Abstract
Mediator is a modular multisubunit complex that functions as a critical coregulator of RNA polymerase II (Pol II) transcription. While it is well accepted that Mediator plays important roles in the assembly and function of the preinitiation complex (PIC), less is known of its potential roles in regulating downstream steps of the transcription cycle. Here we use a combination of genetic and molecular approaches to investigate Mediator regulation of Pol II elongation in the model eukaryote, Saccharomyces cerevisiae. We find that ewe (expression without heat shock element) mutations in conserved Mediator subunits Med7, Med14, Med19, and Med21—all located within or adjacent to the middle module—severely diminish heat-shock–induced expression of the Hsf1-regulated HSP82 gene. Interestingly, these mutations do not impede Pol II recruitment to the gene’s promoter but instead impair its transit through the coding region. This implies that a normal function of Mediator is to regulate a postinitiation step at HSP82. In addition, displacement of histones from promoter and coding regions, a hallmark of activated heat-shock genes, is significantly impaired in the med14 and med21 mutants. Suggestive of a more general role, ewe mutations confer hypersensitivity to the anti-elongation drug 6-azauracil (6-AU) and one of them—med21—impairs Pol II processivity on a GAL1-regulated reporter gene. Taken together, our results suggest that yeast Mediator, acting principally through its middle module, can regulate Pol II elongation at both heat-shock and non–heat-shock genes.
IN eukaryotes, transcription of the DNA template into pre-mRNA by RNA polymerase II (Pol II) occurs in a well-defined, stepwise fashion. First, chromatin, the nucleoprotein complex in which the DNA is packaged, must unfold into a 10 nm, beads-on-a-string filament, and for many genes a nucleosome-free region needs to be created over the core promoter (Venters and Pugh 2009). Both are achieved via activator-mediated recruitment of chromatin modification and remodeling enzymes (reviewed in Li et al. 2007). Once a permissive chromatin template has been created, Pol II and the other general transcription factors then bind the core promoter, where they are assembled into a preinitiation complex (PIC; formally analogous to the closed RNA polymerase complex in prokaryotes). Next, Pol II forms an open complex concomitant with ATP-dependent melting of the DNA strands and initiates transcription following phosphorylation of its C-terminal repeat domain (CTD) at Ser5 residues by the TFIIH kinase, Cdk7. Following synthesis of ∼25–30 nucleotides, Pol II pauses, allowing the nascent RNA to be capped. Finally, Pol II transitions to productive elongation, which requires Ser2 phosphorylation of the CTD. In metazoans, this is catalyzed by P-TEFb and in yeast by Bur1 and Ctk1 (reviewed in Saunders et al. 2006).
A key regulator of many of the above steps is Mediator, an evolutionarily conserved, modular multiprotein complex. Mediator acts as a signal transducer through its interaction with gene-specific regulatory proteins and Pol II to fine tune gene transcription levels. In organisms as diverse as budding yeast and mammals, Mediator adopts a similar overall architecture characterized by the presence of four modules termed head, middle, tail, and kinase (Cai et al. 2009). Although there exists only limited sequence homology between orthologous subunits in mammalian and yeast Mediator, their functions seem to be conserved (Bourbon 2008). The head module is essential for Mediator function as mutations within it abolish transcription in vivo (Thompson and Young 1995). Moreover, in yeast, the head subunit Med17/Srb4 physically interacts with the Pol II subunit, Rpb3, and this interaction is important for genome-wide Pol II recruitment in vivo (Soutourina et al. 2011). The middle module confers structural integrity to Mediator (Baumli et al. 2005). It also facilitates activated transcription in purified systems (Baidoobonso et al. 2007) and has a dual role in both activation and repression in cells (Tabtiang and Herskowitz 1998; Gromoller and Lehming 2000; Singh et al. 2006). The middle module also has been implicated in regulating transcriptional silencing (Zhu et al. 2011a) and interaction with histone tails (Zhu et al. 2011b). The tail module is the principal physical target of gene-specific activators and repressors. The strong conservation of function is illustrated by the fact that the tail subunit, Med15/Gal11, is a target of both mammalian sterol regulatory element-binding protein (SREBP), a sensor of cholesterol and regulator of lipid homeostasis, and a zinc finger-containing activator in yeast that senses fatty acid levels. Finally, the kinase (Cdk8) module is a separable subcomplex that reversibly associates with core Mediator and has been implicated in transcriptional repression (reviewed in Malik and Roeder 2010).
There is considerable evidence that Mediator plays roles in PIC assembly and function (reviewed in Malik and Roeder 2000; Myers and Kornberg 2000; Malik and Roeder 2010), and may serve as a scaffold around which the entire PIC assembles (Bernecky et al. 2011). However, less is known of its potential downstream roles. Several recent studies in mammalian cells implicate a role for Mediator in post-recruitment regulation. First, analysis of the ELK1-regulated Egr1 gene revealed that despite a substantial reduction in Egr1 transcript levels in Med23−/− mouse embryonic stem cells, PIC recruitment was only moderately diminished. Pol II occupancy within the proximal coding region, on the other hand, was drastically reduced (Wang et al. 2005). Second, addition of Mediator facilitates Pol II release from a DRB sensitivity-inducing factor (DSIF)-imposed paused state in vitro (Malik et al. 2007). Third, p53-induced changes in the conformational state of Mediator were found to stimulate stalled, promoter-proximal Pol II molecules to productive elongation both in vitro and in cells (Meyer et al. 2010). Fourth, Cdk8 was observed to be a potent regulator of Pol II elongation at serum-responsive genes in human cells, as its knockdown impaired Pol II elongation and correlated with decreased levels of CTD phosphorylation at Ser2 and Ser5. Moreover, co-immunoprecipitation assays demonstrated the presence of the P-TEFb subunits Cdk9 and cyclin T1 in a Cdk8–Mediator complex isolated from these cells (Donner et al. 2010). Fifth, the N-terminal domain of Med26—located at the junction between middle and tail modules—was observed to act as a docking site for complexes that contain transcription elongation factors such as P-TEFb and members of the ELL/EAF family in human cells (Takahashi et al. 2011). Consistent with this, siRNA-mediated depletion of Med26 resulted in defective recruitment of transcription elongation factors and Pol II CTD phosphorylation at several genes (Takahashi et al. 2011).
Taken together, these mammalian studies indicate that Mediator’s role extends beyond PIC assembly and function, and that Mediator has roles at several postrecruitment steps. While less is known in this regard of yeast Mediator, genetic analyses have implicated a role for head (Med18/Srb5, Med19/Rox3, and Med20/Srb2), middle (Med31/Soh1), and tail subunits (Med16/Sin4, Med15, and Med2) in transcription elongation (Malagon et al. 2004; Gaillard et al. 2009). In addition, a recent genetic/ChIP analysis showed that Mediator plays a role in postrecruitment regulation of both a lexA–TBP-driven reporter gene and a natural gene, CYC1 (Lee et al. 2010). Our laboratory has previously shown that Mediator plays an essential role in governing heat-shock (HS) gene expression in Saccharomyces cerevisiae. Genetic analysis demonstrated that six Mediator subunits, located principally in the middle module, can act as negative regulators of heat-shock protein (HSP) gene basal transcription, and positive regulators of induced HSP gene transcription (Singh et al. 2006). These results suggested that Mediator may collaborate with the evolutionarily conserved transcriptional activator, Hsf1, in controlling the dynamic range of HSP gene expression. In this study, we have extended our investigation into how Mediator regulates HSP gene transcription, with emphasis on its role in regulating Pol II elongation. We find that mutations in subunits identified in our earlier genetic screen severely diminish transcriptional output of the Hsf1-responsive gene, HSP82, yet typically do not impede Pol II recruitment to its promoter. Instead, these mutations impair either Pol II elongation rate or processivity at HSP82, and this effect appears to extend to non–heat-shock (NHS) genes as well.
Materials and Methods
Yeast strain construction
Spontaneous ewe suppressors were derived from strain HS1004 (SLY101 background) and sorted into six complementation groups, termed EWE1–EWE6, as described previously (Singh et al. 2006). As HS1004 is a MATa strain that bears the mutated hsp82-ΔHSE1 allele, we replaced hsp82-ΔHSE1 with wild-type HSP82 to permit RNA expression and ChIP analysis of this gene. This was achieved by mating strain SLY101 (MATα, HSP82) to representative suppressors in complementation groups EWE3–EWE6: J121 (bearing med7-1002); J20 (bearing med21/srb7-1002); J84 (bearing med10/nut2-1002); and B20 (bearing med19/rox3-1002). These suppressors were selected on the basis of the robustness of their ewe phenotype (hsp82-ΔHSE1/lacZ basal expression). Diploids were then sporulated and tetrads dissected; spores containing both HSP82 and the ewe mutation were identified using molecular and genetic methods, creating strains termed SK-J121b, SK-J20b, SK-J84b, and SK-B20c, respectively. The engineered mutation, rgr1-Δ2, was crossed into the HS1004 background as previously described (Singh et al. 2006), creating strain SBK502. These strains were employed in RT–qPCR and ChIP assays.
For spot dilution and GLAM (gene length-dependent accumulation of mRNA) assays, the following HS1004 derivatives were used: J37 (med14-1003); J44 (med7-1002, bears the same mutation as J121); J20 (med21-1002); J84 (med10-1002); and B20 (med19-1002). In addition, the closely related MATα strain, HS1001, was used for the following gene deletions (KAN-MX knockouts): med5Δ, med12Δ, med16Δ, med20Δ, and cdk8Δ. Knockout DNA fragments were generated by PCR using as template genomic DNA isolated from strains in the Res-Gen strain collection (generously provided by Kelly Tatchell). Strain genotypes are presented in Table 1.
Table 1 . Yeast strains.
| Strain | Genotype | Source or reference |
|---|---|---|
| SLY101 | MATα ade- can1-100 cyh2r his3-11,15 leu2-3,112 trp1-1 ura3 | Lee and Gross (1993) |
| JHD401 | SLY101; asf1Δ | This study |
| EAS2001 | SLY101; hsp82-2001; sir4Δ2::HIS3 | Sekinger and Gross (1999) |
| JHD303 | EAS2001; ctk1Δ | This study |
| DCY101 | EAS2001; dst1Δ | This study |
| DCY102 | EAS2001; spt4Δ | This study |
| HS1001 | SLY101; hsp82-ΔHSE1 hsp82-ΔHSE1-HIS3::ura3Δ hsp82-ΔHSE1/lacZ::LEU2 TRP1+ | Singh et al. (2006) |
| HS1004 | HS1001; MATa hsp82-ΔHSE1/HIS3::URA3 leu2::hsp82-ΔHSE1/lacZ::leu2::KAN-MX | Singh et al. (2006) |
| B20 | HS1004; med19-1002 | Singh et al. (2006) |
| J20 | HS1004; med21-1002 | Singh et al. (2006) |
| J37 | HS1004; med14-1003 | Singh et al. (2006) |
| J44 | HS1004; med7-1002 | Singh et al. (2006) |
| J84 | HS1004; med10-1002 | Singh et al. (2006) |
| J121 | HS1004; med7-1002 | Singh et al. (2006) |
| DAD2 | HS1001; med20Δ::KAN-MX | This study |
| YAH01 | HS1001; med12Δ::KAN-MX | This study |
| RRG2 | HS1001; cdk8Δ::KAN-MX | This study |
| HS1005 | HS1001; med16Δ::KAN-MX | This study |
| JHD1 | HS1001; med5Δ::KAN-MX | This study |
| SK-HS1004 | SLY101; hsp82-ΔHSE1-lacZ::leu2Δ::KAN-MX | This study |
| SK-B20c | SK-HS1004; MATa med19-1002 | This study |
| SK-J20b | SK-HS1004; MATa med21-1002 | This study |
| SK-J84b | SK-HS1004; med10-1002 | This study |
| SK-J121b | SK-HS1004; MATa hsp82-ΔHSE1/HIS3::URA3 med7-1002 | This study |
| DY2694 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 rgr1-Δ2::LEU2 | D. J. Stillman |
| SBK500 | SK-HS1004; MATa rgr1-Δ2::LEU2 | This study |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| dst1Δ | BY4741; dst1Δ::KAN-MX | Research Genetics |
Sequence analysis of chromosomal mutations
To amplify the pertinent locus of each ewe suppressor, we employed 100 ng of genomic DNA as template and 1 unit of Phusion DNA polymerase (Finnzymes, F-530). All PCR products were purified using QIAquick gel extraction kit (Qiagen, 28704). Purified PCR products were sequenced on both upper and lower strands using primers spaced ∼200–300 bp apart by Retrogen. For each ewe suppressor, two independent genomic DNA samples were analyzed to obviate the possibility of a PCR-generated artifact. Amplicons and sequencing strategy were as follows: med7, amplified region encompassed −85 to +749 (where +1 = ATG initiation codon), two upper and two lower strand sequencing primers used; med10, −141 to +570, two upper, two lower strand sequencing primers; med14, −142 to + 3420, six upper, six lower strand sequencing primers; med19, −91 to +733, two upper, two lower strand sequencing primers; and med21, −127 to +449, two upper, two lower strand sequencing primers.
β-Galactosidase liquid assays
β-Galactosidase liquid assays were performed as previously described (Singh et al. 2006) on cells either maintained at 30° (NHS) or shifted to 39° for 45 min (HS). The latter were returned to 30° for 20 min to permit efficient export and translation of lacZ mRNA.
RT–qPCR analysis
Cells were cultivated to an A600 of ∼0.6 in a 100-ml culture at 30° and 20-ml aliquots were removed and subjected to an instantaneous 30° to 39° upshift through addition of an equivalent volume of prewarmed (51°) medium for 0, 2, 15, and 60 min. Heat-shock induction was terminated through addition of 20 mM sodium azide, and RNA was isolated as above. Contaminating genomic DNA was removed from each RNA sample by digestion with TURBO DNA-free (Applied Biosystems, AM1907), followed by phenol/chloroform extraction. One half microgram of purified RNA was used in each cDNA synthesis with ProtoScript RT–PCR kit (New England Biolabs, E6400S). Random primers were used in cDNA synthesis. Two percent of the synthesized cDNA was added to each 20-µl real-time PCR reaction, performed using RT2 qPCR SYBR Green/ROX master mix (SABiosciences, 330529) on an Applied Biosystems 7900HT real-time PCR system. A standard curve specific for each amplicon was generated, the quantity of DNA present in each sample was determined, and background signal was subtracted. Primers were designed to target the coding regions of HSP82, SSA4, and SCR1. Their coordinates are: HSP82, +1248 to +1444; SSA4, +1092 to +1150; and SCR1, +21 to +76. For quantification, SCR1 was used to normalize both HSP82 and SSA4 mRNA levels.
Chromatin immunoprecipitation ChIP
End point PCR:
End point (gel-based) PCR ChIP assays (Figure 3, Figure S2, and Figure S3) were performed as previously described (Zhao et al. 2005; Kremer and Gross 2009). To effect immunoprecipitation (IP) of Pol II, an anti-CTD rabbit antiserum (Balakrishnan and Gross 2008) was used; to effect immunoprecipitation of Myc-tagged H4, the 9E10 mouse monoclonal antibody (Santa Cruz Biotechnology) was used. For Pol II ChIPs, net signal at HSP82 was normalized to that at a nontranscribed locus on chromosome V (ARS504). For myc-H4 ChIPs, net signal at HSP82 was normalized to that at the PHO5 promoter.
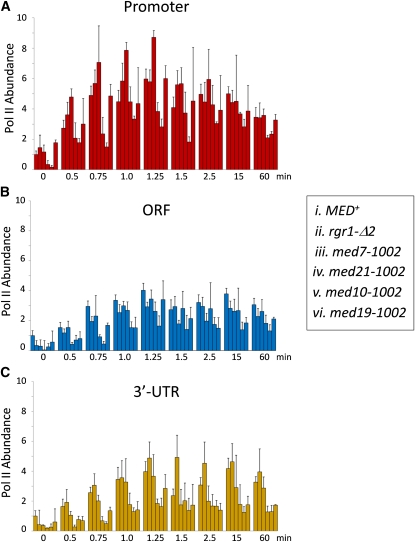
Figure 3 .
ewe mutations differentially affect RNA Pol II abundance at the heat-shock–induced HSP82 gene. In vivo crosslinking analysis of Pol II at the HSP82 promoter (A), ORF (B), and 3′-UTR (C) either prior to or for the indicated times following an instantaneous 39° heat shock. Pol II abundance at HSP82, quantified using an end-point PCR (gel-based) ChIP assay (Kremer and Gross 2009), was normalized to its abundance at a nontranscribed region (ARS504) evaluated in the same multiplex PCR. For each time point, data are presented for WT and the five congenic ewe mutants in the order indicated in the key. For the WT strain, shown are means ± SEM (N = 3 biological samples; PCR = 12); for rgr1-Δ2 and med10-1002, shown are means ± SEM (N = 4; PCR = 4); and for med7-1002, med21-1002, and med19-1002, shown are means ± SD (N = 2; PCR = 2).
Real-time PCR:
For real-time ChIP analysis (Figure 4), 200-ml midlog cell cultures were used in heat-shock time course experiments, and 50-ml aliquots were removed for each time point. Cells were crosslinked with 1% formaldehyde and chromatin was sheared to a mean length of ∼0.5 kb with a Branson 250 sonifier equipped with a microtip. Approximately 2.6 ml of crosslinked chromatin was obtained for each time point and the equivalent of ∼600 µg of chromatin protein (as quantified by the Bradford assay; typically 0.2 ml) was employed for each IP. Immunoprecipitations were conducted through addition of 55 μl of CL-4B protein A sepharose beads and 2 μl of an H3 globular domain-specific antibody (Abcam, ab1791) with incubation at 4° overnight. DNA was purified and dissolved in 50 µl TE, and 2 µl of immunoprecipitated DNA was added to each 20-µl real-time PCR reaction, performed as described above. The quantity of DNA present in each IP was determined using a standard curve specific for each amplicon and background signal arising from beads alone was subtracted. To normalize for variation in sample recovery, abundance of H3 at the PHO5 promoter (which is not activated by heat shock) was determined for each sample. Amplicon coordinates were: HSP82 promoter, −157 to −88; HSP82 ORF, +1248 to +1444; HSP82 3′-UTR, +2134 to +2228; and PHO5 promoter, −197 to −124.
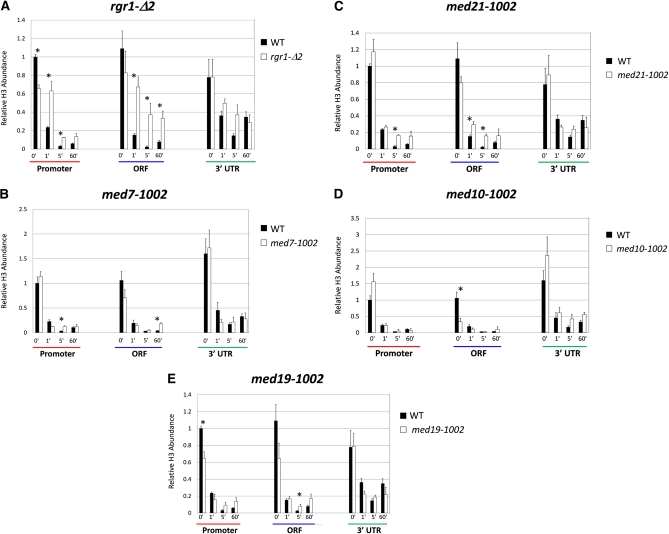
Figure 4 .
Mutations in the Med14/Rgr1 and Med21 subunits of Mediator diminish the extent of histone displacement at HSP82. Kinetics of histone H3 depletion over the promoter, ORF, and 3′-UTR of HSP82 in WT and mutant cells as assayed by ChIP–qPCR at the indicated times following an instantaneous 30° to 39° temperature shift. H3 globular domain abundance at HSP82 was normalized to its abundance at the PHO5 promoter. This was determined by dividing each HSP82 ChIP value (net nanograms DNA immunoprecipitated) by the corresponding PHO5 ChIP value derived from the same biological sample. Note that H3 occupancy of the PHO5 promoter is independent of both heat-shock and ewe strain background (data not shown and Sekinger and Gross 2001). Depicted are means ± SEM (N = 2; qPCR = 4). Asterisks signify a potentially significant difference between the indicated values (P ≤ 0.06; two-tailed t-test; equal variance). (A–E) H3 occupancy in MED+ and the indicated ewe mutant strains. A, C, and E represent one set of immunoprecipitation reactions; B and D represent a second, independently conducted set.
Spot dilution analysis
Spot dilution analysis was conducted essentially as described (Kim et al. 2011). Cultures were applied to SDC −Ura solid medium with or without 50 μg/ml 6-azauracil.
GLAM assay
Wild-type (WT) and mutant strains were transformed with either long transcript plasmid (pSCh212, containing PGAL1-PHO5::lacZ) or short transcript plasmid (pSCh202, containing PGAL1-PHO5) (Morillo-Huesca et al. 2006) (gift of Daniel Ginsburg and Alan Hinnebusch). Cell growth, collection, and acid phosphatase activity were assayed as previously described (Kim et al. 2011).
Results
Previous work has shown that heat-shock–induced gene transcription in S. cerevisiae is resilient to mutations in TFIIA, TFIID, TFIIH, SAGA, the head module of Mediator, and even the Pol II CTD (Moqtaderi et al. 1996; Apone et al. 1998; Lee and Lis 1998; McNeil et al. 1998; Chou et al. 1999). By contrast, such Hsf1-mediated activation is sensitive to mutations in Mediator’s middle and tail modules (Lee et al. 1999; Singh et al. 2006). Here we investigate the nature of the Hsf1 transcriptional defect caused by mutations in the middle module and more generally, the role of Mediator in regulating Pol II elongation in S. cerevisiae. In a previous genetic analysis, we isolated spontaneous mutations in six Mediator subunits—Med7, Med10/Nut2, Med14/Rgr1, Med16/Sin4, Med19/Rox3, and Med21/Srb7—as extragenic suppressors (Singh et al. 2006). These suppressors were selected on the basis of their ability to increase the basal transcription of a chromosomal reporter gene, hsp82-ΔHSE1/lacZ (HSP82 lacking its high-affinity Hsf1 binding site, HSE1, fused to the Escherichia coli lacZ coding region), and were thus termed expression without heat shock element (ewe) mutants. In the present study, we have focused on representative members of complementation groups EWE2–EWE6 that encode subunits located either within or adjacent to the middle module. The EWE1 complementation group is composed of alleles of the nonessential tail subunit, Med16 (Singh et al. 2006), and was not studied here.
We first wished to determine the nature of the mutations borne by these suppressors. As shown in Table 2, the alleles designated med7-1002 and med10-1002 (members of complementation groups EWE3 and EWE5) contain stop codons that result in C-terminal truncations of 25 and 34 amino acids, respectively. Similarly, med19-1002 (EWE6) bears a single nucleotide deletion, resulting in a frameshift and as a consequence, a novel, C-truncated terminus. The fourth spontaneous mutant, med21-1002 (EWE4), bears a T227C point mutation that results in the substitution of proline for leucine at position 76. This mutation is located within the α2 helix of Med21 and a hydrophobic patch that serves as a Med7 binding site (Baumli et al. 2005). Thus, the L76P mutation is predicted to not only break the α2 helix but may also disrupt the Med7–Med21 interaction. Likewise, the stop codon in med7-1002, Q198*, lies within the α3 helix that contacts Med21 through the proteins’ coiled-coil domains (Baumli et al. 2005), suggesting that it too may diminish the stability of the Med7–Med21 subcomplex. Finally, we employed the engineered mutation, rgr1-Δ2, as a representative of the EWE2/MED14 complementation group for ChIP and expression assays. rgr1-Δ2 bears a 336-amino-acid deletion of its C terminus that results in the loss of tail subunits Med3, Med15, and Med16 from isolated Mediator complexes (Jiang et al. 1995; Li et al. 1995).
Table 2 . Sequences and expression phenotypes of ewe mutations.
| EWE Complementation group | Mediator subunit (size) | Allele | Mediator module | Expression phenotypea (hsp82-ΔHSE1/lacZ reporter) | Mutation | |
|---|---|---|---|---|---|---|
| Basal expression | Fold induction | |||||
| Wild type | — | — | — | 1.0 | 10.5 | — |
| EWE2 | Med14/Rgr1 (1082 aa) | rgr1-Δ2 | Spans tail and middle | 140 | 1.2 | 336-amino-acid C-terminal truncationb |
| EWE3 | Med7 (223 aa) | med7-1002 | Middle | 58 | 1.0 | 25-amino-acid C-terminal truncation (C592T converts Q198 to a stop codon) |
| EWE4 | Med21/Srb7 (140 aa) | med21-1002 | Middle | 45 | 1.6 | L76P point mutation (T227C) |
| EWE5 | Med10/Nut2 (158 aa) | med10-1002 | Middle | 130 | 0.84 | 34-amino-acid C-terminal truncation (C371A converts S124 to a stop codon) |
| EWE6 | Med19/Rox3 (220 aa) | med19-1002 | Spans head and middle | 21 | 3.0 | Deletion of A577 causes a frameshift at R193 resulting in a novel, truncated C terminusc |
Basal expression levels (β-galactosidase units) were normalized to the wild-type strain whose expression was set to 1; fold induction represents the quotient of induced expression/basal expression.
Engineered mutation (gift of D. J. Stillman).
C termini: Med19+: KKRKNKSSGS SMATPTHSDS HEDMKRRRLE - Stop. Med19-1002: KKGRTNLEVR WLHQHIVTVM RI - Stop.
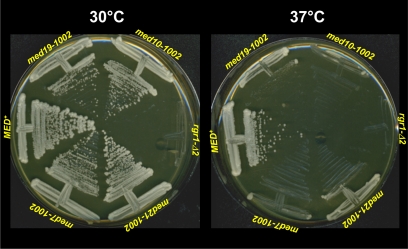
As summarized in Table 2, each ewe mutation has a profound effect on hsp82-ΔHSE1/lacZ basal expression, increasing it 20- to 140-fold. On the other hand, each mutation severely impairs the fold inducibility of hsp82-ΔHSE1/lacZ, consistent with previous observations of other ewe alleles of these genes (Singh et al. 2006). Each mutation also conferred marked temperature sensitivity, while several (med7-1002, med10-1002, and med19-1002) conferred a mild growth defect at 30°; the engineered rgr1-Δ2 mutation caused severe slow growth (see Figure 1). Nonetheless, and important for work described below, a 60 min, 39° heat shock had no discernible effect on the viability of any mutant (see Supporting Information, Figure S1).
Figure 1 .
Growth phenotypes of the MED+ strain SK-HS1004 and congenic ewe mutants employed in this study. Cells were grown on rich medium at the indicated temperature for 2 days.
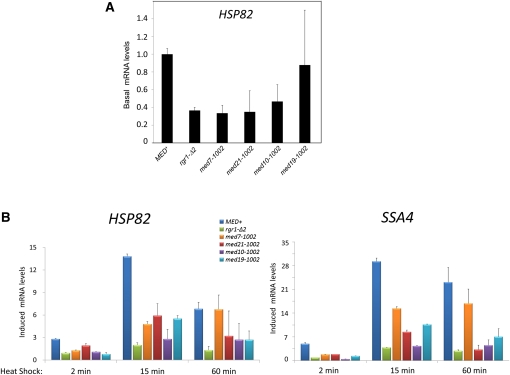
To extend our expression analysis to the wild-type HSP82 gene, we conducted reverse transcription coupled with quantitative PCR (RT–qPCR) on newly constructed strains (see Materials and Methods). As shown in Figure 2A, the spontaneous ewe mutations have a modest effect on basal HSP82 transcription, and may even decrease it. This contrasts with the stimulatory effect of these mutations on the hsp82-ΔHSE1/lacZ reporter, whose promoter is assembled into a repressive dinucleosome (Gross et al. 1993; Venturi et al. 2000) and whose coding region is of bacterial origin. Therefore, the basal expression state of hsp82-ΔHSE1/lacZ may be sensitized to recessive mutations in genes encoding specific subunits of Mediator, possibly as a consequence of the reporter’s unique chromatin structure (see Discussion). In addition, the ewe mutations significantly diminished heat-shock–induced HSP82 mRNA levels at multiple time points (Figure 2B, left). The rgr1-Δ2 mutation, in particular, reduced HSP82 transcript levels (by 70–85%). Similar effects were seen with a second Hsf1-responsive gene, SSA4 (Figure 2B, right).
Figure 2 .
ewe mutations diminish both noninduced and induced HSP82 transcription. (A) RT–qPCR analysis of basal HSP82 mRNA expression levels in the MED+ strain and ewe mutant derivatives. Cells were cultivated at 30° and RNA was isolated. HSP82 mRNA/SCR1 RNA quotients are shown. (SCR1 is a Pol III transcript and served as an internal recovery control.) The MED+ quotient was set to 1.0, and all others were normalized to it. (B) RT–qPCR analysis as in A, except cells were heat-shocked for 2, 15, or 60 min and both HSP82 mRNA/SCR1 RNA and SSA4 mRNA/SCR1 RNA quotients were determined and then normalized to the MED+ T = 0 min sample. For both panels, means ± SEM are depicted (N = 3 or 4).
To gain insight into the basis for the transcriptional defect, we used ChIP to measure Pol II occupancy of the HSP82 promoter and coding region in the context of each ewe mutation. Formaldehyde crosslinking allowed us to take “snapshots” of Pol II occupancy at closely spaced time points during the early stages of heat shock (0.5, 0.75, 1.0, 1.25, 1.5, and 2.5 min) as well as at longer times (15 and 60 min). This analysis revealed that despite the reduction in transcript levels, Pol II recruitment to the promoter was not impeded in the med7, med14 (rgr1-Δ2), and med19 mutants at any time point (Figure 3A; see also Figure S2, black bars, for an alternative depiction of the data). In the med7 mutant, Pol II promoter recruitment may be increased. In addition, Pol II occupancy within the HSP82 coding region was also largely unaffected in the rgr1-Δ2 and med7-1002 mutants (Figure 3, B and C; Figure S2, white and purple bars), raising the possibility that their transcription phenotypes arise from reduced Pol II elongation rate (see Discussion). By contrast, in the med19 and med21 mutants, Pol II occupancy was diminished within the HSP82 transcribed region, particularly toward its 3′-end, suggestive of a defect in polymerase processivity. Finally, in contrast to the others, the med10-1002 mutation substantially reduced Pol II occupancy within the promoter with concomitant reduction within both ORF and 3′-UTR, consistent with a defect in Pol II recruitment. Therefore, ewe mutations affect more than one step in the transcription cycle, and several appear to elicit their effects at a postrecruitment stage.
We next asked whether the ewe mutations affected either the extent or kinetics of nucleosome eviction within HSP82 following heat shock, using histone H3 ChIP as a measure of nucleosome density. Our previous work has shown that mutations in chromatin remodeling (Swi/Snf, Isw1, Isw2, and Chd1) and histone modification (SAGA, NuA3, Set1, Set2, and Paf1) enzymes have little or no effect on this process (Zhao et al. 2005; Kremer and Gross 2009; J. Zhao and D. S. Gross, unpublished observations). In addition, we found that depletion of the histone chaperone Asf1 had no detectable effect (Figure S3A), despite the fact that this protein has previously been shown to play an important role in removing nucleosomes at Pho4- and Gal4-regulated genes (Adkins et al. 2004; Schwabish and Struhl 2006). Knockout of genes encoding transcription elongation factors Ctk1, TFIIS, and Spt4 likewise did not impact histone eviction at HSP82 (Figure S3B). By contrast, the med14 and med21 mutations in Mediator had a marked effect on H3 displacement, and not only at the HSP82 promoter, but also within its ORF (Figure 4, A and C, asterisks). This could be due to impaired nucleosome disassembly, increased reassembly, or a combination of the two. The rgr1-Δ2 chromatin phenotype is particularly notable given that Pol II density at HSP82 is unaffected in this mutant. The med7 and med19 mutations had a lesser, yet discernible, effect on the extent of heat-shock–induced H3 depletion (Figure 4, B and E). Ironically, the med10 mutation did not impair nucleosome eviction (Figure 4D), despite reduced Pol II occupancy within both HSP82 promoter and coding regions. Differences in ewe phenotypes point to the fact that different lesions within Mediator, even those within its middle module, affect transcription, Pol II occupancy, and histone H3 density of activated HSP82 in different ways.
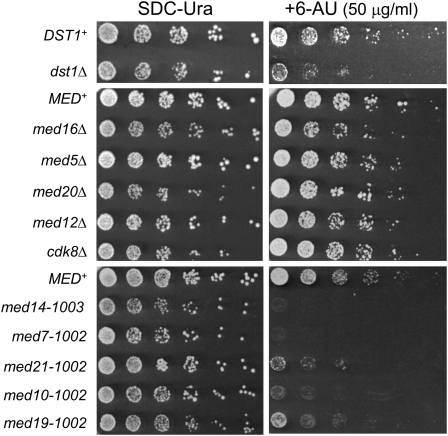
We next investigated whether Mediator mutations affect transcription elongation more generally by comparing the growth of WT and mutant strains at 30° on medium either lacking or containing 6-AU, a drug known to reduce both the elongation rate and processivity of Pol II, secondary to the depletion of cellular UTP and GTP pools (Exinger and Lacroute 1992; Mason and Struhl 2005). As shown in Figure 5, two spontaneous ewe mutants, med14-1003 (bearing a 179-amino-acid C-terminal truncation) and med7-1002, exhibited striking sensitivity to 6-AU (compare to the isogenic MED+ strain, row 9). (The rgr1-Δ2 mutant grew too slowly on control medium to be usable in this assay.) These ewe mutants exceeded the 6-AU sensitivity of a strain lacking TFIIS, an elongation factor that facilitates the release of stalled Pol II molecules (dst1Δ; compare to the isogenic wild-type strain, row 1). Synthetic phenotypes were also observed for the other three ewe mutants, med10-1002, med19-1002, and med21-1002. Interestingly, deletions of several nonessential Mediator subunits, including those from the head (med20Δ), middle (med5Δ), tail (med16Δ), and kinase (cdk8Δ and med12Δ) modules had little or no effect on the 6-AU phenotype (Figure 5, compare rows 4–8 with row 3). These results are consistent with the idea that Ewe subunits of the Mediator middle module, more so than others, play an important role in regulating Pol II elongation in S. cerevisiae.
Figure 5 .
Spontaneous ewe mutants exhibit growth defects on medium containing 6-azauracil. Fivefold serial dilutions of the indicated strains were spotted onto solid media deficient in uracil and containing either no drug (left) or 50 µg/ml 6-AU (right). (Top) Isogenic DST1+ and dst1Δ strains; BY4741 strain background. (Middle) HS1001 derivatives (SLY101 strain background) transformed with plasmid pRS316 (CEN-URA3). The deletion strains med16Δ, med5Δ, med20Δ, med12Δ, and cdk8Δ represent the tail, middle, head, and kinase (twice) modules, respectively. (Bottom) HS1004 derivatives (spontaneous suppressors); SLY101 strain background. Plates were incubated for either 3 or 5 days at 30° (left and right, respectively).
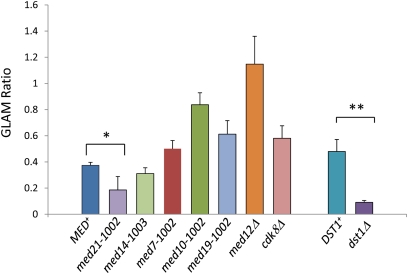
Finally, we wished to investigate more directly whether the ewe mutations impair Pol II elongation, in particular its processivity. To do so, we conducted an in vivo transcription elongation assay that compared the efficiency with which a short, galactose-regulated episomal gene (∼1.5 kb) is transcribed vs. a longer one (∼4.5 kb). The assay is based on comparison of Pho5 acid phosphatase expression arising from two genes sharing the same promoter (GAL1) as well as ORF and terminator (derived from PHO5) but differing in the length of their 3′-UTR (Morillo-Huesca et al. 2006). Previous work has shown that mutations in positive elongation factors (Morillo-Huesca et al. 2006; Ginsburg et al. 2009) or the ectopic expression of a negative elongation factor (Kim et al. 2011) diminish the steady-state levels of long vs. short transcript (termed the GLAM ratio). As shown in Figure 6, whereas the dst1Δ mutation elicited a significant decrease in the GLAM ratio, as expected (Mason and Struhl 2005), none of the spontaneous or engineered Mediator mutations did so, with the exception of med21-1002. Thus, although all ewe mutations elicit a strong 6-AU phenotype, only one has a corresponding effect on Pol II processivity at the episomal PHO5 gene. This assay does not rule out processivity effects on chromosomal genes, whose chromatin might be more stable than the episomal reporter genes, nor does it rule out the possibility that ewe mutations affect Pol II elongation rate.
Figure 6 .
Pol II processivity assay reveals a potential role for Med21. Two sets of isogenic strains, derived from either HS1004 (MED+ cluster) or BY4741 (DST1+ and dst1Δ) and bearing the indicated mutations, were transformed with PGAL1-PHO5 and PGAL1-PHO5::lacZ reporter plasmids, and grown in synthetic medium containing 1% galactose and 1% raffinose to log phase. Cells were harvested and Pho5 expression levels were determined by an acid phosphatase assay. The GLAM ratio represents the acid phosphatase activity of cells expressing the long transcript (PGAL1-PHO5::lacZ) relative to those expressing the short transcript (PGAL1-PHO5). Depicted are mean values ± SEM of three or six independent transformants of each indicated strain/plasmid combination (N = 3 for med14-1003, med21-1002, med19-1002, cdk8Δ, DST1, and dst1Δ; N = 6 for MED+, med7-1002, med10-1002, and med12Δ). *P ≤ 0.05; **P ≤ 0.01.
Discussion
The Mediator middle module plays a role in regulating Pol II elongation
Here we have shown that mutations in conserved Mediator subunits located within or adjacent to the middle module impair the induced expression of the Hsf1-regulated HSP82 heat-shock gene, and in some cases quite severely. Despite this, Pol II occupancy of the HSP82 promoter is generally unaffected, and in the case of med7-1002, actually increased. Thus, a critical function of Mediator—to recruit Pol II to a gene’s promoter (Soutourina et al. 2011)—is typically unimpaired in ewe mutants, at least at HSP82. As Pol II occupancy within the HSP82 coding region is likewise unaffected in the case of the med7 and med14 mutants, these mutations would appear to act by diminishing Pol II elongation rate. Their effect on Pol II elongation bears a strong resemblance to the effect of depleting Cdk8 in HCT116 cells (Donner et al. 2010) since in both cases, impaired Mediator function results in a reduction in transcription without affecting Pol II occupancy. In the med7 mutant, increased Pol II occupancy of the promoter relative to the coding region is consistent with an additional defect in promoter escape. With respect to med19-1002 and med21-1002, an effect on polymerase processivity could be occurring, given reduced Pol II density within the HSP82 coding region relative to the promoter, which is particularly evident toward its 3′-UTR. Finally, med10-1002 may represent an exception to the postrecruitment role of the ewe middle module subunits, since it appears to principally affect Pol II recruitment.
Integrity of the central Med7–Med21 heterodimer within the middle module (Baumli et al. 2005) may be important for yeast Mediator’s postrecruitment function, given that both med7-1002 and med21-1002 exhibit elongation defects. Also consistent is the similar phenotype of med19-1002, given that Med19 has been implicated in stabilizing the association of the middle module with the rest of Mediator (Baidoobonso et al. 2007). A caveat to the above is that our data do not formally rule out the possibility that the Mediator mutations affect mRNA stability instead of, or in addition to, transcription. This consideration is particularly important in light of recent reports of a role for promoter elements in regulating cytoplasmic mRNA decay in S. cerevisiae (Bregman et al. 2011; Trcek et al. 2011). However, a primary role for ewe mutations in regulating HSP82 mRNA stability seems unlikely. First, two ewe mutations, rgr1-Δ2 and med21-1002, significantly diminish H3 eviction within both the promoter and ORF of HSP82 following heat shock, while two others, med7-1002 and med19-1002, elicit more minor, but nonetheless detectable defects in nucleosome disassembly/reassembly. The chromatin phenotypes of these four mutants is consistent with a defect at the level of polymerase elongation (discussed more fully below). Second, with respect to med10-1002, we observed diminished Pol II occupancy within the HSP82 promoter and coding region that paralleled diminished mRNA accumulation. This again is consistent with a transcription defect rather than one in mRNA stabilization.
Given their strong 6-AU phenotypes, the ewe mutations may affect RNA polymerase elongation more generally. And at least one—Med21—may play a role in Pol II processivity at non–heat-shock genes since the L76P mutation caused a reduction in the synthesis of long vs. short PHO5-containing transcripts as deduced from measurement of acid phosphatase levels. Consistent with the general idea that Mediator influences Pol II elongation, others have observed that deletion of the tail subunit Med15 affects Pol II processivity (Gaillard et al. 2009). Additionally, deletion of the head subunit Med18 was shown to reduce Pol II occupancy within the promoters of the CHA1 and INO1 genes yet increase Pol II abundance near their 3′-ends (Mukundan and Ansari 2011). Thus, in contrast to ewe mutations that seem to be linked primarily to Pol II elongation rate and/or processivity, loss of Med18 appears to affect late elongation or termination.
How might the middle module (and in particular, Med7 and Med14) govern the rate with which Pol II traverses the gene? Estimates for the rate of Pol II elongation typically range from 25 to 40 nt/sec, although there are reports of more rapid elongation in certain contexts (reviewed in Ardehali and Lis 2009). Although not tested here, one way Mediator could affect elongation rate is through its modulation of the phosphorylation state of the Pol II CTD, or of one or more elongation factors. This could be achieved directly, through its Cdk8 subunit, or it could be mediated indirectly through other CTD kinases (Donner et al. 2010). The pattern of Ser2, Ser5, and Ser7 phosphorylation on the CTD specifies the cotranscriptional engagement of capping, RNA processing, and polyadenylation enzymes (Akhtar et al. 2009), as well as chromatin modification enzymes (Govind et al. 2010). Thus, Mediator may regulate the CTD phosphorylation pattern that dictates the timing and identity of factors that associate with the transcription elongation complex, and these in turn could affect elongation rate or processivity. Moreover, a collaborator of Mediator may be the SAGA complex. First, SAGA and Mediator exhibit interdependent recruitment at Gcn4-regulated promoters (Qiu et al. 2005) and may well share this interdependence at other Pol II promoters. Second, SAGA has been detected within the coding regions of several Gal4-, Gcn4-, and Hsf1-regulated genes (Govind et al. 2007; Wyce et al. 2007; Kremer and Gross 2009). Therefore, Mediator might control either the Gcn5 histone acetyl transferase (HAT) or Ubp8 ubiquitin protease activities within SAGA by regulating Pol II association of SAGA through CTD phosphorylation. In a similar manner, Mediator could control the Esa1 HAT of NuA4 through Pol II association of NuA4 (Ginsburg et al. 2009).
A role for Mediator in nucleosome disassembly/reassembly
In addition to having a role in Pol II elongation, the Med14 and Med21 subunits facilitate nucleosomal loss at HSP82 following heat shock. This may arise from their role in enhancing nucleosome disassembly or in impairing reassembly (or both). Rapid, gene-wide depletion of nucleosomes is characteristic of induced heat-shock genes from yeast to Drosophila (Zhao et al. 2005; Erkina and Erkine 2006; Petesch and Lis 2008; Kremer and Gross 2009). Poly(ADP)-ribose polymerase (PARP) (or its activity) appears to be important in triggering histone eviction throughout the Drosophila HSP70 gene (Petesch and Lis 2008). While a PARP ortholog has not been identified in yeast, other factors have been shown to be important in transcription-coupled nucleosome eviction in S. cerevisiae. One, the H3/H4 histone chaperone Asf1, mediates histone eviction from activated PHO5 and PHO8 promoters (Adkins et al. 2004) as well as from the galactose-induced GAL1 and GAL10 coding regions (Schwabish and Struhl 2006). Yet we show here that H4 is evicted efficiently at heat-shock–activated HSP82 in an asf1Δ mutant (indistinguishable from the isogenic WT strain), mirroring the effects of ctk1Δ, dst1Δ, and spt4Δ mutations, as well as our earlier findings of snf2Δ, swi1Δ, gcn5Δ, set1Δ, paf1Δ, and sas3Δ mutants (Zhao et al. 2005; Kremer and Gross 2009). Particularly striking is the fact that while inactivation of the Swi/Snf remodeling complex results in a sixfold reduction in HSP82 transcription, only slightly less than that conferred by rgr1-Δ2, rgr1-Δ2 strongly impairs nucleosomal depletion following heat shock, while Swi/Snf mutants have no detectable effect (Zhao et al. 2005). This may be the case because additional ATP-dependent remodeling complexes participate in chromatin remodeling at heat-shock genes (Erkina et al. 2010), while the role played by Med14 (and to a lesser degree, Med21) in regulating nucleosome occupancy is unique.
The chromatin phenotypes of recessive mutations in MED14 and MED21 raise the question of how Mediator might regulate nucleosomal disassembly/reassembly. Particularly so, given that Pol II density is largely unaffected in rgr1-Δ2 and med21-1002 mutants and that Pol II transit has been implicated in mediating nucleosome disassembly within transcribed regions (Teves and Henikoff 2011). Clearly, Pol II density alone does not correlate with H3 depletion within the HSP82 transcribed region. This is consistent with the Pol II and chromatin phenotypes of med10-1002 reported here: substantial reduction in Pol II occupancy yet normal levels of histone displacement. It is also consistent with our earlier observation of an hsp82 promoter mutant termed hsp82-P2, whose induced expression is reduced 85%, and whose nucleosomal displacement is obviated, yet whose Pol II density is unaffected (Zhao et al. 2005). Perhaps this mutant, bearing a double point mutation within HSE1, and the rgr1-Δ2 mutant share a common defect in elongating Pol II. As discussed above, the phosphorylation state of the CTD governs association of various factors with elongating Pol II. These in turn may dictate the efficiency with which nucleosomes are evicted. Thus, in the case of the Mediator mutants rgr1-Δ2 and med21-1002, the Pol II elongation complex might be deficient for nucleosome disassembly (or hyperactive in reassembly) that in turn contributes to the reduction in synthesis of full-length HSP82 mRNA. It is possible that polymerase elongation rate and processivity are respectively impaired in these two mutants, as discussed above. Further work will be necessary to explore these and other potential mechanisms.
The expression phenotype of ewe mutants may stem from multiple factors
Although not the focus of this study, the question arises as to what might be the basis for the substantially increased expression of the hsp82-ΔHSE1/lacZ reporter observed in ewe mutants, as it is not observed at the wild-type HSP82 gene (nor at SSA4 or HSP104; J. Jeon, S. B. Kremer, and D. S. Gross, unpublished observations). It is possible that enhanced basal expression arises from some attribute of the chromatin structure of the fusion gene, whose promoter is assembled into a repressive dinucleosome (Gross et al. 1993; Venturi et al. 2000) and whose coding region is derived from a heterologous, prokaryotic gene. Others have observed that in the context of Mediator mutations (either med16Δ alone or in a med5Δ nut2-1 double mutant), basal expression of HO/lacZ and PHO5/lacZ fusion genes is enhanced, while expression of the intact HO and PHO5 genes remains unaffected (Tabtiang and Herskowitz 1998). Likewise, we have found that, whereas med16Δ strongly derepresses hsp82-ΔHSE1/lacZ, it has no effect on the expression of hsp82-ΔHSE1 itself (H. Singh and D. S. Gross, unpublished observations). This phenomenon is also seen with a second fusion, hsp82-ΔHSE1/HIS3, whose enhanced expression served as the basis for the selection of the ewe mutants (Singh et al. 2006), so it is not unique to lacZ gene fusions. While the basis for enhanced basal expression of fusion genes remains unclear, it could stem from increased transcription initiation secondary to enhanced accessibility of the nucleosomal promoter, or even arise from a post-transcriptional event such as increased mRNA stability. Whatever might be the basis for enhanced basal expression, the dramatic loss in inducibility of the hsp82/lacZ fusion gene in the ewe mutants likely arises from the same defects in Pol II elongation and chromatin remodeling described here for the wild-type gene.
Supplementary Material
Acknowledgments
We thank Jorge Herrera-Diaz, David Chiluiza, and Hesheng Zhang for technical assistance; Kelly Tatchell for yeast strains; and Alan Hinnebusch, Daniel Ginsburg, and Sebastián Chávez for GLAM plasmids. This work was supported by grants awarded to D. S. Gross from the National Science Foundation (MCB-747227 and MCB-1025025).
Footnotes
Communicating editor: K. M. Arndt
Literature Cited
- Adkins M. W., Howar S. R., Tyler J. K., 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14: 657–666. [DOI] [PubMed] [Google Scholar]
- Akhtar M. S., Heidemann M., Tietjen J. R., Zhang D. W., Chapman R. D., et al. , 2009. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell 34: 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apone L. M., Virbasius C. A., Holstege F. C. P., Wang J., Young R. A., et al. , 1998. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol. Cell 2: 653–661. [DOI] [PubMed] [Google Scholar]
- Ardehali M. B., Lis J. T., 2009. Tracking rates of transcription and splicing in vivo. Nat. Struct. Mol. Biol. 16: 1123–1124. [DOI] [PubMed] [Google Scholar]
- Baidoobonso S. M., Guidi B. W., Myers L. C., 2007. Med19(Rox3) regulates intermodule interactions in the Saccharomyces cerevisiae Mediator complex. J. Biol. Chem. 282: 5551–5559. [DOI] [PubMed] [Google Scholar]
- Balakrishnan S. K., Gross D. S., 2008. The tumor suppressor p53 associates with gene coding regions and co-traverses with elongating RNA polymerase II in an in vivo model. Oncogene 27: 2661–2672. [DOI] [PubMed] [Google Scholar]
- Baumli S., Hoeppner S., Cramer P., 2005. A conserved mediator hinge revealed in the structure of the MED7.MED21 (Med7.Srb7) heterodimer. J. Biol. Chem. 280: 18171–18178. [DOI] [PubMed] [Google Scholar]
- Bernecky C., Grob P., Ebmeier C. C., Nogales E., Taatjes D. J., 2011. Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly. PLoS Biol. 9: e1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon H. M., 2008. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36: 3993–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman A., Avraham-Kelbert M., Barkai O., Duek L., Guterman A., et al. , 2011. Promoter elements regulate cytoplasmic mRNA decay. Cell 147: 1473–1483. [DOI] [PubMed] [Google Scholar]
- Cai G., Imasaki T., Takagi Y., Asturias F., 2009. Mediator structural conservation and implications for the regulation mechanism. Structure 17: 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S., Chatterjee S., Lee M., Struhl K., 1999. Transcriptional activation in yeast cells lacking transcription factor IIA. Genetics 153: 1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner A. J., Ebmeier C. C., Taatjes D. J., Espinosa J. M., 2010. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 17: 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkina T. Y., Erkine A. M., 2006. Displacement of histones at promoters of Saccharomyces cerevisiae heat shock genes is differentially associated with histone H3 acetylation. Mol. Cell. Biol. 26: 7587–7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkina T. Y., Zou Y., Freeling S., Vorobyev V. I., Erkine A. M., 2010. Functional interplay between chromatin remodeling complexes RSC, SWI/SNF and ISWI in regulation of yeast heat shock genes. Nucleic Acids Res. 38: 1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exinger F., Lacroute F., 1992. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 22: 9–11. [DOI] [PubMed] [Google Scholar]
- Gaillard H., Tous C., Botet J., Gonzalez-Aguilera C., Quintero M. J., et al. , 2009. Genome-wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4-Not in transcription-coupled repair. PLoS Genet. 5: e1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg D. S., Govind C. K., Hinnebusch A. G., 2009. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol. Cell. Biol. 29: 6473–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind C. K., Zhang F., Qiu H., Hofmeyer K., Hinnebusch A. G., 2007. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25: 31–42. [DOI] [PubMed] [Google Scholar]
- Govind C. K., Qiu H., Ginsburg D. S., Ruan C., Hofmeyer K., et al. , 2010. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell 39: 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromoller A., Lehming N., 2000. Srb7p is a physical and physiological target of Tup1p. EMBO J. 19: 6845–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D. S., Adams C. C., Lee S., Stentz B., 1993. A critical role for heat shock transcription factor in establishing a nucleosome-free region over the TATA-initiation site of the yeast HSP82 heat shock gene. EMBO J. 12: 3931–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. W., Dohrmann P. R., Stillman D. J., 1995. Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcriptional regulation. Genetics 140: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Balakrishnan S. K., Gross D. S., 2011. p53 interacts with RNA polymerase II through its core domain and impairs Pol II processivity in vivo. PLoS ONE 6: e22183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer S. B., Gross D. S., 2009. SAGA and Rpd3 chromatin modification complexes dynamically regulate heat shock gene structure and expression. J. Biol. Chem. 284: 32914–32931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Lis J. T., 1998. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature 393: 389–392. [DOI] [PubMed] [Google Scholar]
- Lee D. K., Kim S., Lis J. T., 1999. Different upstream transcriptional activators have distinct co-activator requirements. Genes Dev. 13: 2934–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Gross D. S., 1993. Conditional silencing: the HMRE mating-type silencer exerts a rapidly reversible position effect on the yeast HSP82 heat shock gene. Mol. Cell. Biol. 13: 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. K., Fletcher A. G., Zhang L., Chen X., Fischbeck J. A., et al. , 2010. Activation of a poised RNAPII-dependent promoter requires both SAGA and mediator. Genetics 184: 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J. L., 2007. The role of chromatin during transcription. Cell 128: 707–719. [DOI] [PubMed] [Google Scholar]
- Li Y., Bjorklund S., Jiang Y. W., Kim Y. J., Lane W. S., et al. , 1995. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 92: 10864–10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagon F., Tong A. H., Shafer B. K., Strathern J. N., 2004. Genetic interactions of DST1 in Saccharomyces cerevisiae suggest a role of TFIIS in the initiation-elongation transition. Genetics 166: 1215–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S., Roeder R. G., 2000. Transcriptional regulation through Mediator-like coactivators in yeast and metzoan cells. Trends Biochem. Sci. 25: 277–283. [DOI] [PubMed] [Google Scholar]
- Malik S., Roeder R. G., 2010. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S., Barrero M. J., Jones T., 2007. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Proc. Natl. Acad. Sci. USA 104: 6182–6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P. B., Struhl K., 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17: 831–840. [DOI] [PubMed] [Google Scholar]
- McNeil J. B., Agah H., Bentley D., 1998. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev. 12: 2510–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Lin S. C., Bernecky C., Gao Y., Taatjes D. J., 2010. p53 activates transcription by directing structural shifts in Mediator. Nat. Struct. Mol. Biol. 17: 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z., Bai Y., Poon D., Weil P. A., Struhl K., 1996. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 382: 188–191. [DOI] [PubMed] [Google Scholar]
- Morillo-Huesca M., Vanti M., Chavez S., 2006. A simple in vivo assay for measuring the efficiency of gene length-dependent processes in yeast mRNA biogenesis. FEBS J. 273: 756–769. [DOI] [PubMed] [Google Scholar]
- Mukundan B., Ansari A., 2011. A novel role for mediator subunit SRB5/MED18 in termination of transcription. J. Biol. Chem. 286: 37053–37057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers L. C., Kornberg R. D., 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69: 729–749. [DOI] [PubMed] [Google Scholar]
- Petesch S. J., Lis J. T., 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134: 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H., Hu C., Zhang F., Hwang G. J., Swanson M. J., et al. , 2005. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol. Cell. Biol. 25: 3461–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A., Core L. J., Lis J. T., 2006. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7: 557–567. [DOI] [PubMed] [Google Scholar]
- Schwabish M. A., Struhl K., 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22: 415–422. [DOI] [PubMed] [Google Scholar]
- Sekinger E. A., Gross D. S., 1999. SIR repression of a yeast heat shock gene: UAS and TATA footprints persist within heterochromatin. EMBO J. 18: 7041–7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger E. A., Gross D. S., 2001. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105: 403–414. [DOI] [PubMed] [Google Scholar]
- Singh H., Erkine A. M., Kremer S. B., Duttweiler H. M., Davis D. A., et al. , 2006. A functional module of yeast Mediator that governs the dynamic range of heat shock gene expression. Genetics 172: 2169–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina J., Wydau S., Ambroise Y., Boschiero C., Werner M., 2011. Direct interaction of RNA polymerase II and Mediator required for transcription in vivo. Science 331: 1451–1454. [DOI] [PubMed] [Google Scholar]
- Tabtiang R. K., Herskowitz I., 1998. Nuclear proteins Nut1p and Nut2p cooperate to negatively regulate a Swi4p-dependent lacZ reporter gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 4707–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Parmely T. J., Sato S., Tomomori-Sato C., Banks C. A., et al. , 2011. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves S. S., Henikoff S., 2011. Heat shock reduces stalled RNA polymerase II and nucleosome turnover genome-wide. Genes Dev. 25: 2387–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. M., Young R. A., 1995. General requirement for RNA polymerase II holoenzymes in vivo. Proc. Natl. Acad. Sci. USA 92: 4587–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T., Larson D. R., Moldon A., Query C. C., Singer R. H., 2011. Single-molecule mRNA decay measurements reveal promoter- regulated mRNA stability in yeast. Cell 147: 1484–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters B. J., Pugh B. F., 2009. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 19: 360–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi C. B., Erkine A. M., Gross D. S., 2000. Cell cycle-dependent binding of yeast heat shock factor to nucleosomes. Mol. Cell. Biol. 20: 6435–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Balamotis M. A., Stevens J. L., Yamaguchi Y., Handa H., et al. , 2005. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell 17: 683–694. [DOI] [PubMed] [Google Scholar]
- Wyce A., Xiao T., Whelan K. A., Kosman C., Walter W., et al. , 2007. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol. Cell 27: 275–288. [DOI] [PubMed] [Google Scholar]
- Zhao J., Herrera-Diaz J., Gross D. S., 2005. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 25: 8985–8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Liu B., Carlsten J. O., Beve J., Nystrom T., et al. , 2011a. Mediator influences telomeric silencing and cellular life span. Mol. Cell. Biol. 31: 2413–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Zhang Y., Bjornsdottir G., Liu Z., Quan A., et al. , 2011b. Histone modifications influence mediator interactions with chromatin. Nucleic Acids Res. 39: 8342–8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.