Abstract
Great progress has been made in understanding the genetic architecture of phenotypic variation, but it is almost entirely focused on how the genotype of an individual affects the phenotype of that same individual. However, in many species the genotype of the mother is a major determinant of the phenotype of her offspring. Therefore, a complete picture of genetic architecture must include these maternal genetic effects, but they can be difficult to identify because maternal and offspring genotypes are correlated and therefore, partially confounded. We present a conceptual framework that overcomes this challenge to separate direct and maternal effects in intact families through an analysis that we call “statistical cross-fostering.” Our approach combines genotype data from mothers and their offspring to remove the confounding effects of the offspring’s own genotype on measures of maternal genetic effects. We formalize our approach in an orthogonal model and apply this model to an experimental population of mice. We identify a set of six maternal genetic effect loci that explain a substantial portion of variation in body size at all ages. This variation would be missed in an approach focused solely on direct genetic effects, but is clearly a major component of genetic architecture. Our approach can easily be adapted to examine maternal effects in different systems, and because it does not require experimental manipulation, it provides a framework that can be used to understand the contribution of maternal genetic effects in both natural and experimental populations.
MATERNAL effects occur when mothers have an indirect causal influence on the expression of traits in their offspring independent of genes passed from mothers to offspring (see Cheverud and Wolf 2009; Wolf and Wade 2009). These maternal effects, which arise from a diversity of factors such as maternally derived mRNA (Berleth et al. 1988), maternal provisioning (Bowen 2009), and maternally determined dispersal (Donohue 1998), have been shown to have important influences on offspring development across a diversity of taxa (Mousseau and Fox 1998; Maestripieri and Mateo 2009). The importance of maternal effects in evolution and ecology has become more broadly recognized (Mousseau and Fox 1998), where maternal effects have been shown to play a role in the evolutionary response to selection (see Kirkpatrick and Lande 1989), mate choice and sexual selection (Wolf et al. 1997, 1999), adaptive evolution (e.g., Badyaev et al. 2002), dynamics of population size (Ginzburg 1998), and niche construction (Odling-Smee et al. 2003). Genetically based maternal effects (“maternal genetic effects”) are of particular importance to many processes because they contribute to the genetic architecture of traits and can, as a result, contribute in nonintuitive ways to evolutionary change (Kirkpatrick and Lande 1989; Cheverud and Wolf 2009). For example, maternal genetic effects can contribute “hidden” variation that can allow for rapid evolution (e.g., Badyaev et al. 2002) and can be the cause of evolutionary time lags, constraints, or momentum (Kirkpatrick and Lande 1989). However, genetic studies of maternal effects have been limited because of the difficulty in applying experimental paradigms, such as cross-fostering, in both natural and experimental populations.
Maternal genetic effects are a form of an indirect genetic effect (Moore et al. 1997; Wolf et al. 1998), where genes in the mother have their phenotypic effect in her offspring. As a result, the phenotypic consequences of maternal genetic effects appear not in mothers, but in their offspring, where the phenotype of offspring is, to some degree, a “property” of the maternal genotype. Therefore, to study maternal genetic effects, one must be able to attribute phenotypic variation in offspring to genetic variation in their mothers, but because mothers and their offspring share half of their genes, the two effects are partially confounded. This confounding can make it difficult or sometimes impossible to distinguish the influence of the maternal genome from the influence of the offspring genome on the expression of offspring traits (but see Wolf et al. 2011).
Many approaches have been developed to overcome the confounding of direct and maternal effects, but existing approaches still suffer from limitations (see Roff 1997 for a review of empirical approaches to separating direct and maternal effects). For example, approaches that rely on asymmetrical patterns of resemblance of relatives in crossing schemes (e.g., Kever and Rotman 1987; Jarvis et al. 2005) cannot directly differentiate maternal effects from other potential causes of asymmetry, such as genomic imprinting (Hager et al. 2008) or uniparental inheritance such as cytoplasmic or sex chromosome inheritance (Wolf and Wade 2009). Other approaches, such as experimental cross-fostering (Bateman 1954; Cox et al. 1959; White et al. 1968), can detect maternal effects but cannot differentiate between maternal genetic and environmental sources unless a more complex design using related mothers is followed (Wilson et al. 2005; Wilson and Festa-Bianchet 2009).
More recently, genomic mapping approaches have been implemented to identify maternal genetic effects by mapping from the maternal genotype to offspring phenotypes. For example, Wolf et al. (2002) used means of cross-fostered litters to identify maternal effects and Casellas et al. (2009) mapped maternal effects in small chromosomal blocks in subcongenic mice by mapping maternal genotype to offspring phenotype using an unspecified linear model. Thus, despite a growing body of information accumulating on maternal effects in general, the technical challenges in design and analysis mean that we still have limited reliable data on maternal genetic effects, especially from populations where experimental manipulation is not possible or long-term pedigree data are not available (but exceptions exist, e.g., Wilson et al. 2005). To overcome these challenges Wolf et al. (2011) used a linear model approach in a population with cross-fostering to separate the independent effects of the maternal and offspring genotypes on the offspring phenotype. Although that approach was fruitful and identified a number of loci affecting prenatal and postnatal growth, it was limited by the fact that no formal statistical framework was available to explore the properties of the analysis or the nature of the results produced.
Here we develop a simple conceptual approach for the identification and characterization of maternal effects that uses genetic information from parents and offspring to achieve what we call “statistical cross-fostering.” This approach uses marker information from parents and their offspring to develop an analysis that accounts for the genomic autocorrelation of mothers and their offspring caused by Mendelian inheritance. These sorts of marker data are becoming widely available and therefore, implementing such an approach could be achieved in most systems. Because this approach does not require any experimental manipulation, it can be applied to natural populations, assuming that appropriate marker information is available. We illustrate this approach using data from an experimental population of mice. We develop a formal statistical model, but stress the conceptual basis for the approach, rather than the specific quantitative techniques used (which can be adapted to the specific nature of other data sets).
A Conceptual Framework for Studying Maternal Genetic Effects
We first present a conceptual model where we illustrate the confounding effects of the offspring and maternal genomes and then use this model to illustrate why cross-fostering allows for the separation and identification of maternal effects. We then illustrate how one can break the correlation between the maternal and offspring genomes without cross-fostering by using subsets of genotypes to achieve what we call statistical cross-fostering. This approach is then formalized into an orthogonal linear model and applied to an experimental population of mice.
We consider a hypothetical population where there are two alleles at a locus, A1 and A2, with frequencies p1 and p2, respectively. For simplicity, we assume that there is random mating and that these genotypes occur in Hardy–Weinberg proportions. Deviations from these assumptions will alter the specific results, but not the general conclusions that we emphasize. We assume that the locus can have a direct effect and a maternal effect. These effects can be characterized using the traditional additive and dominance genotypic values or effects (Falconer and Mackay 1996). Importantly, however, we define the direct and maternal effects in an idealized model where each is defined in the absence of the other effect. This is done because we wish to define a maternal effect as being attributable to the maternal genotype and a direct effect as being attributable the property of the offspring genotype. Because the two are confounded by relatedness, the presence of both effects makes it problematic to clearly separate direct effects from indirect maternal effects in the whole population when both are present. This is not a limitation of the model per se, but rather is a limitation imposed by the biology of maternal effects, and therefore we use the idealized model to illustrate this confounding and then examine how the two can be successfully separated.
For direct effects (in the absence of maternal effects), the additive effect (ao) is defined as half the difference between the mean phenotypes (the “genotypic values”) of the two homozygotes (A1A1 mean minus A2A2 mean), and the dominance effect (do) is defined as the deviation of the heterozygote mean from the midpoint between the two homozygotes (i.e., unweighted mean of the two classes) (see Falconer and Mackay 1996).
Maternal genetic effects (in the absence of direct effects) are defined by the average phenotype of the offspring of mothers of a particular genotype. The additive maternal effect (am) is defined as half the difference in the mean phenotypes of the offspring of the two types of homozygous mothers (again, the A1A1 mean minus A2A2 mean), while the dominance maternal effect (dm) is defined as the deviation of the mean phenotype of offspring that have heterozygous mothers from the midpoint between the mean phenotypes of the offspring that have homozygous mothers.
Maternal genetic effects in intact families
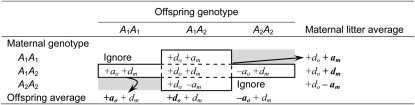
The idealized direct and maternal effects we have defined above can be used to characterize the phenotypes of offspring as a function of their own genotype and the genotype of their mother (see Table 1), where the offspring phenotype is the sum of the direct and maternal effects. In doing so, it is important to note that, because mothers homozygous for one allele cannot have offspring that are homozygous for the alternate allele, there are two maternal–offspring genotype combinations that cannot occur (Table 1). These missing combinations are wholly responsible for the confounding of maternal and offspring effects (Wade 1998) and hence are the main obstacle to studying maternal genetic effects.
Table 1. Expected offspring phenotype as a function of their own genotype and the genotype of their mother.
| Offspring genotype | ||||
|---|---|---|---|---|
| A1A1 | A1A2 | A2A2 | Maternal litter average | |
| Maternal genotype | ||||
| A1A1 | +am +ao | +am +do | — | +am + p2do + p1ao |
| A1A2 | +dm +ao | +dm +do | +dm –ao | +dm + do + ao(p1 – p2) |
| A2A2 | — | –am +do | –am –ao | –am + p1do – p2ao |
| Offspring average | +ao + p2dm + p1am | +do + dm + am(p1 – p2) | –ao + p1dm – p2am | |
Each cell gives the direct (subscripted “o”) and maternal (subscripted “m”) effects that contribute to the phenotype of an offspring as a function of the maternal–offspring genotype combination. The cells with dashes exist under Mendelian inheritance. The expected phenotypes of the offspring associated with each maternal genotype are given as row means and the average phenotype of each offspring genotype is given by the column means. The frequencies of each maternal–offspring genotype combination are shown in parentheses (Wade 1998).
Using the genotypic values of offspring defined as a function of the direct effect of their genotype and the maternal effect of their mother’s genotype, we can examine the mean phenotypes of offspring of the different genotypes or the means of offspring produced by the different types of mothers (these are the row and column means in Table 1). These mean phenotypes clearly demonstrate that the presence of a maternal effect is manifested in the average phenotypes associated with the offspring genotypes and, likewise, the direct effect is manifested in the average phenotypes of the offspring associated with the different maternal genotypes. For example, offspring with the A1A1 genotype have an average phenotype of +ao + p2dm + p1am, illustrating that the additive and dominance maternal effects contribute to the average phenotype of the A1A1 individuals, despite the fact that these effects do not arise from their own genotype, but their contribution is frequency dependent. This frequency dependence occurs because the expected phenotype of A1A1 individuals depends on the probability that they have A1A1 mothers (and hence experience the +am effect) vs. A1A2 mothers (and experience the +dm maternal effect) (see Cheverud and Wolf 2009). Likewise, the average phenotype of the offspring of the A1A1 mothers is +am + p2do + p1ao, showing the symmetrical confounding of direct effects with maternal effects.
These average phenotypes can be used to examine the apparent additive or dominance direct and maternal effects, where these are defined from the marginal means and include the confounding influence of the maternal and offspring genotypes. It is important to keep in mind that the direct (ao and do) and maternal (am and dm) effects are defined in the absence of the other effect, but here we ask, “What would the apparent direct or maternal effect be if both occur?” These are the values we would observe in a population if we measured individual phenotypes or the phenotypes of the offspring of a set of mothers and used them to infer direct and maternal effects, but did not account for the confounding. The apparent additive direct effect (ao(apparent)), calculated as half the difference between the A1A1 homozygote and the A2A2 homozygote (Table 1), would be
| (1) |
(Cheverud and Wolf 2009), which illustrates that the presence of maternal effects contributes to the apparent additive direct effect of the locus. Importantly, maternal effects do not contribute to the apparent dominance effect of the locus (calculated from the marginal means as the deviation of the A1A2 phenotypic mean from the unweighted mean phenotype of the A1A1 and A2A2 genotypes), and so the apparent dominance effect would simply be do, the true effect value. The apparent additive maternal effect am(apparent)), of the locus, when both maternal and direct effects occur, is
| (2) |
Like direct dominance effects, the apparent dominance maternal effect is not confounded with direct effects. Because of the confounding of direct and maternal additive genetic effects, an analysis focused on either direct or maternal effects would detect an apparent effect whenever either is present if the causal effects cannot be identified or controlled for, meaning that empirical analysis of either type of effect in the absence of consideration of the other may produce spurious results (Hager et al. 2008).
Analysis of maternal genetic effects with cross-fostering
The confounding of direct and maternal effects can be removed by cross-fostering offspring genotypes across maternal genotypes randomly, such that offspring experience the different maternal genotypes in proportion to their frequency in the population. This scheme is shown in Table 2, which illustrates the expected phenotypes of offspring with different direct and maternal effects, but where the maternal effect arises from the “foster” mother. Note that there are no empty cells in the cross-fostering design since the constraint of Mendelian inheritance has been removed. From the marginal means (mean phenotypes as a function of the direct or maternal genotype) it is clear that, although maternal effects contribute to the mean phenotype of the different offspring genotypes, they contribute the same value to all genotypes (i.e., they contribute am(p1 – p2) + 2p1p2dm to all offspring genotypes) and therefore do not appear to contribute a direct effect. Likewise, direct effects do not contribute to apparent maternal effects. As a result, the apparent additive direct effect is ao, and the apparent additive maternal effect is am.
Table 2. Expected offspring phenotype under random cross-fostering.
| Offspring genotype | ||||
|---|---|---|---|---|
| A1A1 | A1A2 | A2A2 | Maternal litter average | |
| Foster mother genotype | ||||
| A1A1 | +am +ao | +am +do | +am –ao | +am + ao(p1 – p2) + 2p1p2do |
| A1A2 | +dm +ao | +dm +do | +dm –ao | +dm + ao(p1 – p2) + 2p1p2do |
| A2A2 | –am +ao | –am +do | –am –ao | –am + ao(p1 – p2) + 2p1p2do |
| Offspring average | +ao + am(p1 – p2) + 2p1p2dm | +do + am(p1 – p2) + 2p1p2dm | –ao + am(p1 – p2) + 2p1p2dm | |
Each cell gives the direct (subscripted “o”) and maternal (subscripted “m”) effects that contribute to the phenotype of an offspring as a function of the genotype of the offspring and their foster mother. The expected phenotypes of the offspring of each foster mother genotype are given as row means and the average phenotype of each offspring genotype is given by the column means. The frequencies of each foster mother–offspring genotype combination under random cross-fostering are shown in parentheses below the effects.
It is clear that, by randomizing offspring genotypes across maternal genotypes, one can remove the issue of confounding and study maternal or direct effects without a concern for the other. However, this approach can identify only maternal effects that occur after the time of the cross-fostering and, therefore, many maternal effects will be missed because it may be very difficult or impossible to perform cross-fostering early enough in development, especially when there are complex connections between maternally and offspring-derived tissues (e.g., moving embryos prenatally in mammals or prior to hatching in species developing within eggs or disentangling embryos from maternal components of seeds) (but see Brumby 1960; Cowley et al. 1989; Atchley et al. 1991; Rhees et al. 1999 for exceptions). Furthermore, even when cross-fostering is technically possible, it may not be practical or even allowable when data come from natural populations where one cannot manipulate natural families. Nonetheless, this approach can be used to identify maternal genetic effects arising after the timing of the cross-fostering, such as postnatal effects in mammals (e.g., Wolf et al. 2002). However, it generally means that most information on maternal effects that is derived from cross-fostering schemes comes from the period of external development in species where offspring can be swapped between nests or litters (Price 1998; Maestripieri and Mateo 2009).
Analysis of maternal genetic effects through statistical cross-fostering
When one cannot implement random cross-fostering across maternal genotypes, it may be possible to use intact families and to achieve the statistical separation of direct and maternal effects, as long as marker data are available for both mothers and their offspring. Importantly, the critical issue with regard to the analysis of maternal effects in intact families is the confounding of maternal and offspring genotypes. This confounding cannot be removed without experimental manipulation, but we can use the mother–offspring classes that exist to separately infer direct and maternal effects. The key to this approach is the fact that heterozygous offspring can be produced by all maternal genotypes and heterozygous mothers can produce all types of offspring. Hence, when studying maternal effects one can limit the scope of inference to only heterozygous offspring at a locus and thereby produce a population where genetically variable mothers all have genetically identical offspring (again at that locus, but it is only at that locus that their genomes are confounded). This is illustrated in Figure 1, where the average phenotypes of the different offspring genotypes are simply the phenotypes of the offspring from heterozygous mothers and, likewise, the average phenotypes of the offspring associated with the different genotypic classes of mothers are simply the phenotypic values from the heterozygous offspring. Therefore, the phenotypes are not marginal means (because they are not averaged over different genotype classes), but rather are estimated by directly holding the other effect constant. From the values in Figure 1, it is clear that, as is the case for random cross-fostering across maternal genotypes, maternal effects can contribute to the mean phenotype of all genotypes of offspring, but they contribute the same value to all genotypes. The same is true for the mean phenotype of offspring produced by all genotypes of mothers, where direct effects contribute to the mean, but have the same contribution to all maternal genotypes. Therefore, neither contributes an apparent effect to the other. So the apparent direct effect is, as expected, ao and the apparent maternal effect is, as expected, am (and again, apparent dominance effects are correctly measured as do and dm).
Figure 1.
The confounded cells containing homozygous offspring from homozygous mothers are removed. The phenotypes of the offspring of heterozygous mothers are used as the estimates of the phenotype for each offspring genotype. Likewise, the phenotypes of heterozygous offspring are used as the measures of the phenotypes produced by each maternal genotype.
Thus, by removing the confounded genotype combinations from an analysis, one can construct an idealized analysis at a locus by using genetically identical offspring when testing for a maternal effect and genetically identical mothers when testing for a direct effect. When both are present, this analysis also allows one to identify pleiotropic effects, where a locus has both a direct and a maternal effect. Such pleiotropy may be expected on theoretical grounds (Wolf and Brodie 1998; Wolf 2001) and appears significant in many systems where it has been examined (generally appearing as a genetic correlation between direct and maternal effects) (Roff 1997).
Confounding effects of genomic imprinting
Hager et al. (2008) demonstrated that the presence of an additive maternal genetic effect could lead to the appearance of parent-of-origin–dependent effect (i.e., the scenario where the effect of an allele depends on which parent it is inherited from) that mimics genomic imprinting and, likewise, that the presence of a parent-of-origin–dependent effect caused by genomic imprinting could lead to the appearance of a pattern consistent with an additive maternal genetic effect when the genomic imprinting effect is not accounted for (Casellas et al. 2009; Cheverud and Wolf 2009). Here the parent-of-origin effect is simply defined by a difference in the average phenotypes of reciprocal heterozygotes (i.e., A1A2 and A2A1, with the first allele coming from the father and the second from the mother) (Wolf et al. 2008b). We assume that a true parent-of-origin effect results from genomic imprinting because the reciprocal heterozygotes differ in the parent of origin of the two alleles (it is therefore referred to as the “imprinting effect” hereafter). Again, the confounding of genomic imprinting with maternal effects occurs because the homozygous mothers produce only one of the two possible reciprocal heterozygotes (e.g., heterozygous offspring of A1A1 mothers must have received the A1 allele from their mother). This scenario is illustrated in Table 3.
Table 3. Expected offspring phenotype as a function of their own genotype and the genotype of their mother when there is a genomic imprinting effect.
| Offspring genotype | |||||
|---|---|---|---|---|---|
| A1A1 | A2A1 | A1A2 | A2A2 | Maternal litter average | |
| Maternal genotype | |||||
| A1A1 | +am + ao | +am + do – io | — | — | +am + aop1 + dop2 – iop2 |
| A1A2, A2A1 | dm + ao | dm+ do – io | dm+ do + io | dm – ao | + dm + do+ (ao + io) (p1 – p2) |
| A2A2 | — | — | –am+ do + io | –am – ao | – am – aop2 +dop1 + iop1 |
| Offspring average | amp1 + dmp2 + ao | amp1 + dmp2 +do – io | –amp2 + dmp1+do+ io | –amp2 + dmp1 – ao | |
The format matches Table 1, except that, for the offspring, the reciprocal heterozygotes are separated into two classes that differ in the parent of origin of their alleles (with the paternally inherited allele listed first). The cells with dashes are the ones that would be used in the identification of additive maternal effects in the statistical cross-fostering design (Figure 1). The imprinting effect in these cells is boxed in boldface type.
The imprinting effect is measured by the parameter io, which is defined as half the difference between the reciprocal heterozygote means, with the sign of the effect defined by the paternally inherited allele (Wolf et al. 2008b), meaning that io is defined as the mean of the A1A2 heterozygote (who gets the A1 allele from its father, which is the “plus” allele as defined by the sign of the additive effect above) minus the mean of the A2A1 heterozygote (who gets the A2 allele from its father). From the values in Table 3 it is clear that the heterozygous offspring produced by the A1A1 and A2A2 mothers (Table 3) differ by a factor of –2io. As a result, the additive maternal effect estimated using just these offspring would yield an estimated additive maternal effect of –io. Because this is the approach we advocate above for identifying maternal effects under the statistical cross-fostering design, the presence of genomic imprinting could result in the appearance of a nonexistent maternal effect. Therefore, when identifying maternal-effect loci using statistical cross-fostering, it is important to recognize that the apparent additive maternal effects (but not dominance) may actually be caused by genomic imprinting.
This confounding of maternal effects and genomic imprinting effects does not, however, represent an insurmountable problem because, by using genotype data from mothers and offspring, one can infer the phase of the reciprocal heterozygotes, and therefore, whenever statistical cross-fostering can be achieved, it should also be possible to differentiate between the reciprocal heterozygotes and estimate the imprinting effect. The imprinting effect can be analyzed the same way that additive and dominance effects are under the statistical cross-fostering design. The imprinting effect is estimated as the difference between the phenotypes of the reciprocal heterozygote offspring produced by heterozygous mothers. The phase of the alleles in the mothers does not need to be accounted for since genomic imprinting is reset each generation, and therefore both of the reciprocal heterozygote mothers can produce either of the reciprocal heterozygote offspring. This can be clearly seen in Table 3, where the expected phenotypes of the A2A1 and A1A2 heterozygotes produced by heterozygous mothers (the middle row of Table 3) are –io and +io, respectively. Using the offspring from heterozygous mothers, we would estimate the parent-of-origin effect to be +io. Thus, if genomic imprinting is leading to the appearance of an apparent maternal effect at a locus, we expect the additive maternal effect to be similar in magnitude and opposite in sign to the parent-of-origin effect at the locus. Because these individuals have genetically identical mothers at this locus, the imprinting effect cannot be attributed to a maternal effect (Hager et al. 2008). Consequently, by applying the statistical cross-fostering design to ordered genotypes one should be able to identify the imprinting effects and reconcile the appearance of an additive maternal effect at these loci as being caused by genomic imprinting and, therefore, eliminate these loci as maternal-effect loci.
Thus, when significant additive effects are attributable to true maternal effects, not genomic imprinting, we generally expect to find that the reciprocal heterozygote offspring raised by the different homozygous mothers are phenotypically different, but this difference is not seen among the reciprocal heterozygote offspring raised by heterozygous mothers. In contrast, we expect that genomic imprinting effects should generally make the reciprocal heterozygote offspring phenotypically different, regardless of whether their mothers are homozygotes or heterozygotes. Finally, it is important to keep in mind that it is also possible that a locus could show both imprinting and maternal effects, so that it may not be possible to cleanly distinguish between the two.
An Orthogonal Model of Statistical Cross-Fostering
The statistical cross-fostering scheme can be achieved by simply subdividing the data into separate analyses of direct and maternal effects, as described above (and illustrated in Figure 1), or developed in a single analysis, where these effects are fitted simultaneously as orthogonal components. We present the conceptual framework without providing an explicit statistical model for an analysis because it can be adapted to the specific nature of any given data set. We build on the conceptual framework for the statistical cross-fostering scheme presented above by developing this framework into a linear model where a single analysis is used to simultaneously fit direct and maternal effects as orthogonal components. Because of the potential confounding effects of genomic imprinting effects, we base this model on ordered genotypes in the offspring generation (i.e., matching Table 3). If one were to use unordered genotypes, then the model would be identical, except that the two reciprocal heterozygotes produced by heterozygous mothers would be combined into one cell and the index variable for the parent-of-origin effect would be removed from the design matrix (see below).
To develop a model for direct and maternal effects we assign a set of index values to each of the eight maternal–offspring genotype combinations that can be used in a regression model to provide estimates of the direct and maternal genetic-effect parameters. These index values define the structure of the relationship between the measured phenotypes and measured genotypes in the model and are used, therefore, to estimate the corresponding genetic effects. The index values are chosen to reflect the statistical cross-fostering design described above, but using all of the genotype data from the population in a single analysis. With eight classes of maternal–offspring genotypes we define eight coefficients in the model, with one being the intercept [or reference point, R (Álvarez-Castro and Carlborg 2007)] and the other seven being the three direct-effect parameters (ao, do, and io), two maternal-effect parameters (am and dm), and two new parameters (cmo and ddmo) not present in the framework presented above. The first of these new parameters, cmo, is the confounded effect and the second, ddmo, is the interaction between the dominance effects of the maternal and offspring genomes (see below). These coefficients are shown in Table 4, where each maternal–offspring genotype combination is assigned an index value for each of these eight effects. The matrix in Table 4 defines the genetic design matrix of the model (SMO), which gives the relationship between the maternal–offspring genotype classes and the genetic effects being estimated. This can be formalized by defining the vector of maternal–offspring genotypic values (GMO) (average phenotypes of offspring as a function of their genotype and the genotype of their mother) with the genotypes arrayed as in the first column in Table 4 (i.e., the vector contains eight values, which are the mean phenotypes of the genotype combinations in that column, in the order shown in Table 4) and the vector of genetic effects (EMO) as the row of coefficients in Table 4: , where T denotes the transpose. Thus, it is the genetic design matrix (SMO) that links the vector of genotypic values (GMO) to the vector of genetic effects (EMO):
| (3) |
This design matrix can be inverted to solve for the definition of the genetic effects in terms of the genotypic values, i.e., , and therefore the inverse of the genetic design matrix defines the solution for the genetic effects as a function of the average phenotypes (i.e., genotypic values) associated with each of the maternal–offspring genotype combinations. The inverse of the genetic design matrix is shown in Table 5. The rows can be read as a set of contrasts based on the genotypic values.
Table 4. The statistical cross-fostering design matrix.
| Combination | Coefficients | |||||||
|---|---|---|---|---|---|---|---|---|
| R | ao | do | io | am | dm | cmo | ddmo | |
| M11O11 | 1 | 0 | 0 | 0 | 1 | |||
| M11O21 | 1 | 0 | 0 | 1 | − | 0 | − | |
| M12O11 | 1 | 1 | − | 0 | 0 | 0 | − | |
| M12O21 | 1 | 0 | −1 | 0 | 0 | |||
| M12O12 | 1 | 0 | 1 | 0 | 0 | |||
| M12O22 | 1 | −1 | − | 0 | 0 | 0 | − | |
| M22O12 | 1 | 0 | 0 | −1 | − | 0 | − | |
| M22O22 | 1 | 0 | − | 0 | 0 | − | −1 | |
The rows are labeled to reflect the corresponding maternal–offspring genotype combination, as in Table 3 [given as MijOkl, where Mij is the unordered genotype of the mother (with i and j being the two alleles she has at the focal locus) and Okl is the ordered genotype of the offspring (with again, k and l being the two alleles that the offspring has at the focal locus]. The columns labeled “Coefficients” correspond to the index scores used in a regression model to fit the intercept or reference point for the model (R), the three direct genetic effects (ao, do, and io), and the two maternal effects (am and dm). The last two columns correspond to terms that we do not interpret here because they do not correspond to independent effects of the maternal and offspring genomes. The first of these, cmo, is the confounded effect (see text) and the second, dd, is an interaction between the dominance effects in mothers and their offspring (see Discussion)
Table 5. The inverse of the genetic-effect design matrix shown in Table 4.
| Genotypic values | ||||||||
|---|---|---|---|---|---|---|---|---|
| Genetic effects | M11O11 | M11O21 | M12O11 | M12O21 | M12O12 | M12O22 | M22O12 | M22O22 |
| R | ||||||||
| ao | 0 | 0 | 0 | 0 | − | 0 | 0 | |
| do | − | − | − | − | ||||
| io | 0 | 0 | 0 | − | 0 | 0 | 0 | |
| am | 0 | 0 | 0 | 0 | 0 | − | 0 | |
| dm | − | − | − | − | ||||
| cmo | 0 | 0 | 0 | 0 | 0 | 0 | − | |
| ddmo | − | − | − | − | ||||
The inverse of the genetic-effect design matrix in Table 4 gives the solution to the vector of genetic effects from the linear model. The columns of the matrix are labeled with the genotypic effect and the rows are labeled with the corresponding genetic effect.
From the values in Table 5 we can see that, as expected under the statistical cross-fostering framework, the additive direct effect is based on the homozygous offspring of heterozygous mothers and is defined formally as half the difference between the average phenotypes of these two classes,
| (4) |
while the additive maternal effect is based on the heterozygous offspring of homozygous mothers,
| (5) |
This again highlights the constraint that the additive maternal effect is confounded with the imprinting effect in the offspring because the two classes in this contrast differ in the parent of origin of the alleles (O21 vs. O12). Likewise, the imprinting effect is based on a contrast between the reciprocal heterozygote offspring from heterozygous mothers:
| (6) |
In contrast to additive and parent-of-origin effects, however, it is clear from Table 5 that the dominance effects under this model are actually based on all of the genotypic values—i.e., they are not restricted to a subset of classes in the two contrasts. This is the primary difference between this unified orthogonal model and the conceptual approach to statistical cross-fostering described above. The dominance direct effect is defined as the difference between the average phenotypic value of heterozygous pups and that of homozygous pups,
| (7) |
and dominance maternal effects are defined as the difference between the average phenotypic value of pups with heterozygous mothers and that of pups with homozygous mothers:
| (8) |
The genetic design matrix also yields two terms that cannot be attributed to direct or maternal effects. The first of these we call the confounded effect, cmo, which is a contrast between the two classes of homozygous offspring of homozygous mothers,
| (9) |
It accounts for the cells where the additive direct and maternal effects are fully confounded, and hence this contrast is included only to make the genetic design matrix of full rank. That is, this term could be significant when either the direct or the maternal effect or both direct and maternal effects are present, and it can be nonsignificant even when they are both present because they may be of opposite sign and cancel out each other (e.g., a positive maternal effect of am = +Q and a negative direct effect of ao = −Q could make the phenotypes of the two classes of homozygous offspring of homozygous mothers phenotypically identical).
The genetic design matrix also includes a single interaction term, ddmo, which is an interaction between the dominance direct and maternal effects. It is conceptually analogous to dominance-by-dominance epistasis (Cheverud 2000), except that the interactions are between the maternal and offspring genomes (Wolf 2001) and the interactions occur within a locus, rather than between loci (Wade 1998). It corresponds to a contrast between the combinations where the mother and her offspring have the same genotype (ignoring parent-of-origin of alleles) and where they have different genotypes:
| (10) |
As with the confounded effect, cmo (Equation 9), the dominance interaction effect, ddmo, is included primarily because it allows us to create a full rank orthogonal model. Finally, note that the reference point for this model is simply the grand mean (i.e., is the average of the eight maternal–offspring genotype classes).
The effects in the genetic-effects vector, EMO, can be estimated empirically by regression using the coefficients shown in the genetic-effects design matrix (Table 4) as the independent variables and individual phenotypic values (i.e., the trait values measured for individuals, not genotype classes) as the dependent variables (Cheverud 2000, 2006). That is, although we define the effects above in terms of average phenotypes of the maternal–offspring genotype classes, the effects can be estimated using individual phenotypes rather than the average values of each of the genotype classes. The regression model that yields estimates of the seven genetic effects and the reference point in the model is given as
| (11) |
where Pj(x) is the phenotypic value of individual j with maternal–offspring genotype combination x (where x identifies which of the eight maternal–offspring combinations the individual has), XZ(x) is the genotypic index value (from Table 4) of parameter Z for that maternal–offspring genotype combination (where Z is one of the seven genetic effects being estimated), and rj is the residual from the model for individual j. The form of the model (Equation 11) is analogous to the model used in Wolf et al. (2011) to examine prenatal and postnatal maternal effects, except that the model presented herein is based on an explicit orthogonal model structure that yields estimates of genetic effects corresponding to the specific forms given in Equations 4–10.
To demonstrate the use of this statistical cross-fostering scheme we present an empirical study using an experimental population of mice. We test for direct and maternal effects on size and growth traits, using marker data. We then use several identified loci to illustrate the phenotypes of individuals as a function of their genotype and the genotype of their mothers.
Materials and Methods
Experimental population
We use an experimental population that was derived from a cross between inbred lines of mice that were originally derived by artificial selection for either large (the LG/J line) or small (the SM/J line) body weight at 60 days of age (Goodale 1938; MacArthur 1944; Chai 1956). These lines had been inbred for >120 generations prior to the crossing, making them essentially devoid of within-strain genetic variation. We use the F2 generation of the line cross as our parental generation and their F3 progeny as the “offspring” generation (Kramer et al. 1998; Vaughn et al. 1999). The population was created by mating 10 SM/J males to 10 LG/J females, producing 52 F1 individuals. The F1 animals were randomly mated to produce 510 F2 animals. Approximately 400 of these F2 animals were randomly mated (∼200 males and 200 females) to create the F3 population, so the mothers of these F3 individuals are our focal set for analyses of maternal effects. The F3 population contains a total of 1632 F3 individuals in 200 full-sibling families (of which 1552 were genotyped). Half litters were reciprocally cross-fostered at random between pairs of females that gave birth on the same day (Wolf et al. 2002) so only some families were cross-fostered. In this study we limit our focus to those mice that were not cross-fostered to understand how one might study maternal effects in the absence of experimental manipulation (in Wolf et al. 2011 we use the cross-fostered population to examine how experimental cross-fostering can be used to identify pre- and postnatal maternal effects, but do not develop the statistical cross-fostering framework therein). Therefore, our focal population is the 937 F3 individuals from 194 families who were not cross-fostered. Pups were weaned at 21 days of age and randomly housed with 3 or 4 other same-sex individuals. Previous quantitative genetic analysis suggested that maternal effects contribute a large component of variation for the first few weeks of age in this population (Kramer et al. 1998), but this previous study was unable to differentiate genetic from nongenetic maternal effects and accounted only for postnatal maternal-effect variation.
All animals were weighted weekly from 1 week through 10 weeks of age, using a digital scale with an accuracy of 0.1 g. For consistency with previous work on this population, sex differences in body size and those associated with differences in birth and weaning litter sizes were removed prior to analyses.
Genotypes
Details of the genotype data used in all analyses are given in Wolf et al. (2008b). Briefly, all F2 and F3 individuals were genotyped at 353 autosomal single-nucleotide polymorphism (SNP) loci, using the Illumina Golden-Gate assay, with an average map distance between markers in the F2 generation of 4 cM. A complete list of the markers and their physical and recombinational map positions are given in Supporting Information, Table S2.
To link the experimental population to the model presented above we assign the A1 allele to the LG/J line and the A2 allele to SM/J. Allele frequencies at all loci are close to the expected frequency of 0.5 and all genotypes occur in approximately Hardy–Weinberg proportions (conforming to , 2p1p2, and for the A1A1, A1A2, and A2A2 genotypes).
QTL analysis
We used a linear mixed-models framework to estimate direct and maternal effects associated with marker loci, using the index values in Table 4 in a regression model analogous to Equation 11. This was done by assigning the appropriate index values at each locus on the basis of the SNP genotype of the mother and her offspring. The model shown in Equation 11 was fitted as a mixed model with the fixed effects corresponding to the genotypic index values and with the mother as a random (classification) effect. Therefore, Equation 11 corresponds to the fixed-effects part of the model, but the implementation in our experimental population included an additional random effect. The random effect accounts for the residual relatedness of siblings, who share alleles at other loci and also share common environmental effects, which together produce a phenotypic autocorrelation of siblings that inflates the apparent significance of genetic effects (Lynch and Walsh 1998; Wolf et al. 2008b). This mixed model was fitted using maximum likelihood in the Mixed Procedure of SAS (SAS version 9.1; SAS Institute, Cary, NC). We used a 2-d.f. test that simultaneously tested the am and dm effects together to produce an overall test for maternal effects at a locus. Denominator degrees of freedom were generated by the Satterthwaite approximation (see Littell et al. 2006), which uses the variance structure of the model (i.e., the structure of the random effects) to determine the degrees of freedom. In this model, the Satterthwaite approximation essentially determines the effective sample size for each fixed effect (Ames and Webster 1991; Keselman et al. 1999; Faes et al. 2009) on the basis of family structure, and consequently, the degrees of freedom for the maternal-effect terms are close to the number of mothers while the degrees of freedom for the direct-effect terms are close to the total number of F3 individuals (i.e., the number of offspring) (Wolf et al. 2011). Probabilities from all significance tests were converted to logarithmic probability ratios [LPR = –log10(probability)], which are analogous to the LOD scores that are commonly reported. Proportions of variance explained by QTL (R2) were estimated by calculating the genetic variance contributed by a locus as
| (12) |
and dividing this variance by the total phenotypic variance. Only significant effects at a locus were included in this calculation to avoid artificial inflation.
To facilitate the identification of QTL locations, we used multivariate versions of the model in Equation 11 (see Hager et al. 2009). Weight measurements were divided into three sets, which correspond to different growth phases (Kramer et al. 1998): (1) preweaning weights (weeks 1–3); (2) postweaning weights, where there is rapid weight gain (weeks 4–6); and (3) adult weights, when there is slow growth (weeks 7–10). The multivariate model was fitted using the framework described by Fry (2004), modified to include a multivariate QTL effect. In this model, the weekly weights are treated as repeated measures of a “weight” trait and QTL effects are fitted to this vector of weight traits. Week is included in this model to account for variation in weight through time. The correlation between weekly weights measured within individuals was modeled using the Toeplitz autoregressive structure (Kincaid 2005), which approximates the temporally autocorrelative structure of the weight traits (Kramer et al. 1998). Denominator degrees of freedom were determined using the Kenward–Roger approximation, which is analogous to the Sattherthwaite approximation but is preferred for repeated measures designs (Kenward and Roger 1997; Schaalje et al. 2001).
Maternal-effect QTL (meQTL) locations were first identified using the LPR values from the multivariate models, with the maximum LPR value on a chromosome above the threshold value (see below) taken as evidence of a meQTL on that chromosome. However, once a locus was identified, the location was refined (using the results from the multivariate model) by examining the individual patterns of the additive and dominance maternal effects; if the locus had only one form of maternal effect, then the meQTL position was taken as the location of the LPR peak for the one significant effect alone. This is because random variation in the other term in the model can displace the overall peak away from the peak for the significant effect, but the best evidence for the meQTL location is assumed to be the position that maximizes the fit for the significant effect. We also examined the multivariate genome scan for the possibility of multiple peaks on a chromosome, which could appear as either two or more peaks in the overall test for maternal effects or a difference in the position of the peaks for individual additive and dominance maternal effects underlying a single peak. We found no cases of the former (multiple distinct peaks on a chromosome), and so the analysis of multiple distinct peaks is not discussed further. However, we found a case where one region of a chromosome had distinct separate peaks for additive and dominance maternal effects. In this case, we ran a model that included the maternal effects of both loci to determine whether there was support for a two-locus model. Support for two meQTL was determined using a likelihood-ratio test, with the full model containing the meQTL effects at their individual locations and the reduced model containing a single meQTL at the overall peak location. The difference in the −2 log likelihoods of the two models (reduced model minus full model) is approximately chi-square distributed with the number of degrees of freedom corresponding to the number of additional terms in the full model (i.e., the number of terms dropped in the reduced model). The multiple-QTL model was accepted when it had a significantly better fit than the reduced single-QTL model.
Confidence intervals were defined as a one-LPR drop (on the basis of the multivariate model), which is analogous to the commonly used one-LOD drop (Lynch and Walsh 1998). Maternal-effect loci were named following the convention of mebsX.Y (indicating maternal effects on body size), where X is the chromosome number and Y is the locus number on that chromosome to distinguish between multiple QTL on a chromosome.
Whenever maternal-effect loci with additive effects were identified, we examined whether those loci had imprinting effects of similar magnitude but of opposite sign, which was taken as evidence that the maternal effect appeared because of the occurrence of an imprinting effect (see above and Table 3). Because a locus could potentially show both an imprinting effect and a maternal effect, we devised a test to determine whether the appearance of an additive maternal effect could be attributed to the presence of the imprinting effect, with the possibility that the locus may show both a maternal and an imprinting effect. We took the parameter estimate for the imprinting effect (i) from the analysis of direct effects (where the parent-of-origin effect is not confounded with any other effects) and removed (using the linear model) the variation attributable to the imprinting effect from the heterozygous offspring with homozygous mothers. We then reran the analysis of maternal effects as above and tested whether the maternal-effect terms were still significant after being “corrected” for the imprinting effect. This is a very conservative analysis that assumes that the imprinting effect estimated in the analysis of direct effects is the best estimate of the imprinting effect since it is not confounded with any other effects and then removes this variation from the genotypes where the two effects are confounded, thereby removing the confounding.
Significance testing
Previous analyses have demonstrated that the distribution of significance tests associated with direct effects produced by the mixed model behaves as expected under the null model (Wolf et al. 2008b). However, maternal-effect terms in the model are pseudoreplicated because there are, on average, about four pups from each mother that were included in the analysis, and therefore, each maternal genotype appears in the linear equation about four times. We emphasize that this pseudoreplication is not a shortcoming of the analysis, but rather is a feature of the framework and is easily addressed by adjusting the denominator degrees of freedom of the model to reflect the true level of replication (the individual mothers). This pseudoreplication in the design is accounted for by the use of the Satterthwaite approximation for the denominator degrees of freedom, but could also be approximated by adjusting the denominator degrees of freedom to reflect the number of individual mothers in the analysis (Wolf et al. 2011).
Significance thresholds were determined on the basis of the number of tests in a Bonferroni correction for familywise error, using the Šidák equation, 1 – 0.951/n, where n is the number of tests in the family. We calculated both a genomewise and separate chromosomewise thresholds, using the effective number of markers method as described by Li and Ji (2005). This method uses the correlation between markers to estimate the number of independent tests on each chromosome and over the whole genome (since correlated tests are not independent, using M markers results in a threshold based on Meff because the M markers do not count as M independent tests). Because more recombination events have accumulated in the F3 genotypes compared to the F2 (i.e., there is less linkage disequilibrium in the F3 generation), the number of independent tests is lower for the maternal-effect tests compared to the direct-effect tests. The chromosomewise thresholds are used for QTL discovery because they have been shown to increases the discovery of true positives while avoiding the representation of false positives (Chen and Storey 2006). Significance thresholds are given in Table S3. Once a maternal-effect locus was identified, we used a pointwise threshold (i.e., LPR = 1.3 for P = 0.05) to determine which individual effects were significant at that locus.
Results
The orthogonal model of statistical cross identified seven loci showing significant maternal effects on offspring body size (Table 6). Two of these loci, mebs17.1 and mebs18.1, had significant additive maternal effects and imprinting effects (Table 7) that are of opposite signs, suggesting that the maternal effect may have appeared because of the presence of imprinting. However, the patterns are complex and somewhat ambiguous. For mebs17.1, the imprinting effect is strongest at week 2 and is no longer significant after week 5, while the additive maternal effect peaks at week 6. At week 2 the magnitude of the additive maternal effect is nearly identical to that of the imprinting effect (standardized am = −0.260 and io = 0.255), strongly suggesting that the two arise from an imprinting effect. The temporal pattern would suggest that the locus has an imprinting pattern early and a maternal effect later in life; however, when the imprinting-effect variation is removed, the additive maternal effect is no longer significant at any age (i.e., partialling the imprinting effect even when it is not significant later in life still results in a nonsignificant additive maternal effect). Consequently, the conservative conclusion is that the appearance of a significant additive maternal effect can be attributed to the presence of the imprinting effect. We follow this conclusion in the presentation of the results, but we note that this same locus was previous identified by Wolf et al. (2011) as a maternal-effect locus with a persistent additive postnatal maternal effect similar to the one observed for mebs17.1. Because the postnatal maternal effects identified by Wolf et al. (2011) were detected as the effect of the genome of the foster mother on the phenotypes of her foster pups, they could not have been caused by genomic imprinting (since the nurse mothers and their foster pups were unrelated). Furthermore, this conclusion is supported by the results from Hager et al. (2008), who identified a locus (Wtmge17.1) at this same genomic region as a maternal-effect locus that appears to have an imprinting effect. Therefore, prior evidence suggests that this locus is indeed a maternal-effect locus, but the data used herein do not allow us to differentiate a maternal effect at this locus from an imprinting effect that mimics a maternal effect.
Table 6. Maternal effects of meQTL.
| meQTL | mebs2.1a | mebs3.1 | mebs7.1a | mebs7.2 | mebs8.1a | mebs17.1a | mebs18.1a | R2b | |
|---|---|---|---|---|---|---|---|---|---|
| cM | 44.5 | 25.1 | 57.0 | 76.5 | 48.9 | 9.0 | 0.0 | ||
| Mb | 75.9 | 53.1 | 122.7 | 145.0 | 98.2 | 20.4 | 3.7 | ||
| C.I. (Mb) | 69.2–82.2 | 46.1–63.8 | 117.6–134.5 | 122.7–145.0 | 83.2–114.5 | 8.4–61.3 | 3.7–34.0 | ||
| Traits | Early | +amc | +dm | −dm | −ami | −am | |||
| Mid | +amc | +am | +dm | −dm | −ami | −am | |||
| Late | +amc | +dm | +dm | +am | −dm | −ami | −am | ||
| Week 1 | +am | +dm | −dm | −ami | −amc | 25.1 | |||
| Week 2 | +am | +dm | −dm | −ami | −am | 26.9 | |||
| weaning | Week 3 | +amc | +dm | −dm | −ami | −am | 28.0 | ||
| Week 4 | +amc | +am | +dm | −dm | −ami | −am | 36.3 | ||
| Week 5 | +amc | +dm | +dm | +am | −dm | −ami | −ami | 22.6 | |
| Week 6 | +amc | +am+dm | +dm | +am | −dm | −ami | −ami | 21.8 | |
| Week 7 | +amc | +am+dm | +dm | +am | −dm | −ami | −ami | 22.2 | |
| Week 8 | +amc | +dm | +dm | +am | −dm | −ami | −ami | 21.8 | |
| Week 9 | +am | +dm | +dm | +am | −dm | −ami | −ami | 20.2 | |
| Week 10 | +am | +dm | +dm | +am | −dm | −ami | 20.6 |
The map position in both centimorgans (cM) and megabases (Mb) along with the confidence interval (C.I.) (in Mb) and the effects on each of the 10 weekly weight measurements are given for each locus. The rows labeled “Early,” “Mid,” and “Late” give the significance values from the multivariate tests. Significance tests are in LPR units (where LPR = −log10[p]). Entries in boldface type are significant using the chromosome-level significance threshold. Additive maternal effects that appear to be attributable to an imprinting effect are marked with a superscript “i”. The estimates of all effects and the significance values for all tests are provided in Table S1. The proportions of phenotypic variance accounted for by the set of loci (R2) on each of the weekly weight traits are also included.
Loci also show significant maternal effects in the analyses of Wolf et al. (2011).
Does not include variation from apparent additive maternal effects that were attributed to imprinting effects (i.e., those with a superscript “i”).
Entries are significant using the genome-wide threshold.
Table 7. Direct effects of meQTL.
| meQTL | mebs2.1 | mebs3.1 | mebs8.1 | mebs17.1 | mebs18.1 | R2 | |
|---|---|---|---|---|---|---|---|
| Traits | Week 1 | +io | 0.82 | ||||
| Week 2 | +do | +do | +ioa | 5.15 | |||
| weaning | Week 3 | +do | +io | 2.59 | |||
| Week 4 | +do | +do | +io | 3.54 | |||
| Week 5 | +do | +io | +io | 3.34 | |||
| Week 6 | +do | 0.81 | |||||
| Week 7 | −im | +io | 1.81 | ||||
| Week 8 | −im | +io | 1.84 | ||||
| Week 9 | −im | +io | 2.10 | ||||
| Week 10 | +ao | +io | 1.67 |
The effects on each of the 10 weekly weight measurements are given for each locus. Significance tests are in LPR units (where LPR = −log10[p]). Entries in boldface type are significant using the chromosome-level significance threshold. The estimates of all effects and the significance values for all tests are provided in Table S1 and genomic locations in Table 4. The proportions of phenotypic variance accounted for by the set of loci (R2) on each of the weekly weight traits are also included.
Significant using the genome-wide threshold.
For mebs18.1, there is a strong additive maternal effect in the first few weeks of age (weeks 1–4; see Table 8 for an example of week 1 weight for this locus), which cannot be attributed to an imprinting effect (both because the imprinting effect is not significant and removing the imprinting effect from the maternal effect does not result in a nonsignificant additive maternal effect), but at week 5 a significant imprinting effect appears and from that age onward it appears to account for the additive maternal effect (i.e., partialling the imprinting effect from the additive maternal effect makes the maternal effect nonsignificant). Thus, our conclusion is that there is support for both an imprinting and a maternal effect at mebs18.1, which are temporally separate.
Table 8. Examples of additive and dominance maternal effects.
| Offspring genotype | |||||||
|---|---|---|---|---|---|---|---|
| LL | SL | LS | SS | Maternal litter average | |||
| A. Expected week 1 weights from the mixed model (in standardized units) of offspring as a function of their genotype and the genotype of their mothers at mebs18.1a | |||||||
| Maternal genotype | |||||||
| LL | –0.15 | –0.28 | — | — | –0.21 | ||
| LS,SL | –0.06 | –0.05 | –0.11 | –0.25 | –0.12 | ||
| SS | — | — | 0.41 | 0.28 | 0.34 | ||
| Offspring average | –0.10 | –0.17 | 0.15 | –0.01 | |||
| B. Expected week 4 weights from the mixed model (in standardized units) of offspring as a function of their genotype and the genotype of their mothers at mebs8.1b | |||||||
| Maternal genotype | |||||||
| LL | 0.15 | 0.07 | — | — | 0.11 | ||
| LS,SL | –0.14 | –0.19 | –0.43 | –0.30 | –0.26 | ||
| SS | — | — | 0.29 | 0.15 | 0.22 | ||
| Offspring average | 0.00 | –0.06 | –0.07 | –0.07 | |||
Table cells with an em dash indicate combinations of material and offspring genotypes that cannot occur under Mendelian inheritance.
This locus has a strong additive maternal effect (standardized am = −0.36 from the linear model, but it is calculated as −0.35 from these cell means) and no direct effect.
This locus has a strong dominance maternal effect (standardized am = −0.43 from the linear model, but it is calculated as −0.49 from these cell means) and no direct effect.
Overall, the temporal pattern of maternal effects varies greatly across loci, but all loci have significant maternal effects on several weight measurements (Table 6). This may be partly due to the use of a multivariate model to identify loci, where we use an overall test for effects on a growth phase to identify loci that are, therefore, likely to be pleiotropic across the growth phase. Interestingly, the traits with the fewest significant effects are the three preweaning weights (weeks 1–3), which are each affected by the same set of five of the seven loci. No locus had effects limited to early, preweaning weights and only one locus (mebs18.1) had an effect that peaked before weaning (with a very strong peak effect at week 1; see Table 8). Two loci (mebs3.1 and mebs7.2) had effects limited to postweaning weights and both had effects that peaked very late (at weeks 7 and 10, respectively). Interestingly, four loci show effects through all of the weekly weight measures (although the late effects of mebs18.1 appear to be caused by an imprinting effect), demonstrating that maternal effects that appear early can be persistent through development. The effect estimates, significance values, and percentages of variance explained by the meQTL are provided in Table S1.
The overall percentage of the phenotypic variance explained by the maternal effects of the meQTL (Table 6) peaks at week 4 (the week after weaning). After week 4 the percentage of variance explained declines slowly, but the meQTL continue to explain ∼20% of the variance through week 10. The additive maternal effect of mebs2.1 on week 4 weight explains the most variance of any locus on any trait (nearly 11% of the variance).
The meQTL have relatively minor direct effects (Table 7), and there is a particular paucity of additive effects, with only one locus showing a significant additive direct effect on a single weight trait. Two loci (mebs2.1 and mebs3.1) show overdominance direct effects, while three others (mebs8.1, mebs17.1, and mebs18.1) show significant imprinting effects. In the case of these imprinting effects, the appearance of a significant direct effect could be potentially biased since the imprinting effects contributed to the significance of the maternal effect.
Discussion
Our conceptual framework, which we call statistical cross-fostering, opens the possibility of identifying and characterizing maternal-effect loci in natural and experimental populations in which cross-fostering or other manipulations have not been done or are not possible. This is an important advancement because, although maternal effects are likely to be a major component of the genetic architecture of many traits, they are largely ignored in studies of genetic architecture. Consequently, we currently have a very poor understanding of their contribution to genetic variation and as a result, we have a very biased view of the genetic basis of phenotypic variation. The statistical cross-fostering framework accomplishes the separation of maternal effects from the direct effects of the individual’s own genome by using a set of offspring genotypes that are uncorrelated to the genotypes of their mothers, thereby removing the confounding effects of the two genomes. Conceptually, this process could be achieved by limiting the analysis of maternal effects at a given locus to a set of genetically identical offspring coming from genetically variable mothers while limiting the analysis of direct effects to offspring from genetically identical mothers (Figure 1). We implement this approach using an orthogonal model (Table 4) that achieves the statistical cross-fostering design by removing the confounded combinations (i.e., homozygous mothers having homozygous offspring) from the analysis of additive direct and maternal effects, while using the entire data set in a single model-fitting process. We have applied this framework to an analysis of maternal genetic effects in an experimental population of mice and identified a set of six maternal-effect QTL that explain a major component of phenotypic variation in body size at all ages.
Although the statistical cross-fostering approach opens up new avenues of research on maternal genetic effects, it is important to understand that to achieve this separation of maternal and direct effects requires us to impose assumptions on the analysis. This is because the missing maternal–offspring genotype combinations are necessarily absent from the analysis. We discuss these assumptions and limits, but we strongly emphasize that these are imposed by biological constraints and are not simply statistical limitations. Despite these limits, the ability to detect and characterize maternal-effect loci in systems where it would not otherwise be possible to separate maternal and direct genetic effects should provide significant insights into the genetic basis of phenotypic variation.
Empirical analysis of meQTL
Our statistical cross-fostering approach identified six maternal-effect loci with pleiotropic effects on body weight (see Table 6), demonstrating that maternal effects can be successfully identified without experimental manipulation. Two of these loci have maternal effects limited to the phases of growth after weaning (which occurred at week 3 of age). Overall, the maternal-effect loci explain the most variance in the traits around weaning, with a peak at week 4. This pattern differs from that observed in this population for postnatal maternal effects using variance partitioning in an experimental cross-fostering design (Kramer et al. 1998), where the proportion of variance explained by the foster mother peaked at week 2 and showed a clear decline by week 4. However, it is similar to the overall pattern of maternal genetic effects observed using a reciprocal crossing experiment (Jarvis et al. 2005), which, like our study, combines pre- and postnatal effects and matches the pattern for the sum of pre- and postnatal effects identified in Wolf et al. (2011). Four of the loci have effects that extend through to 10 weeks of age, which is somewhat counterintuitive given that maternal care and all contact with the mothers ended at weaning (week 3). Indeed, two of these loci have effects that are not significant until after weaning, suggesting that early maternal influences may affect developmental and physiological processes that are manifest after the end of actual maternal care. For most of these loci, the effects on adult traits are relatively strong, and together they account for >20% of the variance in body weight from week 5 to week 10. Two loci have additive maternal effects on week 9 weight, which is approximately the target of selection in the LG/J and SM/J strains (Goodale 1938; MacArthur 1944; Chai 1956), with the sign of effect matching the direction of divergence between these lines (i.e., the allele derived from LG/J has a positive maternal effect), suggesting that these maternal effects may have contributed to the evolved difference in week 9 weight between these strains.
Although the absence of cross-fostering prevents us from separating the maternal effects into pre- and postnatal components, the previous results of Wolf et al. (2011) provide some insights into the likely origin of the effects for several loci. Four of the seven loci detected here (mebs2.1, mebs7.1, mebs17.1, and mebs18.1) map very close to and have similar patterns of effect to loci identified using experimental cross-fostered pups by Wolf et al. (2011). The results from Wolf et al. (2011) suggest that mebs2.1 and mebs18.1 have prenatal effects, while mebs7.1 and mebs17.1 have postnatal effects. Interestingly, in our analysis we were unable to support the hypothesis that mebs17.1 is a maternal-effect locus because the pattern of effect could be explained by the presence of genomic imprinting at this locus. However, the results from Wolf et al. (2011) strongly suggest that this locus does indeed have a maternal effect since the effect detected there was postnatal and, therefore, was not confounded with genomic imprinting (see below). This conclusion is further supported by the results from Hager et al. (2008), where they concluded that this same locus has a maternal effect that creates an apparent imprinting effect. In reexamining the results from Wolf et al. (2011) we also find that, although not significant at the threshold used for QTL discovery in that study, mebs8.1 appears to have a dominant prenatal maternal effect on body weight from week 6 to week 10 that is significant at the P < 0.05 level (with LPR values ranging from 1.38 to 1.88 for these traits). Thus, the level of overlap between the loci identified here and those detected in our previous work on maternal effects in this population is surprisingly good given that the effect sizes are relatively small and the two analyses used different groups of pups and different statistical methods. This overlap allows us to conclude that three of the seven loci (mebs2.1, mebs8.1, and mebs18.1) arise from prenatal effects and two (mebs7.1 and mebs17.1) from postnatal effects. This suggests that, because maternal effects can potentially arise from these two very different sources (pre- vs. postnatal), one should consider using experimental cross-fostering at birth in conjunction with our statistical cross-fostering model (which allows for the identification of prenatal effects without prenatal cross-fostering) to identify the origin of maternal effects.
The loci detected here do not correspond to the maternal-effect loci detected in another study by Wolf et al. (2002) that used cross-fostered pups, where the litter means were treated as traits of the nurse mother (so only postnatal effects were examined, and they combined the offspring into a single measure). Although one locus (mebs7.1) does map close to one of the loci identified by Wolf et al. (2002), the patterns of effect are incompatible (with the sign of the dominance effect being opposite). The difference in the results of these studies is likely caused by the much lower power of the Wolf et al. (2002) study, where individual variation and among-litter variation were not accounted for in the model. That study also used a much more diffuse marker map and a smaller number of mothers, which further reduced power.
The maternal-effect loci we identified generally show weak direct effects (Table 7), which is somewhat surprising given that maternal effects tend to be genetically correlated to direct effects (Roff 1997). Previous analyses focused on identifying direct effects for these same traits (Wolf et al. 2008b; Hager et al. 2009) and did not identify any of these same loci, which is not surprising given that the direct effects we identified here are small. The one exception is mebs17.1, which appears to have a strong imprinting effect in our analysis, but this locus was not previously identified as an imprinted locus because, again, the imprinting effect was attributed to a maternal effect (Hager et al. 2008; Wolf et al. 2011). Analyses of body composition traits in these same animals (Cheverud et al. 2008) did identify direct effects for two of these loci (mebs3.1 and mebs7.1), both of which showed an imprinting effect (see Bwi3.1 and Bwi7.1 in Cheverud et al. 2008).
Confounding with imprinting effects
Additive maternal genetic effects are partially confounded with genomic imprinting effects because of the correlation between genotype of the mother and the parent of origin of alleles in the offspring (Hager et al. 2008; Wolf and Wade 2009). This is clear from Table 3, where the imprinting effect (io) is fully confounded with the additive maternal effect (am) in heterozygous offspring, meaning that a locus may appear to have an additive maternal effect when an imprinting effect is present if we limit the analysis of maternal effects to heterozygous offspring. This occurs because, although the two different homozygous mothers produce heterozygous offspring that are genetically identical at a locus, they are not epigenetically equivalent because they necessarily differ in the parent of origin of the two alleles (i.e., the two types of mothers produce the alternative forms of the reciprocal heterozygotes). This possibility can be accounted for because the analysis of direct effects allows one to identify an imprinting effect independent of the maternal effect and thereby determine when maternal effects appear only because of the presence of the imprinting effect. The confounding is essentially asymmetrical, where additive maternal effects are confounded with imprinting effects, but imprinting effects can be examined independent of maternal effects. This is because heterozygous mothers can have either type of heterozygous offspring, but homozygous mothers can produce only one of the two types of reciprocal heterozygotes. Therefore, one can estimate the imprinting effect in the heterozygous offspring of heterozygous mothers (Hager et al. 2008), where the difference between reciprocal heterozygotes must be caused by a imprinting effect, and then remove this effect from the heterozygotes produced by homozygous mothers and determine whether there are still significant additive maternal effects.
In our analysis of meQTL we found evidence for two cases where the additive maternal effect was caused by an imprinting effect at the locus. For one locus, mebs17.1, the apparent additive maternal effect at all ages appears to be caused by an imprinting effect. Although the additive maternal effect at this locus is significant from week 1 to week 9 of age and the imprinting effect is not significant after week 5, the imprinting effect is still large enough to account for the apparent maternal effect. However, as we noted above, the results from Wolf et al. (2011; see also Hager et al. 2008) suggest that this locus does indeed have a maternal effect, despite the fact that we could not distinguish the maternal effect from an imprinting effect here. For the second locus, mebs18.1, the pattern appears to be clearer, where the locus has a very strong additive maternal effect from week 1 to week 4, but from week 5 onward, the appearance of an additive maternal effect can be accounted for by the presence of an imprinting effect. The co-occurrence of genomic imprinting and maternal effects is perhaps not surprising given that several imprinted loci have been shown to affect both growth and parental behavior traits [e.g., Peg3 and Grb10 (Li et al. 1999; Charalambous et al. 2010)].
It is important to keep in mind that this analysis requires one to be able to infer the parent of origin of alleles to identify imprinting effects (Wolf et al. 2008a). Parent-of-origin information can be determined using marker genotype data combined with pedigree information to infer haplotype configurations using various algorithms. In the simplest case, one can genotype the mother, the father, and their offspring to track alleles within families. In our analysis, this was done using the combined genotype data from linked markers from the parents and siblings of an individual in an algorithm that finds the most likely haplotype configuration for the entire set (Li and Jiang 2005). In populations where this cannot be achieved, one can still identify maternal-effect loci, but there will be a chance that any loci with additive maternal effects could be explained by the occurrence of an imprinting effect at that locus (Casellas et al. 2009).
Potential for nonadditivity
An important assumption of analyses that separate maternal and direct effects is the assumption that the two genomes have independent effects that are additive (Wade 1998). However, it is possible that the two genomes interact, where the maternal effect depends on the offspring genotype and vice versa (Gavrilets 1998; Wade 1998; Wolf 2001). For example, if heterozygous offspring respond differently from homozygous offspring to the maternal genotype at the focal locus, then the maternal effects characterized will not apply to the entire population of offspring. Such nonadditivity is analogous to an epistatic interaction between the maternal and the offspring genomes (Wade 1998; Wolf 2001), but within a locus, and means that the effects cannot be cleanly separated into independent influences of the two genomes (Cheverud and Wolf 2009). The missing maternal–offspring genotype combinations make some types of interaction effects confounded with the direct and maternal effects detected using the statistical cross-fostering design, and therefore it is important to recognize the potential for such nonadditivity in the interpretation of effects.
The patterns and implications for the effect of nonadditivity can be clearly seen by drawing an analogy between the maternal–offspring model that includes interactions between the maternal and offspring genomes (Table 9A) and a classic model of two-locus epistasis, where interactions are between two different loci within the same individual (Table 9B) (Cheverud 2000). In the two-locus epistasis model (Table 9A) there are four main effects (additive and dominance effects of the two loci) and four epistatic interactions (pairwise interactions of the additive and dominance effects of the two loci). These effects can be estimated in a regression framework using the phenotypic information from the two-locus genotypes (Cheverud 2000; Álvarez-Castro and Carlborg 2007). However, with the maternal–offspring model, the two missing cells necessarily make some effects confounded, meaning all of the effects cannot be estimated independently in a regression model. The additive-by-additive maternal–offspring interaction effect (i.e., the interaction between the additive direct and additive maternal effects; denoted aamo) is confounded with the additive maternal (am) and direct (ao) effects and in our framework contributes to the confounded effect (cmo) and consequently is not estimated independently (Table 5). Under the statistical cross-fostering approach, the additive-by-dominance (admo) and dominance-by-additive (damo) maternal–offspring interaction effects are confounded with the additive maternal and direct effects, respectively (compare Figure 1 to Table 9B). This constraint is imposed on the analysis as essentially the assumption that heterozygous offspring are representative of all offspring when estimating additive maternal effects and that the offspring of heterozygous mothers are representative of all offspring when estimating additive direct effects. Although this constraint complicates the interpretation of the direct and maternal effects, it still means that, when there is a significant additive maternal effect, the offspring phenotype depends on the genotype of its mother, so there is some form of context-dependent maternal effect (Cheverud and Wolf 2009) and the maternal effect estimated applies only to the subpopulation of heterozygous offspring. The only interaction effect that can be estimated independently is the dominance-by-dominance (ddmo) maternal–offspring interaction effect (Equation 10), which is a measure of the degree to which the dominance maternal effect depends on the offspring’s genotype and vice versa.
Table 9. Relationship between two-locus epistasis and the maternal-effect model.
| Locus B | |||
|---|---|---|---|
| B1B1 | B1B2 | B2B2 | |
| A. Two-locus epistasis with unordered genotypesa | |||
| Locus A | |||
| A1A1 | +aA +aB + aa | +aA + dB + ad | +aA − aB − aa |
| A1A2 | +dA +aB + da | +dA +dB + dd | +dA − aB − da |
| A2A2 | −aA + aB −aa | −aA + dB − ad | −aA − aB + aa |
| Offspring genotype | |||
| A1A1 | A1A2 | A2A2 | |
| B. Maternal-effect modelb | |||
| Maternal genotype | |||
| A1A1 | +am +ao + aamo | +am + do + admo | — |
| A1A2 | +dm +ao + damo | +dm +do + ddmo | +dm − ao − damo |
| A2A2 | — | −am + do − admo | −am − ao + aamo |
With two-locus epistasis with unordered genotypes, epistasis can be measured as the interaction between the additive (aA and aB) and dominance effects of the two loci (dA and dB). There are four forms of epistasis corresponding to the additive-by-additive (aa), additive-by-dominance (ad), dominance-by-additive (da), and dominance-by-dominance (dd) interaction effects (Cheverud 2000).
With the maternal-effect model, the cells are maternal–offspring genotype combinations at a single locus (Wade 1998; Wolf and Hager 2006), so the interactions are between the additive (am) and dominance (dm) maternal effects with the additive (ao) and dominance (do) direct effects at the same locus. There are two missing genotype combinations (dashes) because mothers that are homozygous for one allele cannot have offspring that are homozygous for the other allele.
Although the confounding of nonadditive effects representing interactions between the maternal and offspring genomes at a locus can make the interpretation of maternal effects problematic, it is important to keep in mind again that it is a constraint imposed by the nature of the problem, not the model.
Supplementary Material
Acknowledgments
This research was supported by grants from the Biotechnology and Biological Sciences Research Council (BBSRC) (United Kingdom), an Underwood Fellowship from the BBSRC, and National Institutes of Health grant DK055736.
Footnotes
Communicating editor: G. A. Churchill
Literature Cited
- Álvarez-Castro J. M., Carlborg O., 2007. A unified model for functional and statistical epistasis and its application to quantitative trait loci analysis. Genetics 176: 1151–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames M., Webster J. T., 1991. On estimating approximate degrees of freedom. Am. Stat. 1991: 1. [Google Scholar]
- Atchley W. R., Logsdon T., Cowley D. E., 1991. Uterine effects, epigenetics, and postnatal skeletal development in the mouse. Evolution 45: 891–909. [DOI] [PubMed] [Google Scholar]
- Badyaev A. V., Hill G. E., Beck M. L., Dervan A. A., Duckworth R. A., et al. , 2002. Sex-biased hatching order and adaptive population divergence in a passerine bird. Science 295: 316–318. [DOI] [PubMed] [Google Scholar]
- Bateman N., 1954. The measurement of milk production of mice through preweaning growth of suckling young. Physiol. Zool. 27: 163–173. [Google Scholar]
- Berleth T., Burri M., Thoma G., Bopp D., Richstein S., et al. , 1988. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 7: 1749–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen W. D., 2009. Maternal effects on offspring size and development in pinnipeds, 104–132 Maternal Effects in Mammals, edited by Maestripieri D., Mateo J. J. University of Chicago Press, Chicago. [Google Scholar]
- Brumby P. J., 1960. The influence of the maternal environment on growth in mice. Heredity 14: 1–18. [Google Scholar]
- Casellas J., Farber C. R., Gularte R. J., Haus K. A., Warden C. H., et al. , 2009. Evidence for maternal QTL affecting growth and obesity in adult mice. Mamm. Genome 20: 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai C. K., 1956. Analysis of quantitative inheritance of body size in mice. I. Hybridization and maternal influence. Genetics 41: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charalambous M., Cowley M., Geoghegan F., Smith F. M., Radford E. J., et al. , 2010. Maternally-inherited Grb10 reduces placental size and efficiency. Dev. Biol. 1: 1–8. [DOI] [PubMed] [Google Scholar]
- Chen L., Storey L. D., 2006. Relaxed significance criteria for linkage analysis. Genetics 173: 2371–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud J. M., 2000. Detecting epistasis among quantitative trait loci, 58–81 Epistasis and the Evolutionary Process, edited by Wolf J. B., Brodie E. D. I., Wade M. J. Oxford University Press, New York. [Google Scholar]
- Cheverud J. M., 2006. Genetic architecture of quantitative variation, 288–309 Evolutionary Genetics: Concepts and Case Studies, edited by Fox C. W., Wolf J. B. Oxford University Press, New York. [Google Scholar]
- Cheverud J. M., Wolf J. B., 2009. Genetics and evolutionary consequences of maternal effects, 11–37 Maternal Effects in Mammals, edited by Maestripieri D., Mateo J. J. University of Chicago Press, London. [Google Scholar]
- Cheverud J. M., Hager R., Roseman C., Fawcett G., Wang B., et al. , 2008. Genomic imprinting effects on adult body composition in mice. Proc. Natl. Acad. Sci. USA 105: 4253–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley D. E., Pomp D., Atchley W. R., Eisen E. J., Hawkins-Brown D., 1989. The impact of maternal uterine genotype on postnatal growth and adult body size in mice. Genetics 122: 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D. F., Legates J. E., Cockerham C. C., 1959. Maternal influence on body weight. J. Anim. Sci. 18: 519–527. [Google Scholar]
- Donohue K., 1998. Maternal environmental effects in plants, 137–177 Maternal Effects as Adaptations, edited by Mousseau T. A., Fox C. W. Oxford University Press, New York. [Google Scholar]
- Faes C., Molhenberghs G., Aerts M., Verbeke G., Kenward M. G., 2009. The effective sample size and an alternative small-sample degrees-of-freedom method. Am. Stat. 63: 389–399. [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics, Ed. 4 Longman, Essex, UK. [Google Scholar]
- Fry J. D., 2004. Estimation of genetic variances and covariances by restricted maximum likelihood using Proc Mixed, 11–34 Genetic Analysis of Complex Traits Using SAS, edited by Saxton A. M. SAS Institute, Cary, NC. [Google Scholar]
- Gavrilets S., 1998. One-locus two-allele models with maternal (parental) selection. Genetics 149: 1147–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzburg L. R., 1998. Inertial growth: population dynamics based on maternal effects, 42–53 Maternal Effects as Adaptations, edited by Mousseau T. A., Fox C. W. Oxford University Press, New York. [Google Scholar]
- Goodale H. D., 1938. A study of the inheritance of body weight in the albino mouse by selection. J. Hered. 29: 101–112. [Google Scholar]
- Hager R., Cheverud J. M., Roseman C., Wolf J. B., 2008. Maternal effects as the cause of parent-of-origin effects that mimic genomic imprinting. Genetics 178: 1755– 1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager R., Cheverud J. M., Wolf J. B., 2009. Relative contribution of additive, dominance and imprinting effects to phenotypic variation in body size and growth between divergent selection lines of mice. Evolution 63: 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis J. P., Kenney-Hunt J., Ehrich T. H., Pletscher L. S., Semenkovich C. F., et al. , 2005. Maternal genotype affects adult offspring lipid, obesity, and diabetes phenotypes in LGXSM recombinant inbred strains. J. Lipid Res. 46: 1692–1702. [DOI] [PubMed] [Google Scholar]
- Kenward M. G., Roger J. H., 1997. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics 53: 983–997. [PubMed] [Google Scholar]
- Keselman H. J., Algina J., Kowalchuk R. K., Wolfinger R. D., 1999. The analysis of repeated measurements: a comparison of mixed-model satterthwaite f tests and a nonpooled adjusted degrees of freedom multivariate test. Comm. Stat. Theory Methods 28: 2967–2999. [Google Scholar]
- Kever W. J. M., Rotman G., 1987. Development of ethanol tolerance in relation to the alchohol dehydrogenase locus in Drosophila melamogaster. II. The influence of phenotypic adaptation and maternal effect on survival on alcohol supplemented media. Heredity 58: 239–248. [Google Scholar]
- Kincaid C., 2005. Guidelines for selecting the covariance structure in mixed model analysis, paper no. 198-30 in Proceedings of the Thirtieth Annual SAS Users Group International Conference. SAS Institute, Cary, NC. [Google Scholar]
- Kirkpatrick M., Lande R., 1989. The evolution of maternal characters. Evolution 43: 485–503. [DOI] [PubMed] [Google Scholar]
- Kramer M. G., Vaughn T. T., Pletscher L. S., King-Ellison K., Adams E., et al. , 1998. Genetic variation for body weight gain and composition in the intercross of Large (LG/J) and Small (SM/J) inbred strains of mice. Genet. Mol. Biol. 21: 211–218. [Google Scholar]
- Li J., Ji L., 2005. Adjusting multiple testing in multilocus analysis using the eigenvalues of a correlaton matrix. Heredity 95: 221–227. [DOI] [PubMed] [Google Scholar]
- Li J., Jiang T., 2005. Computing the minimum recombination haplotype configuration from incomplete genotype data on a pedigree by integer linear programming. J. Comput. Biol. 12: 719–739. [DOI] [PubMed] [Google Scholar]
- Li L., Keverne E. B., Aparicio S. A., Ishino F., Barton S. C., et al. , 1999. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science 284: 330–333. [DOI] [PubMed] [Google Scholar]
- Littell R. C., Williken G. A., Stroup W. W., Wolfinger R. D., Schabenberger O., 2006. SAS for Mixed Models. SAS Institute, Cary, NC. [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA. [Google Scholar]
- MacArthur J., 1944. Genetics of body size and related characters. I. Selection of small and large races of the laboratory mouse. Am. Nat. 78: 142–157. [Google Scholar]
- Maestripieri D., Mateo J. J. (Editors), 2009. Maternal Effects in Mammals. University of Chicago Press, Chicago. [Google Scholar]
- Moore A. J., Brodie E. D., III, Wolf J. B., 1997. Interacting phenotypes and the evolutionary process: I. Direct and indirect genetic effects of social interactions. Evolution 51: 1352–1362. [DOI] [PubMed] [Google Scholar]
- Mousseau T. A., Fox C. W., 1998. Maternal Effects as Adaptations. Oxford University Press, New York. [Google Scholar]
- Odling-Smee F. J., Laland K. N., Feldman M. W., 2003. Niche Construction: The Neglected Process in Evolution. Princeton University Press, Princeton, NJ. [Google Scholar]
- Price T., 1998. Maternal and paternal effects in birds, 202–224 Maternal Effects as Adaptations, edited by Mousseau T. A., Fox C. W. Oxford University Press, New York. [Google Scholar]
- Rhees B. K., Ernst C. A., Miao C. H., Atchley W. R., 1999. Uterine and postnatal maternal effects in mice selected for differential rate of early development. Genetics 153: 905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff D. A., 1997. Evolutionary Quantitative Genetics. Chapman & Hall, New York. [Google Scholar]
- Schaalje G. B., McBride J. B., Fellingham G. W., 2001. Approximations to distributions of test statistics in complex mixed linear models using SAS Proc MIXED, paper 26226 in Proceedings of the Twenty-Sixth Annual SAS Users Group International Conference. SAS Institute, Cary, NC. [Google Scholar]
- Vaughn T. T., Pletscher L. S., Peripato A., King-Ellison K., Adams E., et al. , 1999. Mapping quantitative trait loci for murine growth—a closer look at genetic architecture. Genet. Res. 74: 313–322. [DOI] [PubMed] [Google Scholar]
- Wade M. J., 1998. The evolutionary genetics of maternal effects, 5–21 Maternal Effects as Adaptations, edited by Mousseau T. A., Fox C. W. Oxford University Press, New York. [Google Scholar]
- White J. M., Legates J. E., Eisen E. J., 1968. Maternal effects among lines of mice selected for body weight. Genetics 60: 395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. J., Festa-Bianchet M., 2009. Maternal effects in wild ungulates, 83–103 Maternal Effects in Mammals, edited by Maestripieri D., Mateo J. J. University of Chicago Press, Chicago. [Google Scholar]
- Wilson A. J., Coltman D. W., Pemberton J. M., Overall A. D. J., Byrne K. A., et al. , 2005. Maternal genetic effects set the potential for evolution in a free-living vertebrate population. J. Evol. Biol. 18: 405–414. [DOI] [PubMed] [Google Scholar]
- Wolf J. B., 2001. Gene interactions from maternal effects. Evolution 54: 1882–1898. [DOI] [PubMed] [Google Scholar]
- Wolf J. B., Brodie E. D., III, 1998. Coadaptation of parental and offspring characters. Evolution 52: 535–544. [DOI] [PubMed] [Google Scholar]
- Wolf J. B., Hager R., 2006. A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 4: e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J. B., Wade M. J., 2009. What are maternal effects (and what are they not)? Philos. Trans. R. Soc. Lond. B Biol. Sci. 364: 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J. B., Moore A. J., Brodie E. D., III, 1997. The evolution of indicator traits for parental quality: the role of maternal and paternal effects. Am. Nat. 150: 639–649. [DOI] [PubMed] [Google Scholar]
- Wolf J. B., Brodie E. D., III, Cheverud J. M., Moore A. J., Wade M. J., 1998. Evolutionary consequences of indirect genetic effects. Trends Ecol. Evol. 13: 64–69. [DOI] [PubMed] [Google Scholar]
- Wolf J. B., Moore A. J., Brodie E. D., III, 1999. Maternal and paternal effects in sexual selection and the evolution of parental investment. J. Evol. Biol. 12: 1157–1167. [Google Scholar]
- Wolf J. B., Vaughn T. T., Pletscher L. S., Cheverud J. M., 2002. Contribution of maternal effect QTL to genetic architecture of early growth in mice. Heredity 89: 300–310. [DOI] [PubMed] [Google Scholar]
- Wolf J. B., Hager R., Cheverud J. M., 2008a Genomic imprinting effects on complex traits: a phenotype-based perspective. Epigenetics 3: 295–299. [DOI] [PubMed] [Google Scholar]
- Wolf J. B., Cheverud J. M., Roseman C., Hager R., 2008b Genome-wide analysis reveals a complex pattern of genomic imprinting in mice. PLoS Genet. 4: e1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J. B., Leamy L. J., Roseman C. C., Cheverud J. M., 2011. Disentangling prenatal and postnatal maternal genetic effects reveals persistent prenatal effects on offspring growth in mice. Genetics 189: 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.