families. It is shown that severe biased estimation may occur after ignoring half-sib relationships. The methods are tested via Monte Carlo computer simulation. Comparison of alternative methods of estimation of linkage disequilibrium is carried out after genotyping a half-sib family with 36 calves with the Illumina 50K BeadChip.
Theory and Methods
AI is in widespread use in cattle with the most common situation being a sire having a single progeny from a number of dams. Three situations are possible when estimating second-order linkage disequilibrium (disequilibrium considering two loci) between two DNA markers: (a) the sire is a homozygote at the two loci, (b) the sire is a homozygote at one locus and a heterozygote at the other, and (c) the sire is a heterozygote at the two loci. For the following derivations, assumptions are: (1) recombination fraction is known without error, and (2) the linkage phase (combination of alleles at two loci on the two homologous chromosomes in diploid individuals) in the sire is known. The impact of departures from these assumptions is addressed in the Discussion.
Double homozygote sire
Let the sire have genotype TTMM at two SNPs, T/t, and M/m. Offspring might have genotypes TTMM, TTMm, TtMM, TtMm indicating that haplotypes TM, Tm, tM, and tm were inherited from dams, respectively. Therefore, the haplotypes in half-sibs are fully informative and linkage disequilibrium can be estimated directly from haplotype counts. Thus, for alleles T and M at two loci, the disequilibrium can be estimated by substituting haplotype and allelic frequencies into ; ; ; and , where is the frequency of the kth allele, and are the linkage disequilibrium and haplotype frequencies between the kth and tth alleles at the two loci, respectively. In addition to allele frequencies, only one parameter for the linkage disequilibrium, δ, needs to be estimated since ; ; ; and . Estimating disequilibrium by direct counts of haplotype and allele frequencies is also the maximum likelihood estimate of linkage disequilibrium. The sampling variance of the estimates of the disequilibrium parameter for the ith family is derived in Appendix A,
where Li(δ|nG) is the maximum likelihood function of the disequilibrium parameter, δ, conditional to the haplotype counts, nG.
The value of the second derivative with respect to the disequilibrium parameter is
Sire is homozygote at one locus and heterozygote at the other
A full and a reduced model are developed in this section. A full model estimates all unknowns (linkage disequilibrium, and allele frequencies for the marker for which the sire is heterozygote) simultaneously. The reduced model estimate only linkage disequilibrium assuming that allele frequencies are known without error (or estimated in a previous step).
Full model for estimating LD in a homo-heterozygote sire:
Let the sire have genotype TTMm at two SNPs, T/t, and M/m. The likelihood equation for the ith family is
| (1) |
where nj,i are the genotype counts (nG) from offspring from the ith sire family (j = TTMM, TTMm, TTmm, TtMM, TtMm, and Ttmm), and is the probability of the jth genotype among progeny. These probabilities can be obtained after adding the corresponding frequencies for all possible matings (Table 1): ; ; ; ; ; . Equation 1 can be solved by the EM algorithm after making haplotype frequencies equal to their expected values,
| (2) |
where Ni is the size of the ith half-sib family. Equations 2 can be solved iteratively after giving a starting value to the haplotype frequencies and by estimating in each iteration . The starting values used in this study were the product of allele frequencies, so disequilibrium was null (δ = 0).
Table 1. Genotypes in the half-sib offspring from all possible gamete combinations produced from a heterozygote sire at one SNP, M/m, and homozygote at the other SNP, T/t.
| Sire(TM/Tm) | ||
|---|---|---|
| Dam | TM | Tm |
| G freq | 1/2 | 1/2 |
| TM fTM | TTMM | TTMm |
| fTM | fTM | |
| Tm fTm | TTMm | TTmm |
| fTm | fTm | |
| tM ftM | TtMM | TtMm |
| ftM | ftM | |
| Tm ftm | TtMm | Ttmm |
| ftm | ftm | |
G, gametes; freq, frequency.
Reduced model for estimating LD in a homo-heterozygote sire family:
In a reduced model, allele frequencies are not estimated simultaneously with haplotype frequencies but are assumed to be known. The estimate of linkage disequilibrium is
where ,
and
The derivation is given in Appendix B.
The disequilibrium estimated in the reduced model gives slightly different estimates than the disequilibrium estimated using a full model but has the advantage of faster computation when a large number of SNPs are tested. The approximated sampling variance of the estimates of the disequilibrium parameter for the ith family is
where
as derived in Appendix A.
Sire is heterozygote at two SNPs
Equations for a full and a reduced model follow. A full model estimates allele and haplotype frequencies simultaneously whereas a reduced model works first estimating allele frequencies and then haplotype frequencies. The full model has better statistical properties but the reduced model has faster computation and, therefore, is practical for large-scale testing of disequilibria among SNPs.
Full model for estimating LD in a double-heterozygote sire family:
Let the sire have genotype TtMm at two SNPs, T/t, and M/m and linkage phase (TM/tm). As before, nj,i are the genotype counts from offspring from the ith sire family (j = TTMM, TTMm, TTmm, TtMM, TtMm, Ttmm, ttMM, ttMm, and ttmm). The recombination fraction is c, which is assumed to be known without error. The likelihood equation for data of the ith half-sib family is
| (3) |
where the probabilities of offspring genotypes among half-sib offspring are obtained from Table 2: ; ; ; ; ; ; ; ; and .
Table 2. Genotypes and their frequencies among half-sib progeny from a double heterozygote sire.
| Sire (phase TM/tm) | ||||
|---|---|---|---|---|
| Dam | TM | Tm | tM | tm |
| G freq | (1 − c) | c | c | (1-c) |
| TM fTM | TTMM | TTMm | TtMM | TtMm |
| (1 − c) fTM | c fTM | c fTM | (1 − c) fTM | |
| Tm fTm | TTMm | TTmm | TtMm | Ttmm |
| (1 − c) fTm | c fTm | c fTm | (1 − c) fTm | |
| tM ftM | TtMM | TtMm | ttMM | ttMm |
| (1 − c) ftM | c ftM | c ftM | (1 − c) ftM | |
| tm ftm | TtMm | Ttmm | ttMm | ttmm |
| (1 − c) ftm | c ftm | c ftm | (1 − c) ftm | |
| Sire (phase Tm/tM) | ||||
| TM | Tm | tM | tm | |
| G freq | c | (1-c) | (1-c) | c |
| TM fTM | TTMM | TTMm | TtMM | TtMm |
| c fTM | (1-c)fTM | (1 − c)fTM | c fTM | |
| Tm fTm | TTMm | TTmm | TtMm | Ttmm |
| fTm | (1 − c)fTm | (1 − c)fTm | c fTm | |
| tM ftM | TtMM | TtMm | ttMM | ttMm |
| c ftM | (1 − c)ftM | (1 − c)ftM | c ftM | |
| tm ftm | TtMm | Ttmm | ttMm | ttmm |
| c ftm | (1 − c)ftm | (1 − c)ftm | c ftm | |
G, gametes; freq, frequency.
Likelihood Equation 3 can be solved by applying the EM algorithm,
| (4) |
where, as before, Ni is the size of the ith half-sib family. Using initial values of the haplotype frequencies and iterating over Equation 4 will converge to ML estimates of haplotype frequencies. Linkage disequilibrium is estimated by .
If the linkage phase of the sire is Tm/tM then the EM equations are
However, the same results can be obtained by making the following substitutions in Equation 4: by ; by ; by ; by ; by ; and by . Linkage phase can be estimated simultaneously to recombination fraction (Gomez-Raya 2001).
Reduced model for estimating LD in a double-heterozygote sire:
A reduced model can be used after assuming that allele frequencies at the two DNA markers are known without error. It makes easier and faster estimation of linkage disequilibrium and its sampling variance. It can be solved by making use of the EM algorithm as described in Equation 4 but using as input parameters estimates of allele frequencies of M and T (as given by Gomez-Raya 2001):
A solution when c = 0 for the reduced model is a positive root between 0 and 1 of the quadratic: , where , , and . Derivation of the method and an explicit solution for fully linked markers is given in Appendix B.
As shown in Appendix A, the reduced model provides a simpler approximated sampling variance of the estimates of the disequilibrium parameter for the ith family by
This equation can be used as an approximation to the full model with linkage disequilibrium and allele frequencies estimated from that model.
Estimation of LD Across multiple half-sib families
In most instances, genotype information is available for multiple half-sib families (e.g., data from a granddaughter design project). The likelihood equation to estimate LD across half-sib families is
where L(δ, fT, fM|nG) is the likelihood for the ith half-sib family conditional to genotype marker information (nG) and nf is the number of families. Note that depending on the sire genotype, allele frequencies for T and M (double homozygote) or M (homo-heterozygote) do not need to be estimated. The EM algorithm can be applied to multiple families by iterating on the four haplotype frequencies:
| (5) |
where equations for haplotype frequencies for each single family varies depending on the sire genotype. For example,
$
are the equations for haplotype TM if the sire is double homozygote, homo-heterozygote, or double heterozygote, respectively. The frequencies for the other haplotypes are as found in Equations 2 and 4 for homo-heterozygote and double heterozygote sires, respectively. Equation 5 can be solved iteratively after giving a starting value to the haplotype frequencies and by estimating in each iteration and .
The estimation of the sampling variance for linkage disequilibrium in multiple half-sib families can be carried out by:
where second derivatives of the natural logarithm of likelihood varies depending on sire genotype (double homozygote, homo-heterozygote, and double heterozygote) as described in Appendix A.
Hypothesis testing of LD in multiple half-sib families
Testing if linkage disequilibrium is different from 0 can be carried out by a likelihood-ratio test. For the ith half-sib family the likelihood-ratio test is
where and are the likelihoods for the ith family under the null hypothesis (δ = 0) and under the alternative hypothesis with .
A likelihood-ratio test across families is
which is distributed as a χ2 with 1 d.f. Here δ is estimated across all families by the EM algorithm (Equation 5).
Bias in estimating LD in half-sibs after ignoring the family structure
In this section, approximate bias for estimating LD in half-sib families using the method of Excoffier and Slatkin (1995) for unrelated individuals and maternal informative haplotypes is derived algebraically. Only sires that are homo-heterozygotes and double heterozygotes might produce progeny in which haplotypes cannot be fully inferred from the genotypes.
Sire homo-heterozygote: Method of Excoffier and Slatkin (1995) for unrelated individuals:
Assuming genotype TTMm in the sire, the expected frequency of haplotype TM among half-sib progeny can be approximated by
where Ni comes from the contribution of the TM haplotype from the sire and Ni fTM from the contributions of the dams. The total number of haplotypes in the offspring is 2Ni. The approximated expected frequencies of alleles T and M are computed following the same rules:
The expected estimate of the disequilibrium after using the method of Excofier and Slatkin (1995) is
Consequently, the bias after using this method is approximated by
Sire homo-heterozygote: Estimation of LD using informative maternal haplotypes in half-sib families:
Half-sib progeny from heterozygote sires might not be informative. For example, haplotype TM inherited from dams will be informative only in progeny with genotypes TTMM.
Therefore, the expected frequency of haplotype TM among progeny will be estimated by
The estimation of haplotype frequencies and linkage disequilibrium is unbiased when the sire is a homo-heterozygote.
Sire double heterozygote: Method of Excoffier and Slatkin (1995) for unrelated individuals:
Assuming linkage phase TM/tm in the sire, the expected frequency of haplotype TM among half-sib progeny can be approximated by
where Ni(1 − c) and Ni fTM are the sire and dams contributions of haplotype TM among the offspring.
Similarly, the expected frequencies of alleles T and M are approximated by
The expected linkage disequilibrium is
Consequently, the bias for using the method of Excoffier and Slatkin (1995) for unrelated individuals is approximated by
Sire double heterozygote: Estimation of LD using informative maternal haplotypes in half-sib families:
The only informative haplotypes that can be traced up to their mothers are from progeny with genotypes TTMM, TTmm, ttMM, and ttmm. It is because markers are biallelic and only homozygote progeny can be used to trace inheritance when the sire is a heterozygote. If allele frequencies in the dam population are known, then determining the haplotype with the highest probability is feasible. Nevertheless, for intermediate allele frequencies the probability of inheriting either allele is 0.5. For the calculations below, only informative progeny is used.
Assuming linkage phase TM/tm, the expected frequency of informative TM haplotypes among progeny is
The expected values for the frequencies of alleles T and M are
The expected disequilibrium is
For unlinked loci, c = 0.5, the above expression reduces to DTM and the method of informative maternal haplotypes is unbiased.
For 0 < c < 0.5, the bias for using only maternal inherited haplotypes is approximated by
Monte Carlo computer simulation
A Monte Carlo computer simulation was carried out to validate methods for estimating LD proposed in this article as well as to compute power. Three scenarios were simulated corresponding to the three possible situations regarding the genotype of the sire: double homozygote, homo-heterozygote, and double heterozygote. In addition, a multifamily situation was also simulated.
Sire double homozygote:
A random generator from the uniform distribution was used to assign progeny with the haplotypes TM, Tm, tM, and tm according to their probability (frequency): , , , and , where the allele frequencies , , , , and δ were input parameters. If the drawing of the uniform distribution was between 0 and fTM, then the offspring inherited haplotype TM from his dam. If the drawing of the uniform distribution was between fTM and fTM+ fTm then the offspring inherited haplotype Tm from his dams. Assigning offspring to other haplotypes was done following the same rule.
Sire homo-heterozygote:
A random generator from the uniform distribution was used to assign progeny with the genotypes TTMM, TTMm, TTmm, TtMM, TtMm, and Ttmm according to their probability (frequency): , , , , , and . If the drawing of the uniform distribution was between 0 and , then the offspring had genotype TTMM. If the drawing of the uniform distribution was between and + then the offspring genotype was TTMm. Assigning other genotypes to offspring was done following the same rule.
Sire double heterozygote:
A random generator from the uniform distribution was used to assign progeny with the genotypes TTMM, TTMm, TTmm, TtMM, TtMm, Ttmm, ttMM, ttMm, and ttmm according to their probability (frequency): , , , , , , , , and . If the drawing of the uniform distribution was between 0 and , then the offspring had genotype TTMM. If the drawing of the uniform distribution was between and + then the offspring genotype was TTMm. Assigning other genotypes to offspring was performed following the same rule.
Subroutines in Fortran 90 were written to estimate linkage disequilibrium with the half-sib methods (HS) described in this article as well as the method of Excoffier and Slatkin (1995) (ES) for unrelated individuals, and by making use of maternal informative haplotypes (MIH). Family sizes of 36 and 500 were used to test the methods in small and large families. Empirical power was computed by sorting within each simulation set according to the likelihood-ratio estimate and finding the percentage of replicates that gave a value higher than the value of the χ2 with 1 d.f. at a significance level of 0.01.
Multifamily estimation of linkage disequilibrium:
A total of six families with sizes 94, 77, 106, 81, 79, and 100 half-sib progeny resembling the sire Norwegian cattle population were simulated (after pooling selected and culled bulls in Table 1 of Gomez-Raya et al. 2002). The allele frequencies were intermediate, recombination fraction was 0, 0. 25, or 0.50, and linkage disequilibrium ranged from 0 to 0.25. The sires were simulated as if they were coming from a population with the same linkage disequilibrium and allele frequencies as used to generate the half-sib progeny. To do so, the two haplotypes at each sire were generated following the same principles as above with probabilities according to the simulated frequencies: , , , and , in which allele frequencies , , , , and δ were input parameters. Thus, the sire could be a double homozygote, homo-heterozygote, or double heterozygote after assigning the two haplotypes. The half-sib progeny was generated as described in the previous section. Estimation of linkage disequilibrium was carried out using the EM algorithm for multiple families. Empirical power and overall likelihood-ratio test were computed for each simulation set. Each experiment was replicated 10,000 times. A Q-Q plot (using proc qqplot of SAS Inst., Cary, NC) was used to investigate the distribution of LRTjoint under the null hypothesis (simulated δ = 0) in the situation for c = 0.
Genome analyses of LD in a beef cattle half-sib family
A half-sib family consisting of 36 calves from commercial beef cattle at the Gund Ranch in Nevada was used to illustrate and to compare alternative methods for estimation of linkage disequilibrium. The first step was to determine paternity of the calves at the ranch. A set of 25 microsatellites (BMS410, BMS499, BMS650, BMS1244, BMS1634, TGLA227, BMS601, BMS1789, BMS2005, ILSTS081, BMS1315, BMS1226, BMS2573, ILSTS058, TGLA126, CSSM66, SPS115, TGLA53, BM1824, BM2113, ETH3R, TGLA122, INRA023, ETH225, ETH10) was used to assign paternity that was carried out using Cervus software. Total DNA from ear notches of calves and sires was purified using the manufacturer’s instructions (Qiagen, CA). The DNA was diluted with AE buffer to 10 ng/μl and stored at −4° prior to genotyping. Primers were diluted to 50 μM and stored at −4°. A primer mix was prepared containing 2 μl of each 50 μM primer set. Each PCR reaction contained a total volume of 15 μl consisting of 1.5 μl of each primer mix, 2 μl water, 4 μl DNA, and 7.5 μl PCR multiplex mix (Qiagen). Gradients were performed to determine the optimal temperature for primer annealing. Amplification was carried out with a TC-512 Thermal Cycler (Techne). The initial denaturation step was performed at 95° for 15 min, followed by 35 cycles of 30 sec at 94°, 1 min and 30 sec at the optimum annealing temperature, and 1 min at 72° with a final extension of 30 min at 60°. Subsequently, 1 μl of PCR product was added to 199 μl water to make a 1:200 dilution. One microliter of this dilution was added to 10 μl of a formamide solution containing 1 ml formamide and 5 μl of ladder and denatured for 5 min at 95°. Genotyping was performed with the Applied Biosystems (ABI) Prism 3730 DNA analyzer.
The Illumina bovine 50K BeadChip was used with bull 302 and his 36 calves to compare methods for estimating LD in half-sib families. The genotyping was carried out at the Core Lab of the University of Colorado, Denver. Only SNPs with a call rate >0.80 in at least 24 calves and MAF of 0.10 or more were used. The data were also filtered for SNPs that were not consistent for inheritance from sire to progeny. If a SNP was not consistent for one progeny then the SNP information was discarded for the entire family. Only pairs of SNPs within the same chromosome and within a distance of 50 Mb or less were used for estimating linkage disequilibrium. For the double heterozygote sire, recombination fraction and linkage phase was estimated using the methods proposed by Gomez-Raya (2001). Only SNPs with a recombination fraction of 0.30 or less were used for SNPs in which the sire was double heterozygote. Estimation of disequilibrium was performed using the half-sib method as well as the method of Excoffier and Slatkin (1995) and by making use of maternal informative haplotypes. For comparison of alternative estimation methods of linkage disequilibrium the statistic r2 = δ2/{(fT (1 − fT)fM (1 − fM)} was used. This statistic is widely used and ranges from 0 to 1, which facilitates comparison among methods. The absolute value of the difference between estimates of either ES or MIH and estimates HS were also used to evaluate discrepancies between methods.
Results
Table 3 shows simulation results for estimating linkage disequilibrium in a half-sib family from a homo-heterozygote sire with 36 or 500 progeny and dam allele frequencies of 0.5 at both SNPs. For these allele frequencies, the maximum possible linkage disequilibrium, δ, is 0.25. The method proposed in Equation 3 of this article (HS) yields identical estimates to the true (simulated) values of linkage disequilibrium with large family size (500). There was very little bias when the family size is small (36). For the examples in Table 3, the bias using the HS method is <3% . On the other hand, estimates are biased when using the method of Excoffier and Slatkin (1995), which becomes just half of the true disequilibrium at δ = 0.25. The approximation for predicting the expected value for the estimates of linkage disequilibrium using the method of Excoffier and Slatkin (1995) agreed well with the simulation results but tends to underestimate it. On the other hand, the use of maternal informative haplotypes is unbiased, as shown in Table 3 and as proven in the corresponding section of this article.
Table 3. Average estimates of δ in a half-sib family from a homo-heterozygote sire (family size = 36 or 500) with simulated and and varying linkage disequilibrium (δ).
| Estimation method | |||||||
|---|---|---|---|---|---|---|---|
| HS | ES | MIH | |||||
| Family size: Simulated δ | 36 | 500 | 36 | 500 | E(ES) | 36 | 500 |
| 0.000 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 |
| 0.025 | 0.025 | 0.025 | 0.017 | 0.018 | 0.013 | 0.025 | 0.025 |
| 0.050 | 0.049 | 0.050 | 0.034 | 0.036 | 0.025 | 0.050 | 0.050 |
| 0.075 | 0.074 | 0.075 | 0.049 | 0.051 | 0.038 | 0.074 | 0.075 |
| 0.100 | 0.098 | 0.100 | 0.063 | 0.064 | 0.050 | 0.098 | 0.100 |
| 0.125 | 0.122 | 0.125 | 0.075 | 0.077 | 0.063 | 0.122 | 0.125 |
| 0.150 | 0.146 | 0.150 | 0.087 | 0.088 | 0.075 | 0.146 | 0.150 |
| 0.175 | 0.170 | 0.175 | 0.097 | 0.098 | 0.087 | 0.170 | 0.175 |
| 0.200 | 0.195 | 0.200 | 0.106 | 0.107 | 0.100 | 0.194 | 0.200 |
| 0.225 | 0.218 | 0.225 | 0.115 | 0.116 | 0.113 | 0.218 | 0.225 |
| 0.250 | 0.243 | 0.250 | 0.123 | 0.125 | 0.125 | 0.249 | 0.250 |
The number of replicates was 104. HS, Average estimates using the method derived for half-sibs in this article. ES, Average estimates over replicates of linkage disequilibrium using the algorithm of Excoffier and Slatkin (1995). E(ES), Predicted LD using the method of not family structure using the algorithm of Excoffier and Slatkin (1995). MIH, Method of maternal informative haplotypes.
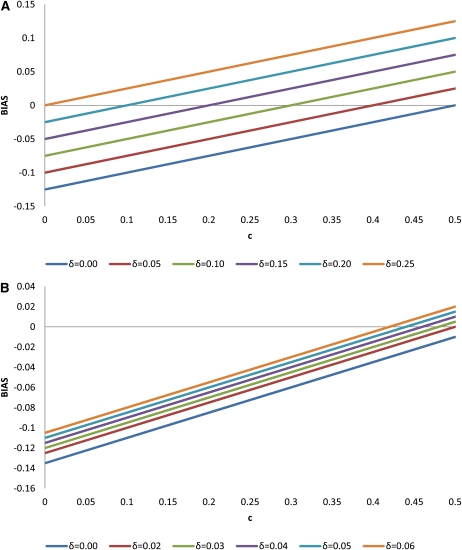
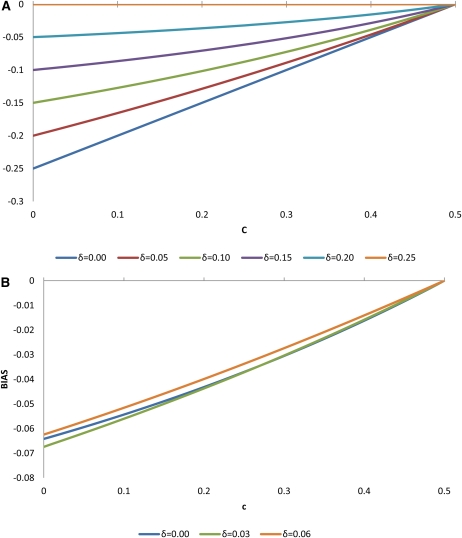
Table 4 shows simulation results for estimating linkage disequilibrium in a half-sib family from a double heterozygote sire for varying recombination fractions and linkage disequilibrium parameters. The allele frequencies at the two loci were 0.5. Each simulation set was analyzed with the EM algorithm developed in this article (HS) as well as the method of ES and by using MIH from dams. The HS method is asymptotically unbiased with average estimates of disequilibria very close to the simulated (true) parameters for large family sizes (500). For small family sizes (36) the estimates of linkage disequilibrium are slightly downward biased. The method of Excoffier and Slatkin (1995) is severely biased upward at low recombination fractions but becomes biased downward at high recombination fractions. The use of only maternal informative haplotypes to estimate disequilibrium is upward biased at low recombination fractions but becomes unbiased when the markers are unlinked. The approximated expected disequilibrium was very close to what was observed in the simulation for both the method of Excoffier and Slatkin (1995) and when using informative maternal haplotypes from dams. Figures 1 and 2 show expected bias in estimating disequilibrium in a half-sib family from a double heterozygote sire for two scenarios regarding allele frequencies at the DNA markers: fT = 0.5, fM = 0.5 and fT = 0.4, fM = 0.1. For a low recombination fraction, bias is negative but becomes positive as recombination fraction increases. The effect is more pronounced for loci at intermediate allele frequencies than for loci with allele frequencies closer to fixation.
Table 4. Average estimates of δ in a half-sib family from a double heterozygote sire (family size = 36 and 500) with simulated and and varying recombination fraction (c) and linkage disequilibrium (δ).
| Family size | Simulated c | ||||||
|---|---|---|---|---|---|---|---|
| 0 | 0.25 | 0.50 | |||||
| Simulated δ | 36 | 500 | 36 | 500 | 36 | 500 | |
| 0.000 | HS | −0.004 | −0.000 | −0.000 | 0.000 | 0.000 | 0.000 |
| ES | 0.106 | 0.108 | 0.057 | 0.059 | 0.000 | 0.000 | |
| E(ES) | 0.125 | 0.063 | 0.000 | ||||
| MIH | 0.220 | 0.250 | 0.110 | 0.125 | 0.000 | 0.000 | |
| E(MIH) | 0.250 | 0.125 | 0.000 | ||||
| 0.100 | HS | 0.092 | 0.099 | 0.094 | 0.099 | 0.094 | 0.100 |
| ES | 0.166 | 0.169 | 0.110 | 0.112 | 0.047 | 0.048 | |
| E(ES) | 0.175 | 0.113 | 0.050 | ||||
| MIH | 0.229 | 0.249 | 0.186 | 0.187 | 0.088 | 0.100 | |
| E(MIH) | 0.250 | 0.188 | 0.100 | ||||
| 0.200 | HS | 0.188 | 0.199 | 0.183 | 0.199 | 0.194 | 0.199 |
| ES | 0.221 | 0.224 | 0.158 | 0.161 | 0.088 | 0.090 | |
| E(ES) | 0.225 | 0.163 | 0.100 | ||||
| MIH | 0.234 | 0.249 | 0.213 | 0.231 | 0.176 | 0.200 | |
| E(MIH) | 0.250 | 0.232 | 0.200 | ||||
The number of replicates was 104. HS, Average estimates using the method derived for half-sibs in this article. ES, Average estimates over replicates of linkage disequilibrium using the algorithm of Excoffier and Slatkin (1995). E(ES), Predicted LD using the method of not family structure using the algorithm of Excoffier and Slatkin (1995). MIH, Method of Maternal Informative Haplotypes. E(MIH), Predicted LD using the method of Maternal Informative Haplotypes.
Figure 1.
Bias in the estimation of LD using the Excoffier and Slatkin (1995) algorithm in a half-sib family from a double heterozygote sire. (A) , , (B) ,
Figure 2.
Bias in the estimation of LD using maternal haplotypes in a half-sib family from a double heterozygote sire. (A) , , (B) , .
In many instances, interest is on the amount of progeny needed for detecting linkage disequilibrium. Empirical power for half-sib families in which the sire was a double homozygote, homo-heterozygote, or double heterozygote is shown in Table 5. The standard deviations among replicates for the same simulation sets are given in Table 6. The simulation results are for varying family sizes and true (simulated) linkage disequilibrium parameters. Disequilibrium (δ) of 0.10 was detected with groups of 100 offspring in most situations. The most powerful situation is when the sire is a homozygote at two loci and all haplotypes are informative. Power in a double heterozygote sire family reduces with genetic distance but it is nearly as powerful as the double homozygote for fully linked loci. Power in a homo-heterozygote sire family is always lower than power in a double homozygote sire family. Standard deviation among replicates follows the same trend as power (Table 6). The double homozygote and the double heterozygote (at c = 0) sire families had the lowest variation (Table 6). Variation among replicates increases with increasing recombination fraction in double heterozygote families. The estimates of disequilibrium for homo-heterozygote had more variation than double homozygote families. There was good agreement between the observed standard deviation among replicates and the average of the estimates of sampling standard deviations of δ obtained in each replicate.
Table 5. Empirical power for estimation of LD in a half-sib families from a double homozygote, homo-heterozygote and a double heterozygote sire for varying family sizes.
| Hetero-hetero: Simulated c | ||||||
|---|---|---|---|---|---|---|
| Simulated δ | Size | Homo-homo | Homo-hetero | 0 | 0.25 | 0.50 |
| 0 | 100 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 |
| 0.025 | 100 | 0.06 | 0.04 | 0.05 | 0.03 | 0.02 |
| 200 | 0.12 | 0.06 | 0.11 | 0.05 | 0.03 | |
| 500 | 0.37 | 0.16 | 0.36 | 0.13 | 0.07 | |
| 1000 | 0.72 | 0.37 | 0.71 | 0.31 | 0.16 | |
| 0.050 | 100 | 0.29 | 0.13 | 0.27 | 0.11 | 0.07 |
| 200 | 0.60 | 0.30 | 0.59 | 0.24 | 0.13 | |
| 500 | 0.98 | 0.73 | 0.97 | 0.64 | 0.38 | |
| 1000 | 1.00 | 0.97 | 1.00 | 0.94 | 0.73 | |
| 0.100 | 100 | 1.00 | 0.64 | 0.93 | 0.51 | 0.32 |
| 200 | 1.00 | 0.93 | 1.00 | 0.87 | 0.63 | |
| 500 | 1.00 | 1.00 | 1.00 | 1.00 | 0.97 | |
| 1000 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
The allele frequencies were and . The significance level was 0.01. The number of replicates was 104.
Table 6. Standard deviation among replicates in the estimation of LD in half-sib families from double homozygote, homo-heterozygote and double heterozygote sire for varying family sizes and and .
| Hetero-hetero: Simulated c | ||||||
|---|---|---|---|---|---|---|
| Simulated δ | Size | Homo-homo | Homo-hetero | 0 | 0.25 | 0.50 |
| 0 | 100 | 0.025 (0.025) | 0.035 (0.035) | 0.025 (0.025) | 0.038 (0.038) | 0.052 (0.049) |
| 0.025 | 100 | 0.024 (0.025) | 0.035 (0.034) | 0.025 (0.025) | 0.038 (0.038) | 0.052 (0.049) |
| 200 | 0.017 (0.018) | 0.025 (0.025) | 0.018 (0.017) | 0.027 (0.027) | 0.035 (0.035) | |
| 500 | 0.011 (0.011) | 0.016 (0.016) | 0.011 (0.011) | 0.017 (0.017) | 0.023 (0.022) | |
| 1000 | 0.008 (0.008) | 0.011 (0.011) | 0.008 (0.008) | 0.012 (0.012) | 0.016 (0.016) | |
| 0.050 | 100 | 0.024 (0.025) | 0.035 (0.034) | 0.024 (0.024) | 0.038 (0.037) | 0.051 (0.048) |
| 200 | 0.017 (0.017) | 0.025 (0.024) | 0.017 (0.017) | 0.027 (0.027) | 0.035 (0.034) | |
| 500 | 0.011 (0.011) | 0.016 (0.016) | 0.011 (0.011) | 0.017 (0.017) | 0.022 (0.022) | |
| 1000 | 0.008 (0.008) | 0.011 (0.011) | 0.008 (0.008) | 0.012 (0.012) | 0.015 (0.015) | |
| 0.100 | 100 | 0.023 (0.021) | 0.033 (0.031) | 0.023 (0.022) | 0.038 (0.037) | 0.048 (0.045) |
| 200 | 0.016 (0.016) | 0.023 (0.022) | 0.016 (0.016) | 0.027 (0.026) | 0.033 (0.032) | |
| 500 | 0.010 (0.010) | 0.015 (0.014) | 0.010 (0.010) | 0.017 (0.017) | 0.021 (0.020) | |
| 1000 | 0.007 (0.007) | 0.010 (0.010) | 0.007 (0.007) | 0.012 (0.012) | 0.015 (0.014) | |
The number of replicates was 104. Values between brackets are average of the estimates of sampling standard deviations of δ obtained in each replicate.
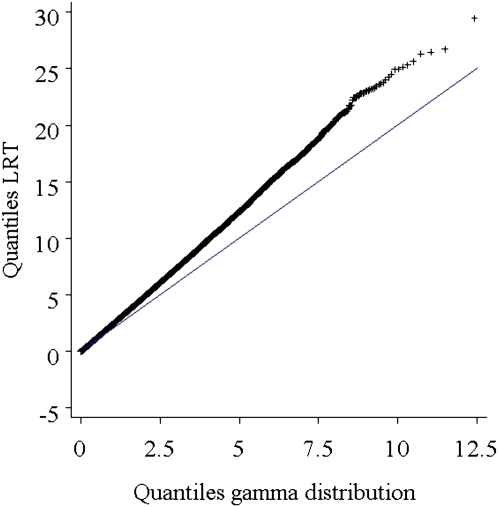
The simulation results for estimating LD using multiple sire families are given in Table 7. There was good agreement between simulated and estimated linkage disequilibrium across the range of simulated disequilibrium parameter and recombination fraction. The range of the absolute difference between estimated and simulated δ was between 0.000 and 0.008. A Q-Q plot of the quantiles of the observed distribution of LRTjoint under the null hypothesis against quantiles from a γ-distribution with shape = 0.5 and scale = 2 is depicted in Figure 3. This gamma distribution is a χ2-distribution with 1 d.f. The cumulative distribution of LRTjoint showed larger variation than a χ2-distribution with 1 d.f.
Table 7. Average estimates of linkage disequilibrium (δ) using the EM algorithm for multiple half-sib families together with statistical power at significance level of 0.01.
| Simulated c | ||||||
|---|---|---|---|---|---|---|
| 0 | 0.25 | 0.50 | ||||
| Simulated δ | δ | Power | δ | Power | δ | Power |
| 0.000 | 0.0000 (0.013) | 0.02 | −0.0001 (0.014) | 0.02 | −0.0001 (0.014) | 0.02 |
| 0.010 | 0.0100 (0.013) | 0.05 | 0.0099 (0.014) | 0.05 | 0.0099 (0.014) | 0.05 |
| 0.020 | 0.0201 (0.013) | 0.20 | 0.0201 (0.014) | 0.17 | 0.0200 (0.014) | 0.16 |
| 0.030 | 0.0301 (0.013) | 0.45 | 0.0301 (0.014) | 0.37 | 0.0301 (0.014) | 0.34 |
| 0.040 | 0.0401 (0.013) | 0.73 | 0.0402 (0.014) | 0.62 | 0.0401 (0.014) | 0.58 |
| 0.050 | 0.0501 (0.012) | 0.91 | 0.0502 (0.014) | 0.83 | 0.0501 (0.014) | 0.79 |
| 0.075 | 0.0751 (0.012) | 1.00 | 0.0751 (0.013) | 0.99 | 0.0752 (0.013) | 0.99 |
| 0.100 | 0.1002 (0.013) | 1.00 | 0.1003 (0.013) | 1.00 | 0.1003 (0.013) | 1.00 |
| 0.125 | 0.1252 (0.010) | 1.00 | 0.1256 (0.012) | 1.00 | 0.1255 (0.012) | 1.00 |
| 0.150 | 0.1502 (0.009) | 1.00 | 0.1506 (0.011) | 1.00 | 0.1506 (0.011) | 1.00 |
| 0.175 | 0.1753 (0.008) | 1.00 | 0.1758 (0.010) | 1.00 | 0.1758 (0.010) | 1.00 |
| 0.200 | 0.1997 (0.007) | 1.00 | 0.2003 (0.008) | 1.00 | 0.2002 (0.008) | 1.00 |
| 0.250 | 0.2457 (0.015) | 1.00 | 0.2443 (0.013) | 1.00 | 0.2453 (0.017) | 1.00 |
The simulated allele frequencies were and . The simulation was carried out for varying recombination fractions (c), linkage disequilibria (δ) and resembling the Norwegian cattle population structure. The number of replicates was 104. The values between brackets are the average of standard deviations of the estimates of δ.
Figure 3.
Q-Q plots of likelihood-ratio test using the multi-half EM algorithm on 106 replicates. Quantiles LRT are the quantiles from simulated data for c = 0 and under the null hypothesis (δ = 0).
Linkage disequilibrium was also estimated in a cattle half-sib family using the Illumina 50K BeadChip. There were 0.00189% inconsistencies between genotypes of sire and calves. Table 8 shows the overall estimates of r2 using HS, ES, and MIH for those situations in which the sire was a homo-heterozygote. There were 314,730 SNP pairs for the entire autosomal genome with average estimates of r2 of 0.115, 0.067, and 0.111 for HS, ES, and MIH methods. The ES method is downward biased since estimates by this method were around half of their value using the half-sib method. The maternal informative haplotype estimates were slightly lower than those obtained by the half-sib method, which might be due to bias because of reduced family size (noninformative offspring is neglected from these analyses).
Table 8. Overall values for estimates of and abs( – ) and abs( – ) for pairs of SNPs for which the sire was homo-heterozygote using alternative methods of estimation: (half-sib), (Excoffier and Slatkin 1995), and (maternal informative haplotypes).
| Chromosome | No. of pairs | abs (− ) | abs (− ) | |||
|---|---|---|---|---|---|---|
| 1 | 23181 | 0.108 | 0.061 | 0.106 | 0.061 | 0.015 |
| 2 | 18656 | 0.108 | 0.067 | 0.105 | 0.062 | 0.016 |
| 3 | 13730 | 0.124 | 0.073 | 0.120 | 0.070 | 0.017 |
| 4 | 16086 | 0.115 | 0.068 | 0.112 | 0.062 | 0.014 |
| 5 | 11164 | 0.132 | 0.077 | 0.129 | 0.075 | 0.016 |
| 6 | 19674 | 0.129 | 0.076 | 0.126 | 0.075 | 0.017 |
| 7 | 11445 | 0.133 | 0.076 | 0.130 | 0.076 | 0.016 |
| 8 | 16238 | 0.111 | 0.062 | 0.107 | 0.061 | 0.015 |
| 9 | 14253 | 0.118 | 0.068 | 0.114 | 0.067 | 0.017 |
| 10 | 14402 | 0.097 | 0.057 | 0.093 | 0.053 | 0.012 |
| 11 | 9344 | 0.132 | 0.077 | 0.128 | 0.076 | 0.016 |
| 12 | 9914 | 0.100 | 0.061 | 0.096 | 0.055 | 0.014 |
| 13 | 11553 | 0.124 | 0.069 | 0.121 | 0.069 | 0.014 |
| 14 | 11315 | 0.121 | 0.067 | 0.117 | 0.069 | 0.015 |
| 15 | 10410 | 0.126 | 0.072 | 0.122 | 0.070 | 0.018 |
| 16 | 7770 | 0.113 | 0.067 | 0.109 | 0.063 | 0.014 |
| 17 | 9220 | 0.101 | 0.065 | 0.098 | 0.056 | 0.013 |
| 18 | 8359 | 0.108 | 0.066 | 0.104 | 0.059 | 0.015 |
| 19 | 9025 | 0.105 | 0.065 | 0.100 | 0.060 | 0.015 |
| 20 | 6660 | 0.109 | 0.063 | 0.105 | 0.059 | 0.014 |
| 21 | 7337 | 0.104 | 0.060 | 0.099 | 0.059 | 0.014 |
| 22 | 6070 | 0.117 | 0.068 | 0.114 | 0.066 | 0.015 |
| 23 | 6754 | 0.106 | 0.064 | 0.103 | 0.061 | 0.016 |
| 24 | 8585 | 0.118 | 0.070 | 0.113 | 0.067 | 0.017 |
| 25 | 7735 | 0.123 | 0.071 | 0.119 | 0.071 | 0.016 |
| 26 | 6339 | 0.115 | 0.069 | 0.111 | 0.064 | 0.017 |
| 27 | 5468 | 0.114 | 0.063 | 0.110 | 0.062 | 0.014 |
| 28 | 7000 | 0.104 | 0.065 | 0.101 | 0.060 | 0.013 |
| 29 | 7043 | 0.101 | 0.059 | 0.097 | 0.057 | 0.013 |
| Overall | 314730 | 0.115 | 0.067 | 0.111 | 0.065 | 0.015 |
abs, the absolute value of the difference.
Table 9 shows overall estimates of r2 using HS, ES, and MIH for SNPs for which the sire was a double heterozygote. There were 208,872 SNP pairs. The results using real data support earlier findings showing that the methods of Excoffier and Slatkin (1995) and maternal informative haplotypes were upward biased. The average estimates of r2 across the genome were of 0.100, 0.267, and 0.925 for HS, ES, and MIH methods.
Table 9. Overall values for estimates of and abs( −) and abs( −) for pairs of SNPs for which the sire was double heterozygote using alternative methods of estimation: (half-sib), (Excoffier and Slatkin 1995), and (maternal informative haplotypes).
| Chromosome | No. of pairs | abs( −) | abs( −) | |||
|---|---|---|---|---|---|---|
| 1 | 19344 | 0.089 | 0.266 | 0.933 | 0.207 | 0.850 |
| 2 | 12033 | 0.092 | 0.269 | 0.945 | 0.207 | 0.859 |
| 3 | 8415 | 0.122 | 0.294 | 0.962 | 0.216 | 0.848 |
| 4 | 10439 | 0.097 | 0.273 | 0.919 | 0.212 | 0.828 |
| 5 | 7380 | 0.115 | 0.287 | 0.920 | 0.217 | 0.814 |
| 6 | 13930 | 0.120 | 0.282 | 0.928 | 0.214 | 0.817 |
| 7 | 8369 | 0.131 | 0.281 | 0.940 | 0.217 | 0.821 |
| 8 | 10948 | 0.096 | 0.269 | 0.950 | 0.210 | 0.862 |
| 9 | 9631 | 0.106 | 0.281 | 0.929 | 0.216 | 0.830 |
| 10 | 7212 | 0.080 | 0.258 | 0.900 | 0.199 | 0.824 |
| 11 | 5530 | 0.115 | 0.283 | 0.929 | 0.221 | 0.823 |
| 12 | 8049 | 0.080 | 0.248 | 0.916 | 0.198 | 0.844 |
| 13 | 7982 | 0.105 | 0.271 | 0.923 | 0.212 | 0.829 |
| 14 | 5828 | 0.111 | 0.265 | 0.923 | 0.207 | 0.823 |
| 15 | 5789 | 0.113 | 0.264 | 0.911 | 0.210 | 0.812 |
| 16 | 5286 | 0.095 | 0.255 | 0.898 | 0.202 | 0.813 |
| 17 | 6340 | 0.091 | 0.270 | 0.916 | 0.209 | 0.830 |
| 18 | 6798 | 0.103 | 0.266 | 0.909 | 0.200 | 0.812 |
| 19 | 5334 | 0.079 | 0.242 | 0.894 | 0.193 | 0.823 |
| 20 | 4127 | 0.088 | 0.258 | 0.959 | 0.204 | 0.878 |
| 21 | 4107 | 0.083 | 0.246 | 0.898 | 0.200 | 0.825 |
| 22 | 4146 | 0.103 | 0.269 | 0.932 | 0.214 | 0.838 |
| 23 | 5161 | 0.094 | 0.262 | 0.912 | 0.204 | 0.828 |
| 24 | 5244 | 0.089 | 0.270 | 0.934 | 0.213 | 0.853 |
| 25 | 4703 | 0.104 | 0.245 | 0.889 | 0.196 | 0.798 |
| 26 | 3970 | 0.098 | 0.260 | 0.910 | 0.203 | 0.821 |
| 27 | 4651 | 0.092 | 0.248 | 0.933 | 0.205 | 0.853 |
| 28 | 4214 | 0.078 | 0.252 | 0.913 | 0.204 | 0.841 |
| 29 | 3912 | 0.082 | 0.239 | 0.922 | 0.191 | 0.847 |
| Overall | 208872 | 0.100 | 0.267 | 0.925 | 0.208 | 0.834 |
abs, the absolute value of the difference.
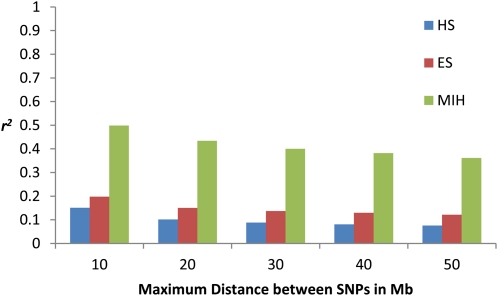
Figure 4 shows average estimates of r2 for the three methods of estimation across the entire genome when the distance between the two SNPs is between 10 and 50 Mb. A total of 829,042 SNPs pairs were tested and the estimates are a pool of all three possible situations regarding sire genotypes: double homozygote, homo-heterozygote, and double heterozygote. This figure shows again that using either ES or MIH methods give estimates upward biased of linkage disequilibria.
Figure 4.
Average values of estimates r2 across the genome using half-sib (HS), Excoffier and Slatkin (ES), and maternal informative haplotypes (MIH) methods for maximum distances between SNPs of 10, 20, 30, 40, and 50 Mb.
Discussion
Early studies estimating linkage disequilibrium in populations with a half-sib structure were carried out using microsatellites (Farnir et al. 2000; Odani et al. 2006). These studies used maternal alleles and estimated the most likely haplotype inherited in sons from dams. Although the methods derived in this article are for biallelic loci such as SNPs, a multiallelic marker can always be reduced to a biallelic one after pooling alleles into two groups. As shown by Gomez-Raya (2001) for a double heterozygote sire, the amount of informative progeny in a half-sib family depends on the recombination fraction between the two markers. Thus, the frequency of informative progeny is c([1 − ft)(1 − fM) + (1 − fT)(1 − fm)], and (1 − c)[(1 − ft)(1 − fm) + (1 − fT)(1 − fM)] for recombinants and nonrecombinants, respectively. Genotypes among offspring that are informative for tracing inheritance from sires are also informative for tracing alleles inherited from dams (with unknown genotypes). For example, for allele frequencies fT = fM = 0.1, the frequency of informative recombinant and nonrecombinant progeny is 0.18c and 0.82(1 − c), which means that the closer the markers are, the lower the proportion of informative recombinants among progeny. Therefore, bias in estimating linkage disequilibrium occurs because of the altered proportion of haplotypes that are informative at varying genetic distances. Nevertheless, Farnir et al. (2000) used not just the informative haplotypes but the most likely haplotype. Sires carrying alleles at low frequency would allow identification of haplotypes with a higher probability, which will reduce the magnitude of the bias as shown in this article. In another study, also investigating linkage disequilibrium with microsatellites in cattle, Tenesa et al. (2003) made use of the method of Excoffier and Slatkin (1995) to estimate linkage disequilibrium. As shown in this article, estimates of LD using that method for unrelated individuals might lead to severe biased estimation when applied in animals with a half-sib structure.
Improvement in the sequencing methods in the last years allowed for the discovery of vast amounts of SNPs in the human and animal genomes (e.g., International Hap Map Consortium 2007). A following step has been the construction of LD maps for the human (Maniatis 2002) and animal genomes (e.g., Khatkar et al. 2006). LD maps are based on: (a) estimation of a linkage disequilibrium parameter, ρ, which has the same maximum absolute value as the statistics D′ of Lewontin (1964), and (b) use of a model of decay of disequilibrium leading to equations of Malecot’s model for isolation by distance (Malecot 1964). Thus, the value of D′ is δ/DMax with DMax = min{ fT (1 − fM), fM (1 − fT)}. Construction of LD maps are carried out estimating ρ between adjacent SNPs and by using composite maximum likelihood for all pairs of adjacent SNPs. Inferences of the decay of disequilibrium over time are made by , where L is a parameter that reflects the residual association at a long distance (d), M is the association at zero distance, and ε is the exponential decline in LD due to recombination over generations. In human genetics, estimation of ρ is performed using unrelated individuals and the Excoffier and Slatkin (1995) algorithm. The construction of LD maps in species with a half-sib family structure like cattle would require methods for the estimation of disequilibrium (δ) as proposed in this article. If δ is biased then ρ should also be biased. If the bias depends upon the distance between the adjacent SNPs as shown here then inferences on population structure and the evolution of the cattle population may not be fully correct. Khatkar et al. (2006) carried out a LD map of bovine chromosome 6 using bulls from the Australian Holstein–Friesian. They estimated average coancestry by 0.012 using available pedigree information. Assuming that pedigrees were complete, coancestry was rather small but still might lead to bias in the estimation of the disequilibria currently present in the Australian dairy population.
The square of the correlation of alleles at two loci (r2) has been widely used in animals with a half-sib structure to estimate linkage disequilibrium (McKay et al. 2007; de Roos et al. 2008; Hayes et al. 2008; Prasad et al. 2008; Sargolzaei et al. 2008; Bovine Hap Map Consortium 2009; Kim and Kirkpatrick 2009; Qanbari et al. 2010). Most of these studies identify phased haplotypes using available information from pedigrees. Haplotypes that could not be phased out were generally ignored. As shown in this article, the proportion of haplotypes that are informative might vary with genetic distance leading to biased estimation of linkage disequilibrium, δ, which would also lead to biased estimates of r2. The magnitude of the bias depends on how much information from pedigrees can be used for phasing haplotypes and on the distances between the SNPs in the LD analyses. Many of the above studies made inferences about the population structure based on r2. However, estimates of r2 might be biased to a different extent for different cattle breeds having a different breeding structure (more or fewer half-sibs families of different sizes). Comparison of r2 estimated without consideration of the breeding structure in different animal populations should be taken with caution. On the other hand, inferences on past population sizes based on Sved’s (1971) equation E(r2) = (1 + 4Nec)−1 (where Ne is the effective population size) might also be inaccurate if r2 has been estimated in half-sib families neglecting noninformative haplotypes.
Assumptions of this study were that linkage phase of the sire and recombination fractions were known without error. The linkage phase can be accurately estimated using the same data if progeny groups are not small (>25) and recombination fraction is not too high (<0.30). For other situations, such as those arising by the use of SNP arrays, linkage phase in the sire can be inferred for each of two adjacent SNPs when they are apart at small distances. Reconstruction of haplotypes for all SNPs for each of the two homologous chromosomes of the sire is then feasible. In the same way, the assumption of known recombination fraction will hold for most situations found in practice when SNPs are adjacent, i.e., c = 0.
The methods developed in this article are for estimation of linkage disequilibrium present in the dam population and contributing to the half-sib progeny since sire haplotypes are ignored in the computations. In most circumstances, this disequilibrium is the most relevant since sires in dairy and beef cattle are likely related and the number of sire haplotypes is rather small. Nevertheless, if many sire families are available, then haplotype frequencies from sires and dams (estimated among half-sib progeny) can be pooled to obtain a joint estimate of linkage disequilibrium across sexes.
The results of the simulations showed that the proposed method for estimating disequilibrium works well for relatively small family sizes and in multifamily situations. The distribution of likelihood-ratio tests when simulating the null hypothesis showed that it had more variation than a χ2 with 1 d.f. This is because likelihood equations for multiple sire families make use of a different number of parameters depending on the sire genotype: double homozygote (δ), homo-heterozygote (δ and fT), and double heterozygote (δ, fT, and fM). In practical terms, resampling or simulation methods may be needed for hypothesis testing. The power figures for multifamily situations are also affected, being lower than those reported for this article.
The methods derived in this article were designed for estimating second-order linkage disequilibrium in half-sib families. The same methods and principles used in this article can be applied to the estimation of third- or higher-order linkage disequilibria. These methods may also be incorporated into a more general situation in which pedigrees are incomplete but much information comes from half-sib families. Nevertheless, if genotype information is available only from males (e.g., genotyping information from a granddaughter design), then little information may be gained by incorporating maternal grandsire in the estimation of haplotype frequencies.
The conclusion of this article is that estimation of linkage disequilibrium in populations with a breeding structure of half-sib families must incorporate that structure in their estimation to provide unbiased estimates of the linkage disequilibrium. Inferences on population structure and evolution of cattle or sheep should be based on linkage disequilibria after accommodating the existing half-sib family structure in these populations.
Acknowledgments
I am very grateful to Dr. David Thain and Jon Wilker for the cattle samples at the Gund ranch and to Veronica Kirchoff for the genotyping of microsatellites. Samples for DNA testing were taken from Nevada Agricultural Experiment Station project no. NEV05339, DNA Paternity Testing and Genetic Improvement of Free Range Beef Cattle. The author is also grateful to Luis Alberto Garcia Cortes and Wendy M. Rauw for criticism of the manuscript. I thank two anonymous reviewers for suggestions that improved the manuscript.
Appendix A: Sampling Variances of the Estimates of Linkage Disequilibrium
The sampling variance of the estimates of the disequilibrium parameter for the ith family is
The denominator of this equation is obtained by taking the second derivative respect to δ for each likelihood equation, which depends on the sires’s genotype.
Sire Double Homozygote
The genotype of the sire is TTMM. To obtain an estimate of the sampling variance it is better to use a full-likelihood equation in which all sources of information are used to estimate δ,
where nTM,i, nTm,i, ntM,i, and ntm,i are the number of offspring inheriting haplotypes TM, Tm, tM, and tm and K is a constant.
Taking natural logarithms in the above equation after ignoring the constant, K, gives The first two derivatives with respect to δ of this equation are
Sire Homo-heterozygote
Let the sire have genotype TTMm at two SNPs, T/t, and M/m. The likelihood equation for the ith family is
where are the probabilities of the jth genotype as described in the text. Ignoring the constant and taking natural logarithm of the above expression gives
The first two derivatives of the above equation with respect to δ are
Note that counts of heterozygous offspring for the marker M/m are not used and, therefore, do not provide information for estimating disequilibrium.
Sire Double Heterozygote
Let the sire have genotype TtMm at two SNPs, T/t, and M/m and linkage phase (TM/tm). As before, nj,i are the genotype counts from offspring from the ith sire family (j = TTMM, TTMm, TTmm, TtMM, TtMm, Ttmm, ttMM, ttMm, and ttmm). The recombination fraction is c, which is assumed to be known without error. The likelihood equation for data of the ith half-sib family is
where is the probability of the jth genotype as described in the text. In the reduced model, ignoring the constant and taking natural logarithm of the above expression gives
The first two derivatives of the above equation with respect to δ are
Appendix B
Reduced Model for Estimating LD in a Homo-heterozygote Sire Family
In a reduced model, allele frequencies are not estimated simultaneously with haplotype frequencies but are assumed to be known. This likelihood equation assuming known allele frequencies in the dam population is
Allele frequencies can be estimated using the same data following Gomez-Raya (2001) by
An explicit solution is obtained for the haplotype frequency of TM after rearranging Equation 2 (text):
The disequilibrium is estimated after substituting , , and into by .
Reduced Model for Estimating LD in a Double Heterozygote Sire
Following Gomez-Raya (2001), allele frequencies of M and T are estimated from the same data by
The maximum likelihood equation assuming known allele frequencies is
which can be solved by making use of the EM algorithm as described in Equation 4 in the text. For fully linked SNPs (e.g., contiguous SNPs in high density arrays) the recombination fraction between the SNPs is 0, and Equation 4 reduces to
After substituting by its value (recall ) and rearranging the above equation, a quadratic is obtained,
where , , and . This is a conventional second-order polynomial with a real solution between 0 and 1. Note that so .
Footnotes
Communicating Editor: H. Zhao
Literature Cited
- Barendse W., Vaiman D., Kemp S. J., Sugimoto Y., Armitage S. M., et al. , 1997. A medium density genetic linkage map of the bovine genome. Mamm. Genome 8: 21–28. [DOI] [PubMed] [Google Scholar]
- Bovine Hap Map Consortium, 2009. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science: 324: 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Y., Lewin H. A., 1995. Linkage information content and efficiency of full-sib and half-sib designs for gene mapping. Theor. Appl. Genet. 90: 699–706. [DOI] [PubMed] [Google Scholar]
- de Roos A. P. W., Hayes B. J., Spelman R. J., Goddard M. E., 2008. Linkage disequilibrium and persistence of phase in Holstein–Friesian, Jersey and Angus cattle. Genetics 179: 1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Slatkin M., 1995. Maximum likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol. 12: 921–927. [DOI] [PubMed] [Google Scholar]
- Farnir F., Coppiettiers W., Arranz J.-J., Berzi P., Cambisano N., et al. , 2000. Extensive genome-wide linkage disequilibrium in cattle. Genome Res. 10: 220–227. [DOI] [PubMed] [Google Scholar]
- Gomez-Raya L., 2001. Biased estimation of the recombination fraction using half-sib families and informative offspring. Genetics 157: 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Raya L., Olsen H. G., Klungland H., Våge D. I., Olsaker I., et al. , 2002. The use of genetic markers to measure genomics response to selection in livestock. Genetics 162: 1381–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B. J., Lien S., Nilsen H., Olsen H. G., Berg P., et al. , 2008. The origin of selection signatures on bovine chromosome 6. Anim. Genet. 39: 105–111. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium, 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes M. S., Keele J. W., Stone R. S., McGaw R. A., Sonstegard T. S., et al. , 1997. A second-generation linkage map of the bovine genome. Genome Res. 7: 235–249. [DOI] [PubMed] [Google Scholar]
- Khatkar M. S., Collins A., Cavanagh J. A., Hawken R. J., Hobbs M., et al. , 2006. A first-generation metric linkage disequilibrium map of bovine chromosome 6. Genetics 174: 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.-S., Kirkpatrick B. W., 2009. Linkage disequilibrium in the North American Holstein population. Anim. Genet. 40: 279–288. [DOI] [PubMed] [Google Scholar]
- Lewontin R. C., 1964. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 49: 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R. Z., Beever J. E., Da Y., Green C. A., Russ I., et al. , 1996. A male linkage map of the cattle (Bos taurus) genome. J. Hered. 87: 261–271. [DOI] [PubMed] [Google Scholar]
- Malecot G., 1964. Les mathématiques de l’hérédité. Masson, Paris. [Google Scholar]
- Maniatis N., Collins A., Xu C. F., McCarthy L. C., Hewett D. R., et al. , 2002. The first linkage disequilibrium (LD) maps: delineation of hot and cold blocks by diplotype analysis. Proc. Natl. Acad. Sci. USA 99: 2228–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay S. D., Schnabel R. D., Murdoch B. M., Matukumalli L. K., Aerts J., et al. , 2007. Whole genome linkage disequilibrium maps in cattle. BMC Genet. 8: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odani M., Narita A., Watanabe T., Yokouchi K., Sugimoto Y., et al. , 2006. Genome-wide linkage disequilibrium in two Japanese beef cattle breeds. Anim. Genet. 37: 139–144. [DOI] [PubMed] [Google Scholar]
- Prasad A., Mckay S. D., Murdoch B., Stothard P., Kolbehdari D., et al. , 2008. Linkage disequilibrium and signatures of selection on chromosomes 19 and 29 in beef and dairy cattle. Anim. Genet. 39: 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qanbari S., Pimentel E. C., Tetens J., Thaller G., Lichtner P., et al. , 2010. The pattern of linkage disequilibrium in German Holstein cattle. Anim. Genet. 41: 346–356. [DOI] [PubMed] [Google Scholar]
- Sargolzaei M., Schenkel F. S., Jansen G. B., Schaeffer L. R., 2008. Extent of linkage disequilibrium in Holstein cattle in North America. J. Dairy Sci. 91: 2106–2117. [DOI] [PubMed] [Google Scholar]
- Sved J., 1971. Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor. Popul. Biol. 2: 125–141. [DOI] [PubMed] [Google Scholar]
- Tenesa A., Knott S. A., Ward D., Smith D., Williams J. L., et al. , 2003. Estimation of linkage disequilibrium in a sample of the United Kingdom dairy cattle population using unphased genotypes. J. Anim. Sci. 81: 617–623. [DOI] [PubMed] [Google Scholar]
- Våge D. I., Olsaker I., Klungl H., Gomez-Raya L., Lien S., 2000. A male genetic map designed for QTL mapping in Norwegian cattle. Acta Agric. Scand. Sect. Anim. Sci. 50: 56–63. [Google Scholar]