Summary
Background and objectives
Familial hypomagnesemia with hypercalciuria and nephrocalcinosis is a rare autosomal recessive renal tubular disease. It is caused by mutations in CLDN16 and CLDN19, encoding claudin-16 and -19, respectively. Familial hypomagnesemia with hypercalciuria and nephrocalcinosis is usually complicated by progressive CKD. The objectives of this study were to describe the clinical and genetic features of familial hypomagnesemia with hypercalciuria and nephrocalcinosis and analyze phenotype–genotype associations in patients with CLDN16 or CLDN19 mutations.
Design, setting, participants, & measurements
Data from 32 genetically confirmed patients (9 patients with CLDN16 and 23 patients with CLDN19 mutations) from 26 unrelated families were retrospectively reviewed.
Results
Diagnosis was based on clinical criteria at a median age of 9.5 years and confirmed by genetic testing at a median age of 15.5 years. In total, 13 CLDN16 or CLDN19 mutations were identified, including 8 novel mutations. A founder effect was detected for the recurrent CLDN16 p.Ala139Val mutation in North African families and the CLDN19 p.Gly20Asp mutation in Spanish and French families. CKD was more frequently observed in patients with CLDN19 mutations: survival without CKD or ESRD was 56% at 20 years of age in CLDN19 versus 100% in CLDN16 mutations (log rank P<0.01). Ocular abnormalities were observed in 91% of patients with CLDN19 mutations and none of the patients with CLDN16 mutations (P<0.01). Treatments seem to have no effect on hypercalciuria and CKD progression.
Conclusions
Patients with CLDN19 mutations may display more severe renal impairment than patients with CLDN16 mutations. Ocular abnormalities were observed only in patients with CLDN19 mutations.
Introduction
Familial hypomagnesemia with hypercalciuria and nephrocalcinosis (FHHNC; Online Mendelian Inheritance in Man 248250) is a rare autosomal recessive disease first described as Michelis-Castrillo syndrome (1). Only 87 patients (51 families) have been described to date in international publications (2,3). It is caused by mutations in the CLDN16 and CLDN19 genes, encoding claudin-16 and -19 (4,5). These two proteins are expressed in the tight junction of the thick ascending limb (TAL) of Henle’s loop and are involved in the paracellular reabsorption of calcium and magnesium (6). Claudin proteins are expressed in various tissues, and claudin-19 has also been detected in the retinal epithelium (7). Mutations of these genes are responsible for urinary loss of magnesium and calcium, resulting in hypomagnesemia, hypercalciuria, nephrolithiasis, and nephrocalcinosis. FHHNC patients suffer from progressive CKD, which may lead to ESRD in the teens or 20s (2). No difference in renal phenotype between patients with mutations in these two genes has yet been described. By contrast, severe ocular involvement (macular coloboma, pigmentary retinitis, nystagmus, or visual loss) has been described in CLDN19 patients, whereas only mild nonspecific ocular involvement (myopia, astigmatism, hypermetropia, or strabismus) has been reported in some CLDN16 patients (3,6,8).
In this study, we aimed to describe the clinical features, molecular genetics, and disease outcome, focusing on renal progression, in a cohort of 32 patients with mutations in the CLDN16 or CLDN19 genes.
Materials and Methods
Patients
Clinical and biochemical data for 32 genetically defined FHHNC patients from 26 families were studied retrospectively. Data collected included demographic, genetic, clinical, and biologic data obtained at diagnosis and during follow-up. Estimated GFR (eGFR) was calculated with the updated Schwartz formula for children and the Modification of Diet in Renal Disease formula for adults (9,10).
Genetic Studies
Detection of Point Mutations.
Genomic DNA was extracted by standard methods from peripheral blood samples after written informed consent had been obtained from the patients or their parents. Mutation analysis was performed based on PCR amplification followed by sequencing as described (11,12) (primers available on request) on an ABI Prism 3730XL DNA Analyzer (Perkin Elmer Applied Biosystems, Foster City, CA). Mutations were interpreted, and the degree of amino acid conservation between orthologs and Grantham distance was assessed with Alamut v.2.0 software (Interactive Biosoftware, Rouen, France; http://www.interactivebiosoftware.com/). Complementary analyses were performed with SIFT (http://www.Blocks.fhcrc.org/sift/SIFT.html), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph/), MutationTaster (http://www.mutationtaster.org/), SNPs&Go (http://snps-and-go.biocomp.unibo.it/snps-and-go/), and MutPred (http://mutpred.mutdb.org/) algorithms.
Quantitative Multiplex PCR for Short Fluorescent Fragments.
We adapted the quantitative multiplex PCR for short fluorescent fragments (QMPSF) method (13) for the detection of large rearrangements of the CLDN19 gene for patients with homozygous mutations and no history of consanguinity for whom parental DNA was not available. Each multiplex PCR included a set of specific primers for each exon (primers available on request), and a fragment from the hydroxymethylbilane synthase gene was the internal control. The forward primer of each pair was 5′ end-labeled with 6-FAM fluorochrome. Amplified DNA fragments were separated by capillary electrophoresis on an ABI Prism 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA). Data were analyzed with GeneMapper Software version 4.0 (Applied Biosystems, Foster City, CA). Each QMPSF reaction was validated with positive and negative reference DNAs.
Haplotype Analysis.
Haplotype analysis was carried out in families harboring two recurrent mutations (p.Ala139Val for CLDN16 and p.Gly20Asp for CLDN19) to determine whether these families were descended from a common ancestor. Polymorphic microsatellite markers flanking each locus were amplified by PCR, subjected to automated capillary electrophoresis, and sized with GeneMapper v 4.0 software. We used the markers D3S1294, D3S1314, and D3S1288 (GenBank accession numbers Z16944, Z17168, and Z16855) for CLDN16 with the following genetic map: D3S1294, 0.4 cM; D3S1314, 0.01 cM; CLDN16, 0.1 cM; D3S1288. For CLDN19, we used markers D1S463, D1S193, and D1S447 (GenBank accession numbers Z23403, Z16490.1, and Z23291.1) with the following genetic map: D1S463, 0.5 cM; D1S193, 0.18 cM; CLDN 19, 0.75 cM; D1S447.
Statistical Analyses
Results were expressed as median and interquartile range (IQR) for continuous variables and percentages for categorical variables. Comparative analyses of qualitative variables were carried out with the chi-squared or Fisher exact test, whereas comparisons of quantitative variables were based on t or Wilcoxon test in cases of skewed distribution. CKD-free survival was estimated from birth with a time to failure Kaplan–Meier method. The endpoint was the first measurement of eGFR below 60 ml/min per 1.73 m2. Data were censored at the last available measurement. ESRD-free survival was also estimated. Log rank tests were used to compare subgroups. A P value<0.05 was considered statistically significant. Statistical analyses were performed with SAS 9.1 (SAS Institute, Cary, NC).
Results
Demographics
Thirty-two patients (14 male and 18 female) from 26 unrelated families were included. Seventeen of the families were Caucasian, seven were from North Africa, one was from sub-Saharan Africa, and one was from the French West Indies. One family included three affected siblings (family 12), and four families included two affected siblings (families 2, 9, 11, and 23).
Genetic Investigations
Molecular studies revealed CLDN16 mutations in the probands of eight families and CLDN19 mutations in the probands of 18 families (Tables 1 and 2). Patients 1, 2, 5, 17, and 18 have been described elsewhere (11,12,14).
Table 1.
Mutations of the CLDN16 gene in familial hypomagnesemia, hypercalciuria, and nephrocalcinosis patients
| Patient | Region/Country of Origin | Consanguinity | Gender | Nucleotidea | Protein | Exon/Intron | Status | Reference (Patient/Mutation) |
|---|---|---|---|---|---|---|---|---|
| 1 | Portugal | Yes | Female | c.485G>T | p.Gly162Val | 3 | Homozygous | 11/11 |
| 2.1 | Morocco | Yes | Female | c.416C>T | p.Ala139Val | 2 | Homozygous | 11/11 |
| 2.2 | Morocco | Yes | Male | c.416C>T | p.Ala139Val | 2 | Homozygous | 11/11 |
| 3 | North Africa | ? | Male | c.239G>A | p.Cys80Tyr | 1 | Homozygous | This study/this study |
| 4 | Martinique | No | Female | c.547A>G | p.Lys183Glu | 3 | Homozygous | This study/this study |
| 5 | Algeria | Yes | Female | c.416C>T | p.Ala139Val | 2 | Homozygous | 11/14 |
| 6 | Algeria | ? | Male | c.416C>T | p.Ala139Val | 2 | Homozygous | This study/11 |
| 7 | Tunisia | Yes | Male | c.445C>T | p.Arg149X | 3 | Homozygous | This study/4 |
| 8 | Morocco | Yes | Female | c.697G>C | p.Gly233Arg | 4 | Homozygous | This study/this study |
Sequences are numbered according to the cDNA sequence (GenBank accession number NM_006580). The A of the ATG initiator codon is denoted nucleotide 1.
Table 2.
Mutations of the CLDN19 gene in familial hypomagnesemia, hypercalciuria, and nephrocalcinosis patients
| Patient | Region/Country of Origin | Consanguinity | Gender | Nucleotidea | Protein | Exon/Intron | Status | Reference (Patient/Mutation) |
|---|---|---|---|---|---|---|---|---|
| 9.1 | Tunisia | Yes | Female | E1_E4del | 1–4 | Homozygous | This study/this study | |
| 9.2 | Tunisia | Yes | Male | E1_E4del | 1–4 | Homozygous | This study/this study | |
| 10 | France | No | Male | c.59G>A | p.Gly20Asp | 1 | Homozygous | This study/5 |
| 11.1 | France | Yes | Female | c.169C>T | p.Gln57X | 1 | Homozygous | This study/this study |
| 11.2 | France | Yes | Male | |||||
| 12.1 | Senegal | No | Male | c.83C>T | p.Pro28Leu | 1 | Homozygous | This study/this study |
| 12.2 | Senegal | No | Male | c.83C>T | p.Pro28Leu | 1 | Homozygous | This study/this study |
| 12.3 | Senegal | No | Female | c.83C>T | p.Pro28Leu | 1 | Homozygous | This study/this study |
| 13 | Spain | No | Male | c.59G>A | p.Gly20Asp | 1 | Homozygous | This study/5 |
| 14 | France | No | Male | c.54G>A | p.Trp18X | 1 | Homozygous | This study/this study |
| 15 | France | No | Male | c.59G>A | p.Gly20Asp | 1 | Homozygous | This study/5 |
| 16 | France | No | Female | c.59G>A | p.Gly20Asp | 1 | Homozygous | This study/5 |
| 17 | France | No | Female | c.59G>A | p.Gly20Asp/p.Val44Met | 1/1 | Compound heterozygous | 12/5 |
| 17 | France | No | Female | c.130G>A | p.Gly20Asp/p.Val44Met | 1/1 | Compound heterozygous | 12/12 |
| 18 | France | No | Female | c.59G>A | p.Gly20Asp | 1 | Homozygous | 12/5 |
| 19 | France | No | Female | c.59G>A | p.Gly20Asp | 1 | Homozygous | This study/5 |
| 20 | France | No | Female | c.59G>A | p.Gly20Asp | 1 | Homozygous | This study/5 |
| 21 | France | No | Female | c.59G>A | p.Gly20Asp | 1 | Homozygous | This study/5 |
| 22 | France | No | Male | c.59G>A | p.Gly20Asp | 1 | Homozygous | This study/5 |
| 23.1 | France | No | Female | c.59G>A | p.Gly20Asp | 1 | Homozygous | This study/5 |
| 23.2 | France | No | Male | c.59G>A | p.Gly20Asp | 1 | Homozygous | This study/5 |
| 24 | Spain | No | Female | c.59G>A | p.Gly20Asp | 1 | Homozygous | This study/5 |
| 25 | France | No | Female | c.59G>A/c.403_406del | p.Gly20Asp/p.Thr135LeufsX9 | 1/3 | Compound heterozygous | This study/5 |
| 26 | France | No | Female | c.59G>A | p.Gly20Asp | 1 | Homozygous | This study/5 |
Sequences are numbered according to the cDNA sequence (GenBank accession number NM_148960). The A of the ATG initiator codon is denoted nucleotide 1.
CLDN16 Mutations.
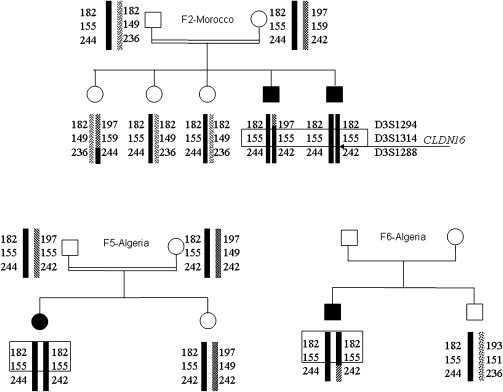
Six different mutations of the CLDN16 gene were detected (five missense and one nonsense) (Table 1). Three of the missense mutations (p.Cys80Tyr, pLys183Glu, and p.Gly233Arg) had never been described. They were not found in 100 control chromosomes and were predicted to be deleterious by at least three of the in silico methods (Supplemental Table 1). All patients were homozygous for the detected mutation. Patients from three North African families harbored the same missense mutation (p.Ala139Val), and an analysis of microsatellite markers showed that they shared the same haplotype at the disease locus (Figure 1), suggesting a common ancestor.
Figure 1.
Haplotypes of three North African families carrying the p.Ala139Val mutation. Families 2 and 5 were consanguineous; family 6 has no history of consanguinity, but the mutation was homozygous.
CLDN19 Mutations.
Seven different mutations of the CLDN19 gene were detected (three missense, two nonsense, one frame shift, and one large deletion) (Table 2). Only two of the missense mutations have been described before. The probands of two families were compound heterozygous. All the other patients were homozygous, although most had no history of consanguinity. The known mutation, p.Gly20Asp, was particularly frequent in our group of patients (homozygous in 13 probands and heterozygous and associated with a second mutation in 2 probands). The parents in families 10, 20, and 26 were heterozygous, whereas QMPSF ruled out heterozygous deletions in the other families. The p.Gly20Asp mutation has been described as a founder effect in Spanish families. These results and the detection of this mutation in 2 Spanish and 12 Southwest France probands are consistent with such a founder effect. An analysis of microsatellite markers also provided support for this hypothesis: most of the patients carrying this mutation had the same haplotype at the disease locus or at least at the closest D1S193 marker (Figure 2).
Figure 2.
Haplotypes of 14 nonconsanguineous French and Spanish families carrying the p.Gly20Asp mutation. In probands of families 17 and 25, this mutation was heterozygous and associated with a second mutation; the probands of all the other families were homozygous.
Clinical and Biologic Presentation at Diagnosis
In this cohort of 32 patients, a family history of FHHNC or other renal symptoms related to the disease (i.e., renal failure, lithiasis, or tubulopathy) was present in 50% of cases. The main symptoms at diagnosis included urinary tract infections, nephrolithiasis, nephrocalcinosis, and polyuria/polydipsia (Table 3). Age at onset ranged from 1 to 14 years, with a median age at onset of 7 years. Median ages at clinical and genetic diagnosis were 9.5 and 15.5 years, respectively. No significant difference in clinical presentation was observed between patients with CLDN16 and CLDN19 mutations (Table 3).
Table 3.
Clinical characteristics of patients with CLDN16 and CLDN19 mutations
| Demographic Characteristics | CLDN16 (n=9) | CLDN19 (n=23) | P Value |
|---|---|---|---|
| Sex ratio (male/female) | 4/5 | 10/13 | 1.00 |
| Age at first symptoms (years) | 7.0 (3–14) | 8.0 (1–13)2 | 0.83 |
| Age at clinical diagnosis (years) | 7.0 (3–14) | 10.0 (1–21) | 0.51 |
| Age at genetic diagnosis (years) | 11.0 (3–23) | 19.0 (10–28) | 0.12 |
| Initial signs | |||
| nephrocalcinosis n (%) | 5 (55.5) | 6 (26.1) | 0.40 |
| nephrolithiasis n (%) | 1 (11.1) | 4 (17.4) | 1.00 |
| urinary tract infections n (%) | 3 (33.3) | 4 (17.4) | 0.37 |
| abdominal pain n (%) | 2 (22.2) | 2 (8.7) | 0.55 |
| polyuria-polydipsia n (%) | 2 (22.2) | 4 (17.4) | 1.00 |
| enuresis n (%) | 0 | 3 (13.0) | 0.54 |
| high BP n (%) | 0 | 1 (4.3) | 1.00 |
| asymptomatic n (%) | 1 (11.1) | 3 (13.0) | 1.00 |
| Clinical progression | |||
| nephrocalcinosis n (%) | 9 (100) | 22 (100)1 | 1.00 |
| nephrolithiasis n (%) | 1 (12.5)1 | 8 (42.1)4 | 0.20 |
| ocular impairment n (%) | 0 | 21 (91.3)1 | <0.01 |
| neurologic impairment n (%) | 2 (22.2) | 1 (4.3) | 1.00 |
| high BP n (%) | 2 (22.2) | 8 (34.8) | 0.68 |
| failure to thrive n (%) | 1 (14.7)2 | 6 (37.5)8 | 0.61 |
Values are expressed as percentages with the exception of age, which is expressed as median and interquartile range. Superscript values correspond to the numbers of missing data.
Hypomagnesemia, hypercalciuria, and nephrocalcinosis were present in all patients at diagnosis (Table 4). Hypomagnesemia and hypercalciuria were of similar severity in the two groups. In addition to nephrocalcinosis, nine patients (28%) presented nephrolithiasis. Median eGFR at diagnosis was also similar in the two groups. Most of the patients had elevated serum parathyroid hormone levels despite a moderate reduction in eGFR (Table 4). Ocular abnormalities were present at diagnosis in 21 of 23 patients (91%) with CLDN19 mutations. Ocular findings included myopia (n=14), pigmentary retinitis (n=5), macular coloboma (n=2), strabismus (n=3), astigmatism (n=2), and nystagmus (n=1). No ocular disease was found in one 10-year-old patient, and no data were available for the remaining patient with a CLDN19 mutation. No ocular abnormality was found in patients with CLDN16 mutations.
Table 4.
Biologic characteristics of patients with CLDN16 and CLDN19 mutations
| CLDN16 (n=9) | CLDN19 (n=23) | P Value | |
|---|---|---|---|
| Initial biologic findings | |||
| serum magnesium concentration (mmol/L) | 0.49 (0.39–0.60)2 | 0.59 (0.50–0.60)6 | 0.15 |
| urinary calcium to creatinine ratio (mmol/mmol) | 0.8 (0.3–5.3)1 | 1.0 (0.8–1.7)3 | 0.40 |
| serum creatinine concentration (µmol/L) | 67.0 (51.0–76.0)1 | 102.0 (60.0–121.0)3 | 0.07 |
| GFR (ml/min per 1.73 m2) | 71.0 (59.0–92.0)1 | 66.0 (57.0–77.0)3 | 0.46 |
| GFR≥60 ml/min per 1.73 m2 | 7 (77.8) | 14 (60.9) | 0.53a |
| 15<GFR<60 ml/min per 1.73 m2 | 2 (22.2) | 5 (21.7) | 0.53a |
| ESRD | 0 | 4 (17.4) | 0.53a |
| PTH concentration (pg/ml) | 107 (107–110)7 | 134 (115–166)9 | 0.44 |
| Biologic findings at last follow-up | |||
| serum magnesium concentration (mmol/L) | 0.54 (0.38–0.52)1 | 0.58 (0.52–0.60)3 | 0.46 |
| urinary calcium to creatinine ratio (mmol/mmol) | 0.7 (0.3–1.4)5 | 0.7 (0.4–1.3)8 | 0.84 |
| GFR (ml/min per 1.73 m2; n=17) | 82.0 (52.0–93.0)5 | 51.0 (43.0–73.0)10 | 0.10 |
| GFR ≥ 60 ml/min per 1.73 m2 | 6 (66.6) | 9 (39.1) | 0.40a |
| 15<GFR<60 ml/min per 1.73 m2 | 2 (22.2) | 8 (34.8) | 0.40a |
| ESRD | 1 (11.1) | 6 (26.1) | 0.40a |
Values are expressed as medians and interquartile ranges or percentages. Superscript values indicate the numbers of missing data. PTH, parathyroid hormone.
Chi-squared test for the three groups.
Clinical and Biologic Follow-Up
Median follow-up time since clinical diagnosis was 7.9 years (IQR=1.4–10.3) in patients with CLDN16 mutations and 3.2 years (IQR=0.3–14.5) in patients with CLDN19 mutations. Age at last follow-up was 12.4 (IQR=4.3–24.0) and 16.8 (IQR=10.5–27.9) years, respectively. Detailed follow-up information was available for 6 patients with CLDN16 mutations and 15 patients with CLDN19 mutations (Table 4).
At last follow-up, median eGFR was higher in CLDN16 patients (82 ml/min per 1.73 m2) than CLDN19 patients (51 ml/min per 1.73 m2), although this difference was not statistically significant (P=0.10). CKD, defined as an eGFR<60 ml/min per 1.73 m2 was found in 3 (33%) patients with CLDN16 mutations and 14 (61%) patients with CLDN19 mutations.
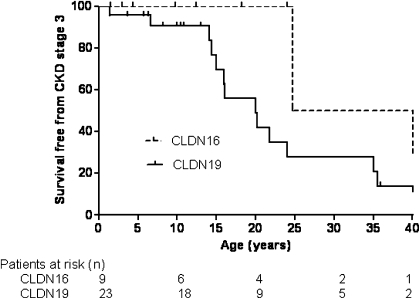
Survival analysis suggested that patients with CLDN19 mutations had a higher risk of progression to CKD and poorer renal survival than patients with CLDN16 mutations. Indeed, CKD-free survival was 100%, 100%, and 50% at 10, 20, and 30 years of age in the CLDN16 group. By contrast, CKD-free survival was 91%, 56%, and 27% at 10, 20, and 30 years of age in patients with CLDN19 mutations (Figure 3) (P<0.01). One of nine patients with CLDN16 mutations and CKD reached ESRD during follow-up at the age of 42 years; 6 of 23 patients with CLDN19 mutations presented ESRD at a median age of 25 years (IQR=20–40).
Figure 3.
Survival without reaching CKD stage 3 by genotype.
High BP occurred in 10 patients (31%) at a median eGFR of 63 ml/min per 1.73 m2 (IQR=24–72). Mild proteinuria (median=350 mg/d, IQR=180–500) was detected in one-half of the patients for whom data were available (3 of 6 with CLDN16 and 7 of 15 with CLDN19 mutations) at a relatively late-stage CKD (median eGFR=42 ml/min per 1.73 m2). Rates of both high BP and proteinuria did not differ between the two groups.
No significant change in magnesemia and calciuria was observed between diagnosis and last follow-up in either of the two groups, and hypercalciuria persisted even at advanced stages of CKD. Patients were treated with magnesium supplements (n=16), thiazide diuretics (n=10), or indomethacine (n=7). However, urinary calcium and serum magnesium concentrations were not significantly affected by either treatment in either group.
Neurologic manifestations, mainly exercise intolerance with electromyological alteration, were present in two previously described patients (12). Electrophysiological studies were not systematically carried out for the patients without clinical manifestations. In addition, one patient presented hypomagnesemia-related seizures.
Discussion
FHHNC is a rare, genetically heterogeneous disease. Most patients described to date have mutations in the CLDN16 gene. We describe here a large cohort of patients with FHHNC mostly from France, and two-thirds had CLDN19 mutations. There were three main findings of our study: (1) FHHNC is often diagnosed late, (2) ocular impairment occurred exclusively in patients with CLDN19 mutations, and (3) patients with CLDN16 and CLDN19 mutations may have different renal prognoses, with a higher risk of CKD and ESRD in patients with CLDN19 mutations. These data contribute to improvements in the phenotypic and genotypic characterization of FHHNC.
The molecular genetic analysis detected two mutated alleles in all patients and expanded the spectrum of known disease-causing mutations of these two genes. Six different mutations of CLDN16 were detected (five missense and one nonsense); three of the missense mutations were previously unknown (p.Cys80Tyr, p.Lys183Glu, and p.Gly233Arg). These mutations affect the first and third transmembrane domains and the second extracellular loop. They result in the replacement of highly conserved amino acids and are probably pathogenic given their predicted biologic consequences (Supplemental Table 1). The haplotype and geographic origins of three families harboring the recurrent known p.Ala139Val mutation were consistent with a founder effect. For CLDN19, five of seven mutations detected were previously unknown: one missense (p.Pro28Leu affecting the first transmembrane domain), two nonsense (p.Trp18X and p.Gln57X), one frame shift (p.Thr135LeufsX9), and one large deletion (E1_E4del). All the previously described mutations of this gene were missense (5). Most of the novel mutations described here result in the production of unstable mRNAs or truncated proteins. Interestingly, the previously described p.Gly20Asp mutation was identified in 15 patients from 14 different families. All of these patients came from southwest France or Spain. The haplotype data were consistent with a founder effect, which was previously identified in Spanish families (5).
FHHNC was diagnosed on the basis of the presence of hypomagnesemia, hypercalciuria, and nephrocalcinosis in all 32 patients. Median time to diagnosis was 2.5 years, suggesting that physicians were insufficiently aware of this rare disease. FHHNC is frequently complicated by progressive renal failure. In the cohort of patients with CLDN16 mutations described in the work by Konrad et al. (2), ESRD was reported in 30% of patients at 15 years and 50% of patients at 20 years. In our study, approximately one-third of patients already had CKD at diagnosis, and more than 20% had reached ESRD at last follow-up at a median age of about 15 years, which is consistent with the findings of Konrad et al. (2). However, the risk of ESRD in patients with CLDN19 mutations was two times the risk of patients with CLDN16 mutations.
The pathogenesis of progressive renal failure in patients with FHHNC remains unclear. Progressive tubulointerstitial nephropathy is probably not directly caused by hypercalciuria and nephrocalcinosis, because no correlation was found between the degree of nephrocalcinosis and CKD progression. Interestingly, Japanese Black cattle harboring large deletions of the CLDN16 gene have chronic interstitial nephritis and CKD (15,16). In the first pathologic study of this bovine model, the work by Sasaki et al. (17) described lesions classed as “renal tubular dysplasia” with a secondary decrease in the number of nephrons. The work by Okada et al. (18) described a decrease in the number of glomeruli with immature tubules, secondary interstitial fibrosis, and lymphocytic infiltration, resulting in abnormal nephron development. Therefore, it has been suggested that defective claudin-16 function disruption may occur very early in the development of tubular tight junctions (2). Unfortunately, no equivalent data are available for humans. However, we can speculate that the early onset of ESRD may result from abnormal renal development complicated by fibrosis and nephrocalcinosis (19).
How can we account for the difference in renal outcome between patients with CLDN16 and CLDN19 mutations without differences in BP or protein excretion? The work by Konrad et al. (2) showed that some mutations of the CLDN16 gene (such as Leu151Phe) resulted in residual function, with a slower progression to renal failure. Five of six CLDN16 mutations described in this study are missense that could have a similar residual function. Concerning CLDN19 mutations, no genotype–phenotype correlation has been published. In our study, 15 of 23 patients with CLDN19 mutations had the recurrent p.Gly20Asp mutation that results in the production of a protein retained within the cell (5). Five other patients had nonsense, large deletion, or frame shift mutations. Thus, phenotype severity with higher risk of ESRD could be explained by complete loss of function mutations detected in our cohort.
Claudin-16 and -19 have also been shown to act in synergy in the reabsorption of calcium and magnesium in TAL (20,21): claudin-16 increases cation permeability, whereas claudin-19 decreases anion permeability. A direct interaction between these two proteins seems to be important for the maintenance of cation selectivity. It has been suggested that the phenotype of patients with mutated claudin-19 may result in part from the defective functioning of claudin-16 (22).
It has also been suggested that other claudins may be involved in paracellular reabsorption and regulation or interact with claudin-16 and -19. TAL also expresses claudin-3, -10b, -11, and -14, and distal tubule expresses claudin-3 and -8. Two works suggest that claudin-14 could play a role in urinary calcium excretion: in Cldn11/Cldn14 double mutant mice, a mild increase in urinary calcium and magnesium excretion was observed (23), and in Icelandic and Dutch populations, an association between two synonymous single nucleotide polymorphisms of claudin-14 gene and a higher risk to develop kidney stones and reduced bone mineral density was described (24). Nevertheless, the mechanism and the nephron segment involved in these observations have not been established. Regarding a possible interaction between claudins, in vitro studies have shown that the loss of either claudin-16 or -19 has no effect on the junctional localization of claudin-10 and -18 (21). Furthermore, phenotypic variability was recently shown in a kindred with three affected members harboring a CLDN16 mutation (25), suggesting the possible involvement of other genes or epigenetic factors.
Mutations of CLDN19 are also associated with extrarenal manifestations, such as ocular abnormalities. Several claudins are expressed in human cornea and retina (26,27). Claudin-19 is the prominent claudin expressed in fetal retinal pigment epithelium, and in minor proportion, claudin-3 is expressed; claudin-16 expression in this epithelium seems to be extremely low. Knockdown of claudin-19 by small interfering RNA abolishes the transepithelial electrical resistance, showing the importance of this protein in the permeability characteristics of this epithelium (28). Neurologic manifestations were previously described in two patients with CLDN19 mutations included in this study (12). Extrarenal symptoms may be accounted for by the expression in other tissues of claudin-19 described in the Cldn19-null mouse, which presents peripheral nervous system deficits caused by the absence of claudin-19 in the tight junction of the Schwann cells of peripheral myelinated axons (29).
Patients have been treated with thiazides and indomethacin to decrease hypercalciuria. Indomethacin decreases sodium reabsorption in the proximal tubule and may also increase the passive paracellular reabsorption of calcium (30). In this study, these treatments were found to have no effect on hypercalciuria, but data were obtained for only a limited number of patients.
The limitations of our study include its retrospective design, the multicentric nature of the population, and missing follow-up data. The relatively small population and the small number of events because of the rarity of the disease limit the power of this study to detect differences in disease expression between the two groups.
This study of 32 patients with FHHNC led to the identification of novel and recurrent mutations in the two genes known to be involved in this disease. Phenotypic analysis showed a similar clinical presentation in patients with mutations in either of these two genes except ocular abnormalities, which were found only in patients with CLDN19 mutations. The follow-up data also suggested that patients with CLDN19 mutations have a higher risk of progression to CKD and a poorer renal outcome.
Disclosures
None.
Acknowledgments
We thank the patients who participated in this study and their families for their cooperation. We thank all the physicians from the 14 French and 2 Spanish centers involved in this study and all the staff of the genetic laboratory at Georges Pompidou Hospital. We also thank the following physicians: Stéphane Decramer (CHU de Toulouse, France), Marie-Pierre Lavocat (CHU de Saint Etienne, France), Jean Sarlangue (CHU de Bordeaux, France), Alberto Vidal (CHU de Albacete, Spain), and Teresa Vendrell (Hospital Vall d’Hebron, Barcelona, Spain).
This study was made possible by funds from The French Ministry of Health (Plan Maladies Rares) and European Community FP7 EUNEFRON 201590.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
This article contains supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.12841211/-/DCSupplemental.
References
- 1.Michelis MF, Drash AL, Linarelli LG, De Rubertis FR, Davis BB: Decreased bicarbonate threshold and renal magnesium wasting in a sibship with distal renal tubular acidosis. (Evaluation of the pathophysiological role of parathyroid hormone). Metabolism 21: 905–920, 1972 [DOI] [PubMed] [Google Scholar]
- 2.Konrad M, Hou J, Weber S, Dötsch J, Kari JA, Seeman T, Kuwertz-Bröking E, Peco-Antic A, Tasic V, Dittrich K, Alshaya HO, von Vigier RO, Gallati S, Goodenough DA, Schaller A: CLDN16 genotype predicts renal decline in familial hypomagnesemia with hypercalciuria and nephrocalcinosis. J Am Soc Nephrol 19: 171–181, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benigno V, Canonica CS, Bettinelli A, von Vigier RO, Truttmann AC, Bianchetti MG: Hypomagnesaemia-hypercalciuria-nephrocalcinosis: A report of nine cases and a review. Nephrol Dial Transplant 15: 605–610, 2000 [DOI] [PubMed] [Google Scholar]
- 4.Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, Praga M, Casari G, Bettinelli A, Colussi G, Rodriguez-Soriano J, McCredie D, Milford D, Sanjad S, Lifton RP: Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science 285: 103–106, 1999 [DOI] [PubMed] [Google Scholar]
- 5.Konrad M, Schaller A, Seelow D, Pandey AV, Waldegger S, Lesslauer A, Vitzthum H, Suzuki Y, Luk JM, Becker C, Schlingmann KP, Schmid M, Rodriguez-Soriano J, Ariceta G, Cano F, Enriquez R, Juppner H, Bakkaloglu SA, Hediger MA, Gallati S, Neuhauss SC, Nurnberg P, Weber S: Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet 79: 949–957, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haisch L, Almeida JR, Abreu da Silva PR, Schlingmann KP, Konrad M: The role of tight junctions in paracellular ion transport in the renal tubule: Lessons learned from a rare inherited tubular disorder. Am J Kidney Dis 57: 320–330, 2011 [DOI] [PubMed] [Google Scholar]
- 7.Angelow S, Ahlstrom R, Yu AS: Biology of claudins. Am J Physiol Renal Physiol 295: F867–F876, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber S, Schneider L, Peters M, Misselwitz J, Rönnefarth G, Böswald M, Bonzel KE, Seeman T, Suláková T, Kuwertz-Bröking E, Gregoric A, Palcoux JB, Tasic V, Manz F, Schärer K, Seyberth HW, Konrad M: Novel paracellin-1 mutations in 25 families with familial hypomagnesemia with hypercalciuria and nephrocalcinosis. J Am Soc Nephrol 12: 1872–1881, 2001 [DOI] [PubMed] [Google Scholar]
- 9.Schwartz GJ, Muñoz A, Schneider MF, Mak RH, Kaskel F, Warady BA, Furth SL: New equations to estimate GFR in children with CKD. J Am Soc Nephrol 20: 629–637, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levey AS, Coresh J, Greene T, Marsh J, Stevens LA, Kusek JW, Van Lente F, Chronic Kidney Disease Epidemiology Collaboration : Expressing the Modification of Diet in Renal Disease Study equation for estimating glomerular filtration rate with standardized serum creatinine values. Clin Chem 53: 766–772, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Blanchard A, Jeunemaitre X, Coudol P, Dechaux M, Froissart M, May A, Demontis R, Fournier A, Paillard M, Houillier P: Paracellin-1 is critical for magnesium and calcium reabsorption in the human thick ascending limb of Henle. Kidney Int 59: 2206–2215, 2001 [DOI] [PubMed] [Google Scholar]
- 12.Faguer S, Chauveau D, Cintas P, Tack I, Cointault O, Rostaing L, Vargas-Poussou R, Ribes D: Renal, ocular, and neuromuscular involvements in patients with CLDN19 mutations. Clin J Am Soc Nephrol 6: 355–360, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houdayer C, Gauthier-Villars M, Laugé A, Pagès-Berhouet S, Dehainault C, Caux-Moncoutier V, Karczynski P, Tosi M, Doz F, Desjardins L, Couturier J, Stoppa-Lyonnet D: Comprehensive screening for constitutional RB1 mutations by DHPLC and QMPSF. Hum Mutat 23: 193–202, 2004 [DOI] [PubMed] [Google Scholar]
- 14.Vargas-Poussou R, Cochat P, Le Pottier N, Roncelin I, Liutkus A, Blanchard A, Jeunemaître X: Report of a family with two different hereditary diseases leading to early nephrocalcinosis. Pediatr Nephrol 23: 149–153, 2008 [DOI] [PubMed] [Google Scholar]
- 15.Hirano T, Kobayashi N, Itoh T, Takasuga A, Nakamaru T, Hirotsune S, Sugimoto Y: Null mutation of PCLN-1/Claudin-16 results in bovine chronic interstitial nephritis. Genome Res 10: 659–663, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirano T, Hirotsune S, Sasaki S, Kikuchi T, Sugimoto Y: A new deletion mutation in bovine Claudin-16 (CL-16) deficiency and diagnosis. Anim Genet 33: 118–122, 2002 [DOI] [PubMed] [Google Scholar]
- 17.Sasaki Y, Kitagawa H, Kitoh K, Okura Y, Suzuki K, Mizukoshi M, Ohba Y, Masegi T: Pathological changes of renal tubular dysplasia in Japanese black cattle. Vet Rec 150: 628–632, 2002 [DOI] [PubMed] [Google Scholar]
- 18.Okada K, Ishikawa N, Fujimori K, Goryo M, Ikeda M, Sasaki J, Watanabe D, Takasuga A, Hirano T, Sugimoto Y: Abnormal development of nephrons in claudin-16-defective Japanese black cattle. J Vet Med Sci 67: 171–178, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Sugiyama A, Ozaki K, Miyazaki, Tanabe Y, Takeuchi T, Narama I: Renal dysplasia unrelated to claudin-16 deficiency in Japanese Black cattle. J Comp Pathol 137: 71–77, 2007 [DOI] [PubMed] [Google Scholar]
- 20.Hou J, Renigunta A, Konrad M, Gomes AS, Schneeberger EE, Paul DL, Waldegger S, Goodenough DA: Claudin-16 and claudin-19 interact and form a cation-selective tight junction complex. J Clin Invest 118: 619–628, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou J, Renigunta A, Gomes AS, Hou M, Paul DL, Waldegger S, Goodenough DA: Claudin-16 and claudin-19 interaction is required for their assembly into tight junctions and for renal reabsorption of magnesium. Proc Natl Acad Sci USA 106: 15350–15355, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amasheh S, Fromm M, Günzel D: Claudins of intestine and nephron—a correlation of molecular tight junction structure and barrier function. Acta Physiol (Oxf) 201: 133–140, 2011 [DOI] [PubMed] [Google Scholar]
- 23.Elkouby-Naor L, Abassi Z, Lagziel A, Gow A, Ben-Yosef T: Double gene deletion reveals lack of cooperation between claudin 11 and claudin 14 tight junction proteins. Cell Tissue Res 333: 427–438, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorleifsson G, Holm H, Edvardsson V, Walters GB, Styrkarsdottir U, Gudbjartsson DF, Sulem P, Halldorsson BV, de Vegt F, d’Ancona FC, den Heijer M, Franzson L, Christiansen C, Alexandersen P, Rafnar T, Kristjansson K, Sigurdsson G, Kiemeney LA, Bodvarsson M, Indridason OS, Palsson R, Kong A, Thorsteinsdottir U, Stefansson K: Sequence variants in the CLDN14 gene associate with kidney stones and bone mineral density. Nat Genet 41: 926–930, 2009 [DOI] [PubMed] [Google Scholar]
- 25.Seeley HH, Loomba-Albrecht LA, Nagel M, Butani L, Bremer AA: Familial hypomagnesemia with hypercalciuria and nephrocalcinosis in three siblings having the same genetic lesion but different clinical presentations [published online ahead of print June 1, 2011]. World J Pediatr doi:10.1007/s12519-011-0295-3 [DOI] [PubMed] [Google Scholar]
- 26.Ban Y, Dota A, Cooper LJ, Fullwood NJ, Nakamura T, Tsuzuki M, Mochida C, Kinoshita S: Tight junction-related protein expression and distribution in human corneal epithelium. Exp Eye Res 76: 663–669, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Rizzolo LJ, Peng S, Luo Y, Xiao W: Integration of tight junctions and claudins with the barrier functions of the retinal pigment epithelium. Prog Retin Eye Res 30: 296–323, 2011 [DOI] [PubMed] [Google Scholar]
- 28.Peng S, Rao VS, Adelman RA, Rizzolo LJ: Claudin-19 and the barrier properties of the human retinal pigment epithelium. Invest Ophthalmol Vis Sci 52: 1392–1403, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto T, Morita K, Takemoto D, Takeuchi K, Kitano Y, Miyakawa T, Nakayama K, Okamura Y, Sasaki H, Miyachi Y, Furuse M, Tsukita S: Tight junctions in Schwann cells of peripheral myelinated axons: A lesson from claudin-19-deficient mice. J Cell Biol 169: 527–538, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monnens L, Starremans P, Bindels R: Great strides in the understanding of renal magnesium and calcium reabsorption. Nephrol Dial Transplant 15: 568–571, 2000 [DOI] [PubMed] [Google Scholar]