Abstract
Aberrant O-glycosylation in the hinge region of IgA1 characterizes IgA nephropathy. The mechanisms underlying this abnormal glycosylation are not well understood, but reduced expression of the enzyme core 1, β1,3-galactosyltransferase 1 (C1GALT1) may contribute. In this study, high-throughput microRNA (miRNA) profiling identified 37 miRNAs differentially expressed in PBMCs of patients with IgA nephropathy compared with healthy persons. Among them, we observed upregulation of miR-148b, which potentially targets C1GALT1. Patients with IgA nephropathy exhibited lower C1GALT1 expression, which negatively correlated with miR-148b expression. Transfection of PBMCs from healthy persons with a miR-148b mimic reduced endogenous C1GALT1 mRNA levels threefold. Conversely, loss of miR-148b function in PBMCs of patients with IgA nephropathy increased C1GALT1 mRNA and protein levels to those observed in healthy persons. Moreover, we found that upregulation of miR-148b directly correlated with levels of galactose-deficient IgA1. In vitro, we used an IgA1-producing cell line to confirm that miR-148b modulates IgA1 O-glycosylation and the levels of secreted galactose-deficient IgA1. Taken together, these data suggest a role for miRNAs in the pathogenesis of IgA nephropathy. Abnormal expression of miR-148b may explain the aberrant glycosylation of IgA1, providing a potential pharmacologic target for IgA nephropathy.
IgA nephropathy (IgAN) is considered the most common form of primary GN throughout the world, and about 40% of patients develop ESRD.1,2
The initial event in the pathogenesis of IgAN is the mesangial deposition of IgA1, aberrantly glycosylated because the hinge-region O-linked glycans lack galactose.3–6 Different studies suggest that this alteration could lead to IgA1 self-aggregation,7 IgA1-IgG immune complex formation,8,9 and complex defective clearance, with sequential deposition in the glomeruli.10
In humans, each IgA1 heavy-chain hinge region has three to five 5 O-linked glycans that are built by stepwise addition of monosaccharides, beginning with the addition of N-acetylgalactosamine by the enzyme N-acetylgalactosaminyltransferase 2 and continuing with the addition of galactose by the enzyme core 1, β1,3-galactosyltransferase 1 (C1GALT1).11–14
IgAN is a complex multifactorial disease whose pathogenic mechanism is still unknown. Several investigators have sought to identify specific genetic markers associated with the development and progression of this disease;15 however, few studies have specifically described intracellular mechanisms associated with disease development.16 Recently, our group identified new mechanisms associated with the pathogenesis of IgAN and showed that WNT-β-catenin and PI3K/Akt pathways were highly activated in patients with IgAN.17 The basis for the abnormal glycosylation in IgAN is still unknown, but some studies suggest that C1GALT1 could be involved because of its altered expression.18
On the other hand, the potential role of microRNAs (miRNAs) in the IgAN pathogenesis has been poorly investigated. After the discovery of miRNAs, great effort has focused on determining their biologic functions and their relevance to diseases. In fact, deregulation of miRNAs has been associated with several disease states, including kidney diseases.19
To our knowledge, this study is the first to evaluate the global miRNA expression profile of IgAN patients′ PBMCs, which are directly involved in the disease.20 We defined the miRNA signature in patients with IgAN and showed that miR-148b regulating C1GALT1 explains the abnormal glycosylation process in IgAN. These results support an important and unreported role of this miRNA in the pathogenesis of IgAN and suggest a pharmacologic rationale for the potential use of synthetic miRNA inhibitors to attenuate IgA1 deglycosylation in the disease.
Results
Identification of Differentially Expressed miRNAs in Patients with IgAN
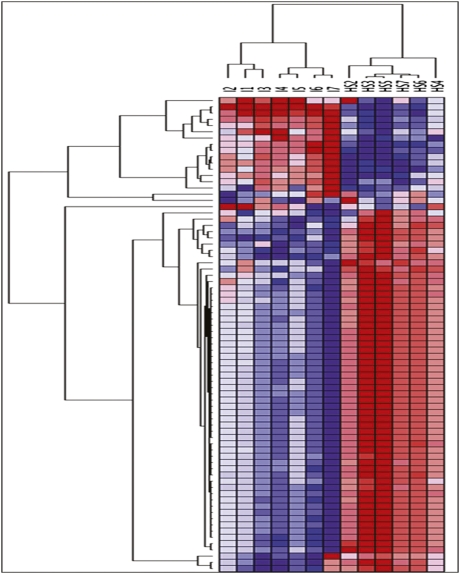
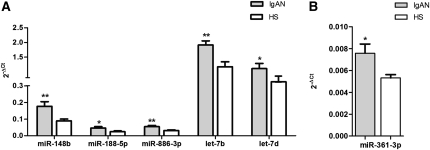
The role of miRNA expression in the pathogenesis of IgAN has not been well explored. To identify miRNAs differentially expressed in IgAN, we analyzed their global expression profile in PBMCs of seven patients with IgAN and seven healthy participants. Among 723 human miRNAs represented on the microarrays, 147 were expressed in each sample. Unsupervised hierarchical clustering analysis generated a tree with the IgAN and healthy participants clearly separated into two groups (Figure 1). This separation was further confirmed by displaying the relationships among miRNA expression patterns using principal component analysis (Supplemental Figure 1). After we applied a fold change threshold > 2 (false discovery rate [FDR]< 0.01), 35 miRNAs were found to be significantly upregulated and 2 were significantly downregulated in IgAN (Supplemental Table 1). To validate microarray results, we performed quantitative real-time PCR (qRT-PCR) for miR-148b, miR-188-5p, miR-361-3p, miR-886-3p, let-7b, and let-7d on miRNAs isolated from PBMCs of an independent set of 10 patients with IgAN and 10 healthy persons with the same clinical and demographic characteristics as those in the population used for microarray experiments (Table 1). The expression of all analyzed miRNAs was significantly higher in patients with IgAN, thereby confirming microarray results (Figure 2, A and B).
Figure 1.
Unsupervised hierarchical clustering of miRNA expression profile. miRNA expression pattern of PBMCs of seven patients with IgAN and seven healthy subjects (HSs) were examined using Agilent array composed of 723 human and 76 human viral miRNAs. A total of 147 miRNAs were expressed in all samples, discriminating patients with IgAN from HSs (P<0.0001; FDR<0.01). Two principal clusters were identified on the basis of differential miRNA expression.
Table 1.
Demographic and clinical features of patients and healthy participants
| Variable | Initial Sample Cohort | Disease Controls | Validation Sample Cohort | ||||
|---|---|---|---|---|---|---|---|
| IgAN | Healthy Participants | MPGN-I | FSGS | HSPN | IgAN | Healthy Participants | |
| Participants (n) | 25 | 25 | 3 | 5 | 10 | 50 | 50 |
| Men/women (n/n) | 16∕9 | 15∕10 | 2/1 | 3/2 | 5/5 | 38/12 | 40/10 |
| Age (yr) | 37.2±10.2 | 36.1±10.6 | 35±9.6 | 42±12.3 | 8.5±3 | 36±12.3 | 43±10.6 |
| Serum creatinine (mg/dl) | 0.9±0.2 | 0.8±0.3 | 0.7±0.05 | 0.8±0.4 | 0.5±0.1 | 1.00±0.5 | 0.9±0.3 |
| Estimate GFR (ml/min per 1.73 m2) | 118±22.3 | 106±13.1 | 122±10 | 116.8±11 | ND | 120±11 | 114±12.3 |
| Proteinuria (g/24 h) | 0.3±0.1 | 0.1±0.2 | 2.7±1.2 | 2.05±0.9 | ND | 0.8±0.2 | 0.06±0.02 |
| Systolic BP (mmHg) | 121±20.4 | 120±10 | 133±11.5 | 126±15 | 118±16 | 124±13.3 | 119±9 |
| Diastolic BP (mmHg) | 72±9 | 74±8 | 80±0.5 | 79±6.5 | 67±8 | 77.2±9.7 | 74±9 |
| Total IgA (ng/μl) | 0.10±0.004 | 0.12±0.01 | ND | ND | 0.15±0.01 | 0.14±0.002 | 0.13±0.001 |
Unless otherwise noted, values are expressed as mean ± SD. IgAN, IgA nephropathy; MPGN-I, membranoproliferative GN type I; FSGS, focal segmental glomerulosclerosis; HSPN, Henoch-Schönlein purpura nephritis; ND, not determined.
Figure 2.
Differential expression validation. Validation of differential expression of miR-148b, miR-188-5p, miR-886-3p, let-7b, and let-7d (A) and miR-361-3p (B) in an independent set of PBMCs from 10 patients with IgAN and 10 healthy subjects (HSs). Expression levels were quantified using qRT-PCR. The miRNA relative expressions were normalized to the expression of U6. Expression levels of miR-148b, miR-188-5p, miR-886-3p, let-7b, let-7d, and miR-361-3p were found to be significantly higher in patients with IgAN than in HSs. The histograms represent the mean ± SEM. *P<0.03; **P<0.01.
In silico Analysis of miRNA Targets
To study the molecular mechanisms in which the miRNAs are involved, we performed a bioinformatic analysis to predict target genes of the 35 upregulated miRNAs. To reduce the number of false-positive results, we used four different algorithms and listed only putative target genes predicted by at least two of them. On the basis of the results of bioinformatics analysis, we found that one of the potential targets of miR-148b was the gene C1GALT1, which plays an important role in the pathogenesis of IgAN. Of note, other miR-148b putative target genes were inversin (INVS) and phosphatase and tensin homologue (PTEN) (Supplemental Table 2), two genes that we found downregulated in patients with IgAN.17
Ingenuity Pathway Analysis (IPA) software was then used to evaluate the biologic interaction among miRNAs. We uploaded the 35 upregulated miRNAs in IPA, and two networks were identified. When we merged each of them with the network resulting from the gene expression profile published in our previous work,17 we found that miRNAs were strongly interconnected with the mRNA network (Supplemental Figure 2). In particular, let-7d directly regulated PTEN, miR-361 regulated INVS, miR-148b regulated both INVS and PTEN, and three miRNAs (let-7d, let-7a, miR-98) indirectly regulate AKT through RET.
Identification of miR-148b as a Regulator of C1GALT1 Expression
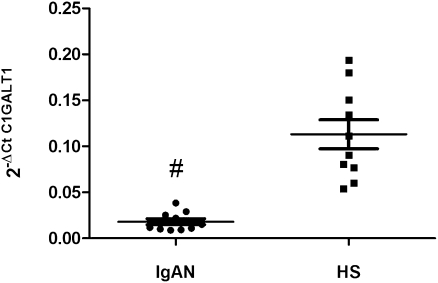
C1GALT1 is known as a directly involved in IgAN; however, the basis for its decreased function in the disease is unknown. Therefore, we evaluated the C1GALT1 mRNA expression levels by qRT-PCR in the same set of RNA samples used in the microarray validation. We found that C1GALT1 levels were significantly lower in patients with IgAN than in the healthy participants (P<0.0001; Figure 3).
Figure 3.
C1GALT1 gene expression levels evaluated by real-time PCR in 10 patients with IgAN and 10 healthy subjects (HSs). C1GALT1 expression levels were significantly lower in patients with IgAN than in HSs. C1GALT1 expression levels were normalized on the housekeeping gene β-actin. The scatter plots represent the mean ± SEM. #P<0.0001.
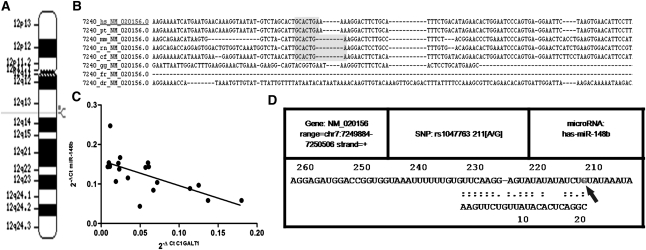
The in silico analysis showed that the miR-148b increase could be the cause of the C1GALT1 reduction. In fact, sequence alignment of human miR-148b with 3′-untranslated region (UTR) C1GALT1 identified a binding site (nucleotides 1355–1361 in human C1GALT1, chr 12q13; Figure 4A) that is well conserved among different species (Figure 4B).
Figure 4.
miR-148b targets C1GALT1. (A) Genomic localization of miR-148b in chromosome 12q13. (B) Sequence alignment of the miR-148b base-pairing sites in the 3′-UTR of C1GALT1 mRNA showing that the regions complementary to miR-148b are highly conserved among human, chimp, mouse, rat, dog, and chicken. The “seed” sequences of miR-148b complementary to C1GALT1 are shown in gray. (C) Linear correlation between the expression of C1GALT1 and the expression of miR-148b in PBMCs of 10 patients with IgAN and 10 healthy subjects (HSs). C1GALT1 mRNA levels inversely correlated with miR-148b expression levels (R2=0.4; P<0.01). (D) Output of bioinformatic analysis performed by MicroSNiPer algorithms showing that the 1365G/A polymorphism (rs1047763) affects miR-148b binding sites in the 3′-UTR of C1GALT1. The 1365G allele (arrow) enhances miR-148b binding, whereas the 1365A allele does not appear in the output as a possible binding site for miR-148b.
To study whether the downregulation of C1GALT1 in patients with IgAN was attributable to increased miR-148b levels, we tested the correlation between C1GALT1 mRNA and miR-148b levels in patients with IgAN and the healthy participants. A negative correlation was observed (R2=0.4; P<0.01; Figure 4C): samples with higher miR-148b expression levels showed lower levels of C1GALT1 mRNA.
The inverse correlation observed between the levels of miR-148b and C1GALT1 mRNA suggests that this gene is probably a target of miR-148b.
Moreover, we performed a bioinformatic analysis to estimate the effect of some single-nucleotide polymorphisms (SNPs) on putative miRNA targets interrogating the 3′-UTR and predicting whether a SNP within the target site will disrupt/eliminate or enhance/create a miRNA binding site. Our results revealed that the 1365G/A polymorphism (rs1047763) in the 3′-UTR of C1GALT1, which is associated with IgAN,21,22 affects miR-148b binding sites in this gene. In fact, the biocomputational analysis showed that the 1365G allele enhances miR-148b binding (Figure 4D).
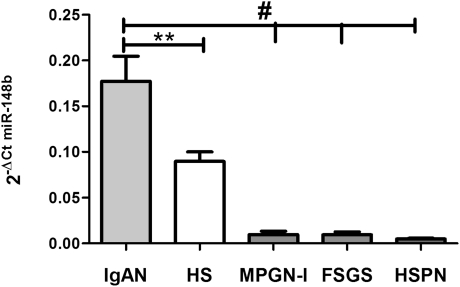
To study whether the upregulated expression of miR-148b was specific to IgAN, we checked the miR-148b expression in PBMCs from controls with three additional diseases: 3 patients with membranoproliferative GN type I , 5 with focal segmental glomerulosclerosis, and 10 with Henoch–Schönlein purpura nephritis. miR-148b expression in these diseases has been compared with that of IgAN and controls. We found that the miR-148b was again higher in patients with IgAN (P<0.0001) than in those with the other three diseases (Figure 5), confirming that higher miR-148b levels are typical of IgAN.
Figure 5.
miR-148b expression levels evaluated by real-time PCR in PBMCs from 3 patients with membranoproliferative GN type I (MPGN-I), 5 with focal segmental glomerulosclerosis (FSGS), and 10 with Henoch-Schönlein purpura nephritis (HSPN). miR-148b expression in these diseases and in healthy subjects (HSs) has been compared with that in IgAN. We found that miR-148b levels were higher in patients with IgAN than in those with MPGN-I, FSGS, and HSPN and in HSs. The miRNA relative expressions were normalized to the expression of U6. The histograms represent the mean ± SEM. **P<0.01; #P<0.0001.
miR-148b Downregulates C1GALT1 mRNA Expression
The nature of target of miR-148b and its mode of action in PBMCs have not been functionally identified; thus, we tested whether miR-148b was able to modulate the expression of C1GALT1. For this purpose, we performed some transient transfection experiments ex vivo using PBMCs from an independent group of four patients with IgAN and four healthy persons.
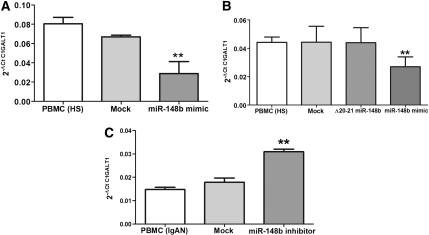
We increased the amount of the endogenous miR-148b within PBMCs isolated from four healthy persons, transfecting short RNA sequences that mimic the action of the miRNA, to simulate the situation found in patients with IgAN. PBMCs of the four healthy persons, transfected with 25nM miR-148b mimic, showed a three-fold reduction in endogenous C1GALT1 mRNA levels (P<0.03; Figure 6A).
Figure 6.
miR-148b regulates C1GALT1 mRNA expression. (A) C1GALT1 expression levels were analyzed by real-time PCR in PBMCs of healthy subjects (HSs) after transfection with miR-148b mimic. Increasing the amount of miR-148b within the PBMCs of HSs resulted in a three-fold reduction in endogenous C1GALT1 mRNA levels. (B) C1GALT1 expression levels were analyzed by real-time PCR in PBMCs of five other HSs after transfection with the mutated miR-148b mimic, Δ20-21, and miR-148b mimic wild-type. C1GALT1 mRNA expression decreased only after transfection with the wild-type miR-148b mimic. (C) C1GALT1 expression levels were analyzed by real-time PCR in PBMCs of patients with IgAN after transfection with miR-148b inhibitor. Silencing the activity of miR-148b within IgAN PBMCs led to a three-fold increase in endogenous C1GALT1 mRNA levels. “Mock” indicates mock-transfected cells going through the transfection processes without addition of mimic/inhibitor miRNA. Expression data were normalized on the housekeeping gene β-actin. Data are representative of four independent experiments for A and C and five independent experiments for B (mean ± SEM). **P<0.01.
To provide evidence for a direct effect of miR-148b on C1GALT1 expression, we transfected PBMCs isolated from five other healthy persons with both a miR-148b mimic and a mutated miR-148b mimic (the Δ20-21 miR-148b, with a substitution from UG to AC in position 20-21 [5′→3′]). We found that the C1GALT1 mRNA expression decreased only after transfection with the wild-type miR-148b mimic (Figure 6B).
To further confirm that C1GALT1 mRNA was targeted by miR-148b, we performed the reverse experiment, in which we used synthetic hairpin miRNA inhibitors to silence the activity of miR-148b in PBMCs isolated from four patients with IgAN. The miRNA inhibitors were chemically modified acids designed to bind and to inhibit endogenous miR-148b. The transient transfection of IgAN PBMCs with 250 nM of miR-148b inhibitor led to a three-fold increase in endogenous C1GALT1 mRNA levels (P<0.01, Figure 6C). In line with our computational analysis, we showed that miR-148b did not affect Cosmc chaperone, which is involved in stability and activity of C1GALT123 (Supplemental Figure 3).
miR-148b Downregulates C1GALT1 Protein Expression
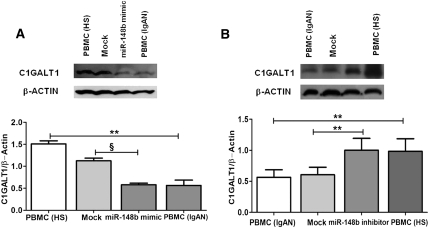
We used the same strategy to functionally inhibit or enhance the mature form of miR-148b in PBMCs of patients with IgAN and healthy persons, respectively, with the aim of establishing whether C1GALT1 protein expression was also effectively controlled by miR-148b. PBMCs of five healthy persons transiently transfected with 25 nM of miR-148b mimic resulted in a remarkable reduction in C1GALT1 protein expression (2.2-fold; P<0.002) as revealed by Western blot. Of note, the increase in the miR-148b level in PBMCs of healthy persons resulted in the same C1GALT1 protein expression levels as in PBMCs of patients with IgAN (Figure 7A). On the contrary, in the reverse experiment, PBMCs of five patients with IgAN transfected with 250 nM of miR-148b inhibitor showed a significant increase in C1GALT1 protein production (1.7-fold increase; P<0.01). Surprisingly, C1GALT1 protein levels in patients with IgAN increased to the same levels as in healthy persons (Figure 7B). Altogether our results demonstrate that miR-148b targets C1GALT1, which is responsible for a decreased activity of the gene in patients with IgAN.
Figure 7.
miR-148b regulates C1GALT1 expression at protein level. (A) Transfection of PBMCs of healthy participant with 25 nM miR-148b mimic resulted in a 2.2-fold reduction in C1GALT1 protein expression. Increasing the miR-148b levels within PBMCs of healthy participants resulted in the same C1GALT1 protein levels as in IgAN PBMCs, as shown by Western blot. (B) Western blot representing protein levels of C1GALT1 in IgAN PBMCs after transfection with 250 nM miR-148b inhibitor. A significant increase in C1GALT1 protein production was shown in IgAN PBMCs transfected with miR-148b inhibitor (1.7-fold increase). miR-148b loss of function led to the same C1GALT1 protein levels as in PBMCs of healthy participants. “Mock” indicates mock-transfected cells going through the transfection processes without addition of mimic/inhibitor miRNA. In both experiments, β-actin was used as endogenous control. Data are representative of five independent experiments (mean ± SEM). **P<0.01, §P≤ 0.001.
miR-148b Expression and Altered IgA1 Glycosylation Relationship
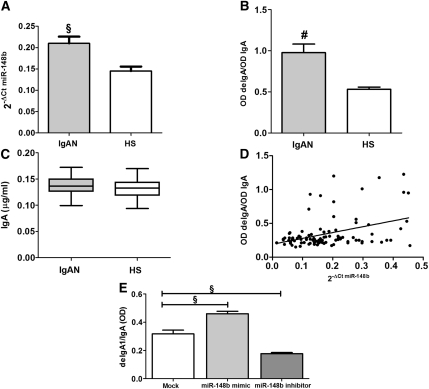
To further validate the relationship between miR-148b expression and altered IgA1 glycosylation, we enrolled an independent cohort of 50 patients with IgAN and 50 healthy persons. Again, miR-148b expression levels, evaluated by qRT-PCR, were significantly higher in PBMCs of patients with IgAN than in those of healthy persons (P<0.001; Figure 8A).
Figure 8.
miR-148b regulates Gal-deficient IgA1 levels. (A) The miR-148b expression levels evaluated by real-time PCR in 50 patients with IgAN and 50 healthy subjects (HSs). miR-148b expression levels were significantly higher in patients with IgAN than in HSs. miR-148b expression levels were normalized to the expression of U6. The histograms represent the mean ± SEM. §P<0.001. (B) shows the serum levels of Gal-deficient IgA1 in patients with IgAN and HSs. The Gal-deficient IgA1 was significantly higher in sera obtained from the patients with IgAN than in sera from HSs. The relative lectin binding per unit IgA1 was calculated as the OD value of lectin over the OD value of total IgA. (C) The serum level of total IgA in patients with IgAN and HSs determined by ELISA. IgA levels in sera obtained from 50 patients with IgAN and 50 HSs were similar. The histograms represent the mean ±SEM. #P<0.0001. (D) Linear correlation between the expression of miR-148b and the Gal-deficient IgA1 levels in 50 patients with IgAN and 50 HSs. miR-148b levels directly correlated with Gal-deficient IgA1 levels (r=0.4,; P<0.0001). (E) The Gal-deficient IgA1 levels in supernatants obtained from DAKIKI cells after transfection with miR-148b mimic and inhibitor. A significant increase in Gal-deficient IgA1 production was shown in DAKIKI cells transfected with 50 nM miR-148b mimic (1.5-fold increase). On the contrary, the transfection with 500 nM miR-148b inhibitor led to a significant reduction in Gal-deficient IgA1 levels (1.8-fold). The relative lectin binding per unit IgA1 was calculated as the OD value of lectin over the OD value of total IgA. The histograms represent the mean ± SEM. §P<0.001.
We next evaluated the galactose-deficient (Gal-deficient) IgA1 in serum of the same patients with IgAN and healthy persons using helix aspersa agglutinin (HAA) lectin binding assay. We found that the Gal-deficient IgA1 was significantly higher in patients with IgAN (mean OD ± SD, 0.98±0.1) than in healthy persons (mean OD, 0.53±0.02; P<0.0001) (Figure 8B), whereas total IgA levels were similar in the two groups (0.137±0.002 and 0.132±0.002 µg/ml, respectively) (Figure 8C). Pearson correlation analysis between miR-148b expression level and Gal-deficient IgA1 level showed a significant positive correlation (r = 0.4; P<0.0001; Figure 8D), supporting our data on C1GALT1 regulation by miR-148b and explaining the abnormal increase in Gal-deficient IgA1 consequent to higher miR-148b levels in patients with IgAN. To confirm the direct cause-and-effect relationship between miR-148b expression and Gal-deficient IgA production, we used as a model the human B lymphoma DAKIKI cells producing IgA1.24 We transfected DAKIKI cells with miR-148b mimic and inhibitor and evaluated changes in the Gal-deficient IgA1 levels by means of HAA lectin binding assay. DAKIKI cells transfected with 50 nM of miR-148b mimic resulted in a remarkable increase in Gal-deficient IgA1 levels (1.5-fold increase; P<0.002; Figure 8E). In contrast, in the reverse experiment, DAKIKI cells transfected with 500 nM of miR-148b inhibitor showed a significant reduction in Gal-deficient IgA1 levels (1.8-fold; P<0.01; Figure 8E). These data further confirmed the role of miR-148b in IgA1 O-glycosylation regulation.
Discussion
IgAN is the most common primary form of GN worldwide,25 leading to ESRD in 40% of patients. There is a consensus opinion that aberrant glycosylation of IgA1 is directly involved in the pathogenesis of the disease.3–5,10,26 This abnormality is manifested by a deficiency of galactose in the hinge-region O-linked glycans of IgA1. Biosynthesis of these glycans begins with the addition of N-acetylgalactosamine by the enzyme N-acetylgalactosaminyltransferase 2 and continues with the addition of galactose by the enzyme C1GALT1.11–14
The mechanisms leading to this aberrant glycosylation have been studied extensively; however, to date, the nature of abnormally O-glycosylated IgA1 is still obscure. Kudo et al. demonstrated an important role for C1GALT1 in the molecular basis for the aberrant IgA1 glycosylation in IgAN.18 However, it is controversial whether IgA1 is undergalactosylated as a consequence of functional changes in C1GALT1 activity or as a consequence of its reduced expression.14,27 Moreover, the molecular mechanisms behind the impaired functioning of C1GALT1 are again unknown. Accumulating evidence suggests the involvement of miRNAs in the pathogenesis of some kidney diseases, such as acute kidney rejection, lupus nephritis, and renal ischemia reperfusion injury.28–30 However, miRNAs potentially involved in the pathogenesis of IgAN have been partially studied in renal tissue.31
Starting from a microarray miRNA expression profile in PBMCs of patients with IgAN and using several bioinformatic approaches, we identified 37 miRNAs that are differentially regulated in IgAN patients compared with healthy persons, along with their candidate target genes.
Furthermore, we overlapped modulated miRNAs with previously identified gene pathways involved in IgAN17 to identify candidate target genes involved in the pathogenesis. This analysis revealed that the miRNAs and genes differentially expressed in IgAN were strongly interconnected, particularly the six miRNAs whose expression we validated by qRT-PCR.
Among 37 miRNAs significantly deregulated in patients with IgAN, miR-148b aroused our interest because its putative target genes were C1GALT1, INVS, and PTEN, three genes downregulated in these patients.11,17 We found a reduced expression of C1GALT1 in patients with IgAN, and we showed that miR-148b expression negatively correlated with the C1GALT1 expression levels in the same patients.
In the last years, two large-scale case-control association studies pointed out the role of C1GALT1 in the pathogenesis of IgAN, reporting some C1GALT1 genetic variants associated with the genetic susceptibility to IgAN in the Italian and Chinese populations.21,22 Both studies identified a positive association between the susceptibility to IgAN and a C1GALT1 SNP at the position 1365 G/A (rs1047763) in the 3′-UTR regulatory region. Furthermore, a correlation between this SNP and a reduced C1GALT1 expression in homozygous 1365G/G individuals was shown.22
Our bioinformatic analysis revealed that rs1047763 SNP is located within the miR-148b binding site on the C1GALT1 3′-UTR region, and the binding affinity of miR-148b is greater when the 1365G-variant allele is present. These results could explain the lower C1GALT1 expression levels in patients with IgAN carrying the 1365G/G genotype.22
Supporting our bioinformatic analysis, we found that C1GALT1 mRNA levels were significantly lower in patients with IgAN. Moreover, using the strategy to functionally enhance or inhibit the mature form of miR-148b, we confirmed biologically, in an ex vivo experiment, that miR-148b targets C1GALT1 and can modulate its mRNA and protein levels. Of note, the loss of function of miR-148b in PBMCs isolated from patients with IgAN led to C1GALT1 protein levels similar to those observed generally in healthy persons. In contrast, miR-148b overexpression in PBMCs isolated from healthy persons led to a reduction in C1GALT1 protein to levels similar to those expressed in patients with IgAN.
Moreover, patients with IgAN who had higher serum levels of Gal-deficient IgA1 also showed higher levels of miR-148b. In contrast, lower levels of serum Gal-deficient IgA1 in healthy persons were associated with low miR-148b levels. This finding supports our data on C1GALT1 regulation by miR-148b and explains the abnormal increase in Gal-deficient IgA1 due to high miR-148b levels in patients with IgAN. The role of miR-148b in IgA1 O-glycosylation regulation was also confirmed by transfection experiments in DAKIKI human B lymphoma cells showing that miR-148b modulated levels of secreted Gal-deficient IgA1.
On the basis of our results, we can hypothesize that in IgAN, lower levels of C1GALT1 due to the inhibiting effect of overexpressed miR-148b may lead to an increase in circulating deglycosylated IgA1.8,9,32–34 These galactose-deficient IgA1 molecules are predisposed to aggregation and formation of immune complexes;8 they are recognized by naturally occurring circulating antibodies.9 Consequently, these complexes are less effectively cleared from the circulation35 and efficiently bind to mesangial cells, inducing cellular proliferation, overproduction of extracellular matrix, and synthesis of inflammatory cytokines that can initiate or perpetuate the course of GN.36–38
Our study has one important limitation. We did not study the effect of miR-148b on the process of IgA1 glycosylation in an experimental model of IgAN because IgA1 is present exclusively in humans and hominoid primates.39,40 However, our study points overall to a new regulatory mechanism of IgAN that can explain the aberrant glycosylation of IgA1 responsible for the pathogenesis of the disease. Moreover, the miR-148b expression levels may be used to develop a method to diagnose the disease through a noninvasive technique, which may replace, in the future, the renal biopsy as a diagnostic approach.
In conclusion, our study shows, for the first time, a global miRNA expression profile in PBMCs of patients with IgAN and a miRNA signature. We also provide evidence for a previously unknown key regulatory mechanism of IgAN by discovering the deregulation of miR-148b, which could explain the aberrant glycosylation of IgA1 in the disease. These findings could also have therapeutic implications because the inhibition of miR-148b expression reverses the lower levels of C1GALT1 typical of IgAN. Therefore, miR-148b levels may be manipulated to provide useful new therapeutic approaches to the disease.
Concise Methods
Sample Collection
A total of 75 patients with IgAN, 75 healthy participants, 10 patients with Henoch-Schönlein purpura nephritis, 3 patients with membranoproliferative GN type I, and 5 patients with focal segmental glomerulosclerosis were included in this study. Written informed consent was obtained from each patient and healthy participant.
The study was initially conducted on a cohort of 25 patients with IgAN and 25 healthy participants. Seven in each group were included in the microarray experiment, 10 participants from each group were used for microarray validation, and 8 participants from each group were used for the in vitro transfections. Patients with IgAN had normal renal function; patients with severe renal damage were excluded from the study. Each patient was selected according to the following demographic and clinical features: age (18–50 years), moderate histologic lesions (G2 according to the Schena classification41), creatinine clearance >90 ml/min per 1.73 m2 (evaluated by the Cockcroft-Gault formula), and clinical follow-up (at least 7 years). The healthy participants were selected on the basis of their demographic characteristics and overlapped completely with IgAN group. All healthy participants had a negative result on a urine test for blood and proteins. All patients with diabetes, chronic lung diseases, neoplasm, or inflammatory diseases and patients receiving antibiotics, corticosteroids, and nonsteroidal anti-inflammatory agents were excluded from the study. No patients had symptomatic coronary artery diseases or a family history of premature cardiovascular diseases.
To validate some of the results obtained, we collected blood samples from an independent cohort of 50 patients with IgAN and 50 healthy persons matched in their demographic and clinical features with the participants included in the first group.
Ten patients with Henoch-Schönlein purpura nephritis, three with membranoproliferative GN type I, and five with focal segmental glomerulosclerosis were selected as disease controls.
All participants were from the Italian population. The study was carried out according to the principles of the Declaration of Helsinki and was approved by our institutional ethics review board.
The main demographic and clinical features of our patients and controls included in the study are summarized in Table 1. There were no statistically significant differences between the IgAN and healthy groups for any measure considered.
PBMCs were isolated by density separation over a Ficoll-Hypaque (Ficoll-Paque Plus, GE Healthcare) gradient (460 g for 30 minutes). PBMCs were washed three times with PBS (pH 7.4)/1 mM EDTA (Sigma). Cells were then counted, and their viability was determined by trypan blue exclusion.
Serum was collected at the same clinical time point, and whole blood was centrifuged at 3000 rpm for 10 minutes at room temperature.
miRNA Isolation
Isolated PBMCs were used for total RNA extraction by means of miRNeasy Mini kit (Qiagen) according to the manufacturer’s protocol. Deoxyribonuclease treatment was carried out to remove any contaminating DNA (RNase-Free DNase Set, Qiagen).
Total RNA, including small RNA fractions, was then eluted in ribonuclease-free water. The RNA concentration was determined with NanoDrop Spectrophotometer (Nanodrop Technologies). The miRNA quality was assessed using Agilent 2100 Bioanalyzer (Agilent Technologies), and only samples with RNA integrity number > 8.5 were used.
miRNA Microarray
miRNA microarray analysis was performed using the Agilent Human miRNA Microarrays V2, which were based on Sanger miRBase release 10.1, according to the manufacturer's protocol. Briefly, 600 ng of total RNA isolated from PBMCs of seven patients with IgAN and seven healthy participants were first dephosphorylated with a calf intestine alkaline phosphatase treatment for 30 minutes at 37°C before labeling. Samples were diluted with DMSO, denatured for 10 minutes at 100°C, and labeled using pCp-Cy3 in T4 RNA ligation buffer.
The labeled RNA was hybridized, washed, stained, and scanned with an Agilent microarray scanner (G2565BA, Agilent). Microarray data analysis was performed using Agilent Feature Extraction Software 9.5.1.1.
Microarray data are available under accession number GSE25590 at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/).
Statistical Analyses and Bioinformatics
For microarray analysis, the raw expression signals were log-transformed, normalized, and filtered according to the median corrected signal of all the miRNAs with an intensity >100 (which is considered as expressed), which resulted in the selection of 147 miRNAs out of the original set of 723 miRNAs. The preprocessed microarray data were imported into the R language for statistical analysis computing (http://www.r-project.org). miRNAs displaying differential expression between patients with IgAN and healthy participants were detected using a two-sample t test. Probe sets were sorted after a P value showed a statistically significant difference and were adjusted to account for multiple testing using the FDR method of Storey and Tibshirani.42 Only genes that were significantly (FDR<0.01) modulated in patients with IgAN compared with healthy participants were considered for further analysis. Significance of differential expression, as determined by the enrichment analysis, was permutated 1000 times. To determine the features that are most closely correlated with IgAN phenotype, we applied a most stringent filter with FDR<0.01 and a fold change > 2. Two-dimensional hierarchical clustering was performed using Spotfire decision site 8.0 (http://spotfire.tibco.com). To assess biologic relationships among genes, we used Ingenuity Pathway Analysis software (IPA, Ingenuity System, Redwood City, CA). IPA computes a score for each network according to the fit of the set of supplied focus miRNAs/genes. These scores indicate the likelihood that focus genes would belong to a network versus those obtained by chance. miRNA targets were predicted by means of miRBase 13.0 (http://microrna.sanger.ac.uk),43 TargetScan 5.1 (http://www.targetscan.org),44 PicTar (http://pictar.org),45 and miRWalk (http://www.ma.uni-heidelberg.de) algorithms. Potential targets were chosen overlapping results from the four algorithms and selecting targets of genes predicted by at least two of the algorithms; the targets were selected by a score cutoff computed by a weighted sum of a number of sequence and context features of the predicted miRNA:mRNA duplex. Bioinformatic analysis to estimate the effect of SNPs on putative miRNA targets was carried out by microSNiPer software (Clinical Brain Disorders Branch, National Institutes of Health, Bethesda, MD; http://cbdb.nimh.nih.gov/microsniper)46 interrogating the 3′-UTR and predicting whether a SNP within the target site will disrupt/eliminate or enhance/create a microRNA binding site. A banded Smith-Waterman dynamic program was used along the best-matched regions. The algorithm output only those miRNAs/seeds in which the alignment was shifted because of a change in the alleles.
A two-tailed t test was used to assess differences in biologic features between patients with IgAN and healthy participants. A Pearson correlation test was used to study continuous variables. All values were expressed as the mean ± SEM of data obtained from at least three independent experiments. Results were considered statistically significant at P<0.05.
qRT-PCR
Total RNA, including small RNA fractions, was reverse transcribed with miScript Reverse Transcription Kit (Qiagen) following the manufacturer’s instructions.
Real-time RT-PCR for the quantification of a subset of miRNAs (miR-148b, miR-188-5p, let-7d, let-7b, miR-361-3p, and miR-886-3p, plus an endogenous control) was carried out with miScript Primer Assays and miScript SYBR Green PCR Kit from Qiagen. Real-time PCR amplification reactions were performed in triplicate in 25 μl of final volume via SYBR Green chemistry on iCycler (Bio-Rad).
Normalization was performed with a small nucleolar RNA U6 endogenous control. Comparative real-time PCR was performed in triplicate, including no-template controls. Relative expression was calculated using the 2-ΔCt method.
For C1GALT1 and Cosmc expression analysis, total RNA was reverse-transcribed with QuantiTect Reverse Transcription Kit (Qiagen) following the manufacturer’s instructions. qRT-PCR amplification reactions were performed in triplicate in 25-μl final volumes using SYBR Green chemistry on an iCycler. qRT-PCR was performed using the QuantiTect Primer Assay and the QuantiFast SYBR Green PCR mix (Qiagen). Genes were amplified according to the manufacturer’s directions. The β-actin gene amplification was used as a reference standard to normalize the target signal.
miR-148b Mimic and Inhibitor Transfection
Isolated PBMCs were cultured at 2 × 106 cells per well in a 12-well plate with RPMI-1640 supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate, 1 mM nonessential amino acids, 25 mM HEPES buffer, and 10% heat-inactivated FBS. The transfection of miRNAs mimic and inhibitor was carried out using TransIT-TKO Transfection Reagent (Mirus) in accordance with manufacturer's procedure. For each transfection, 25 nM of miR-148b mimic (Qiagen), 250 nM of miR-148b inhibitor (Qiagen), and 25 nM of a mutated miR-148b mimic (Δ20-21 miR-148b, with a substitution from UG to AC in position 20-21 [5′→3′]; Qiagen) were used. In transfection experiments, a mock-transfection control was performed by putting cells through the transfection procedure without adding miRNA. The validated nonsilencing siRNA sequence AllStars Negative Control siRNA (50 nM, Qiagen) was used as negative control and Syn-hsa-miR-1 miScript miRNA mimic (25 nM, Qiagen) and anti–hsa-miR-1 miScript miRNA inhibitor (250 nM, Qiagen) were used as positive controls for miR-148b mimic and miR-148b inhibitor transfection, respectively. Each transfection experiment was done in triplicate. After transfection, cells were incubated for 24 hours or 48 hours at 37°C in 5% CO2 and used, respectively, for total RNA extraction and protein extraction.
For analysis of IgA1 glycosylation, the surface IgA1-positive human B lymphoma cell line, DAKIKI, was purchased from ATCC.24,47 Cells were cultured at 1 × 105 cells per well in a 12-well plate with RPMI 1640 medium containing 10% heat-inactivated FBS, 1 mM sodium pyruvate, and 2 mM glutamine. The transfection of miRNAs mimic and inhibitor was carried out using TransIT-TKO Transfection Reagent (Mirus) in accordance with the manufacturer's procedure. For each transfection, 50 nM of miR-148b mimic (Qiagen) and 500 nM of miR-148b inhibitor (Qiagen) were used. Each transfection experiment was done in triplicate. After transfection, cells were incubated for 72 hours at 37°C in 5% CO2.
Western Blot Analysis
The amount of C1GALT1, after transfection, was determined by Western blotting analysis. Total protein extract were prepared with lysis buffer containing 150 mM NaCl, 50 mM Tris-HCl (pH 8), 1% NonidetP-40, 0.1% sodium deoxycholate, and 0.1% SDS, plus proteinase inhibitors. The protein concentration was determined by the Bradford assay (BioRad). Eighty micrograms of each protein lysate was separated on a 10% SDS-PAGE and transferred to polyvinylidene difluoride membrane (Millipore). The membranes were incubated in 5% nonfat milk powder diluted in PBS containing 0.1% Tween-20 for 2 hours at room temperature and probed with a mouse monoclonal anti-C1GALT1 antibody (Santa Cruz Biotechnology) in blocking buffer overnight at 4°C. Finally, membranes were incubated with secondary antibody of horseradish peroxidase conjugated goat anti-mouse IgG (BioRad). Immunocomplexes were detected with enhanced chemiluminescence method (GE Healthcare). The same membranes were stripped and reprobed with anti–β-actin monoclonal antibody (Sigma).
Images of autoradiography were acquired using a scanner EPSON Perfection 2580 Photo (EPSON) and quantified by Image J 1.34 Software (http://rsb.info.nih.gov/ij). The ratio between intensities of C1GALT1 and β-actin bands was used to normalize C1GALT1 expression in each sample.
Determination of Serum IgA
IgA content in serum from each participant was measured in duplicate using ELISA. Briefly, high-adsorption polystyrene 96-microwell plates (Corning Inc.) were coated overnight with 5 μg of F(ab′)2 fragment goat anti-human IgA antibody per ml (Jackson ImmunoResearch Laboratories) in PBS at 4°C. Plates were blocked with 1% BSA in PBS containing 0.05% Tween-20 for 90 minutes at room temperature. Samples diluted in blocking buffer and standard human IgA (Calbiochem) were added to each well and then incubated for 90 minutes at room temperature. The captured IgA was then detected with biotin-labeled F(ab′)2 fragment of goat IgG anti-human IgA (Biosource). The binding was measured after addition of avidin–horseradish peroxidase conjugate (ExtrAvidin; Sigma-Aldrich), and the reaction was developed with the peroxidase chromogenic substrate o-phenylenediamine–H2O2 (Sigma-Aldrich). The color reaction was stopped with 2 N H2SO4, and the OD at 490 nm was determined in a microplate reader (GDV, Italy; model DV 990 B/V6). IgA1 concentration in unknown samples was determined by interpolation of the respective OD into the appropriate standard curve.
Detection of Gal-deficient IgA1
Gal-deficient IgA1 was detected by binding of the lectin, HAA (Sigma-Aldrich), which is specific for terminal GalNAc, as reported elsewhere.48,49 High-adsorption polystyrene 96-microwell plates (Corning Inc.) were coated overnight with 3 μg of F(ab′)2 fragment goat anti-human IgA antibody per ml (Jackson ImmunoResearch Laboratories) in PBS at 4°C. Plates were blocked for 3 hours at room temperature with 1% BSA/PBS containing 0.1% Tween-20. Samples diluted in blocking buffer were added to each well and incubated overnight at 4°C. The captured IgA was subsequently desialylated by treatment for 3 hours at 37°C with 20 mU/ml neuraminidase from Vibrio cholerae (Sigma-Aldrich) in 10 mM sodium acetate buffer (pH 5).7 Samples were then incubated for 3 hours at 37°C with 2 μg of GalNAc-specific biotinylated HAA lectin per ml (Sigma-Aldrich) diluted in blocking buffer. The lectin binding was detected with avidin–horseradish peroxidase conjugate (ExtrAvidin) diluted in blocking buffer, and the reaction was developed with the peroxidase chromogenic substrate o-phenylenediamine-H2O2 (Sigma-Aldrich). The color reaction was stopped with 2 N H2SO4, and the OD at 490 nm was determined in a microplate reader (GDV, Italy; model DV 990 B/V6).
The relative lectin binding per unit IgA1 was calculated as the OD value of lectin over the OD density value of IgA concentration.
Disclosures
An Italian patent entitled “Method and kit for the diagnosis of IgA nephropathy” (patent no. MI2010A002007) was issued on October 28, 2010, to the University of Bari.
Acknowledgments
We are grateful to the patients with IgAN and healthy blood donor participants for their cooperation in this study. We express our appreciation to Dr. Gabriella Aceto and Dr. Anna Rita Cappiello for help with collecting clinical samples from patients with Henoch-Schönlein purpura nephritis.
This work was supported by grants from the MiUR (COFIN-PRIN 2006069815) and Regione Puglia (BISIMANE project, H31D08000030007).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2011060567/-/DCSupplemental.
See related editorial, “microRNA-Induced IgA Nephropathy,” on pages 765–766.
References
- 1.Radford MG, Jr, Donadio JV, Jr, Bergstralh EJ, Grande JP: Predicting renal outcome in IgA nephropathy. J Am Soc Nephrol 8: 199–207, 1997 [DOI] [PubMed] [Google Scholar]
- 2.Schena FP, Coppo R: IgA nephropathies. In: Oxford Textbook of Clinical Nephrology, 3rd ed., edited by Davison AM, Cameron JS, Grünfeld JP, Ponticelli C, Ritz E, Winearls CG, van T Persele C, Oxford, Oxford University Press, 2005, pp 469–501 [Google Scholar]
- 3.Conley ME, Cooper MD, Michael AF: Selective deposition of immunoglobulin A1 in immunoglobulin A nephropathy, anaphylactoid purpura nephritis, and systemic lupus erythematosus. J Clin Invest 66: 1432–1436, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barratt J, Feehally J: IgA nephropathy. J Am Soc Nephrol 16: 2088–2097, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Coppo R, Amore A: Aberrant glycosylation in IgA nephropathy (IgAN). Kidney Int 65: 1544–1547, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Novak J, Julian BA, Tomana M, Mesteck J: Progress in molecular and genetic studies of IgA nephropathy. J Clin Immunol 21: 310–327, 2001 [DOI] [PubMed] [Google Scholar]
- 7.Kokubo T, Hiki Y, Iwase H, Tanaka A, Toma K, Hotta K, Kobayashi Y: Protective role of IgA1 glycans against IgA1 self-aggregation and adhesion to extracellular matrix proteins. J Am Soc Nephrol 9: 2048–2054, 1998 [DOI] [PubMed] [Google Scholar]
- 8.Tomana M, Matousovic K, Julian BA, Radl J, Konecny K, Mestecky J: Galactose-deficient IgA1 in sera of IgA nephropathy patients is present in complexes with IgG. Kidney Int 52: 509–516, 1997 [DOI] [PubMed] [Google Scholar]
- 9.Tomana M, Novak J, Julian BA, Matousovic K, Konecny K, Mestecky J: Circulating immune complexes in IgA nephropathy consist of IgA1 with galactose-deficient hinge region and antiglycan antibodies. J Clin Invest 104: 73–81, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mestecky J, Tomana M, Crowley-Nowick PA, Moldoveanu Z, Julian BA, Jackson S: Defective galactosylation and clearance of IgA1 molecules as a possible etiopathogenic factor in IgA nephropathy. Contrib Nephrol 104: 172–182, 1993 [DOI] [PubMed] [Google Scholar]
- 11.Suzuki H, Moldoveanu Z, Hall S, Brown R, Vu HL, Novak L, Julian BA, Tomana M, Wyatt RJ, Edberg JC, Alarcón GS, Kimberly RP, Tomino Y, Mestecky J, Novak J: IgA1-secreting cell lines from patients with IgA nephropathy produce aberrantly glycosylated IgA1. J Clin Invest 118: 629–639, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eijgenraam JW, van Kooten C: IgA1 glycosylation in IgA nephropathy: As sweet as it can be. Kidney Int 73: 1106–1108, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Buck KS, Smith AC, Molyneux K, El-Barbary H, Feehally J, Barratt J: B-cell O-galactosyltransferase activity, and expression of O-glycosylation genes in bone marrow in IgA nephropathy. Kidney Int 73: 1128–1136, 2008 [DOI] [PubMed] [Google Scholar]
- 14.Qin W, Zhou Q, Yang LC, Li Z, Su BH, Luo H, Fan JM: Peripheral B lymphocyte beta1,3-galactosyltransferase and chaperone expression in immunoglobulin A nephropathy. J Intern Med 258: 467–477, 2005 [DOI] [PubMed] [Google Scholar]
- 15.Beerman I, Novak J, Wyatt RJ, Julian BA, Gharavi AG: The genetics of IgA nephropathy. Nat Clin Pract Nephrol 3: 325–338, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Lai AS, Lai KN: Molecular basis of IgA nephropathy. Curr Mol Med 5: 475–487, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Cox SN, Sallustio F, Serino G, Pontrelli P, Verrienti R, Pesce F, Torres DD, Ancona N, Stifanelli P, Zaza G, Schena FP: Altered modulation of WNT-beta-catenin and PI3K/Akt pathways in IgA nephropathy. Kidney Int 78: 396–407, 2010 [DOI] [PubMed] [Google Scholar]
- 18.Kudo T, Iwai T, Kubota T, Iwasaki H, Takayma Y, Hiruma T, Inaba N, Zhang Y, Gotoh M, Togayachi A, Narimatsu H: Molecular cloning and characterization of a novel UDP-Gal:GalNAc(alpha) peptide beta 1,3-galactosyltransferase (C1Gal-T2), an enzyme synthesizing a core 1 structure of O-glycan. J Biol Chem 277: 47724–47731, 2002 [DOI] [PubMed] [Google Scholar]
- 19.Kato M, Arce L, Natarajan R: MicroRNAs and their role in progressive kidney diseases. Clin J Am Soc Nephrol 4: 1255–1266, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barratt J, Smith AC, Molyneux K, Feehally J: Immunopathogenesis of IgAN. Semin Immunopathol 29: 427–443, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Pirulli D, Crovella S, Ulivi S, Zadro C, Bertok S, Rendine S, Scolari F, Foramitti M, Ravani P, Roccatello D, Savoldi S, Cerullo G, Lanzilotta SG, Bisceglia L, Zelante L, Floege J, Alexopoulos E, Kirmizis D, Ghiggeri GM, Frascà G, Schena FP, Amoroso A, European IgAN Consortium : Genetic variant of C1GalT1 contributes to the susceptibility to IgA nephropathy. J Nephrol 22: 152–159, 2009 [PubMed] [Google Scholar]
- 22.Li GS, Zhang H, Lv JC, Shen Y, Wang HY: Variants of C1GALT1 gene are associated with the genetic susceptibility to IgA nephropathy. Kidney Int 71: 448–453, 2007 [DOI] [PubMed] [Google Scholar]
- 23.Ju T, Cummings RD: A unique molecular chaperone Cosmc required for activity of the mammalian core 1 β 3-galactosyltransferase. Proc Natl Acad Sci USA 99: 16613–16618, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada K, Kobayashi N, Ikeda T, Suzuki Y, Tsuge T, Horikoshi S, Emancipator SN, Tomino Y: Down-regulation of core 1 beta1,3-galactosyltransferase and Cosmc by Th2 cytokine alters O-glycosylation of IgA1. Nephrol Dial Transplant 25: 3890–3897, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Julian BA, Waldo FB, Rifai A, Mestecky J: IgA nephropathy, the most common glomerulonephritis worldwide. A neglected disease in the United States? Am J Med 84: 129–132, 1988 [DOI] [PubMed] [Google Scholar]
- 26.Julian BA, Novak J: IgA nephropathy: An update. Curr Opin Nephrol Hypertens 13: 171–179, 2004 [DOI] [PubMed] [Google Scholar]
- 27.Allen AC, Topham PS, Harper SJ, Feehally J: Leucocyte beta 1,3 galactosyltransferase activity in IgA nephropathy. Nephrol Dial Transplant 12: 701–706, 1997 [DOI] [PubMed] [Google Scholar]
- 28.Anglicheau D, Sharma VK, Ding R, Hummel A, Snopkowski C, Dadhania D, Seshan SV, Suthanthiran M: MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci USA 106: 5330–5335, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Te JL, Dozmorov IM, Guthridge JM, Nguyen KL, Cavett JW, Kelly JA, Bruner GR, Harley JB, Ojwang JO: Identification of unique microRNA signature associated with lupus nephritis. PLoS ONE 5: e10344, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Godwin JG, Ge X, Stephan K, Jurisch A, Tullius SG, Iacomini J: Identification of a microRNA signature of renal ischemia reperfusion injury. Proc Natl Acad Sci USA 107: 14339–14344, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, Szeto CC: Intrarenal expression of microRNAs in patients with IgA nephropathy. Lab Invest 90: 98–103, 2010 [DOI] [PubMed] [Google Scholar]
- 32.Allen AC, Harper SJ, Feehally J: Galactosylation of N- and O-linked carbohydrate moieties of IgA1 and IgG in IgA nephropathy. Clin Exp Immunol 100: 470–474, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen AC, Bailey EM, Brenchley PE, Buck KS, Barratt J, Feehally J: Mesangial IgA1 in IgA nephropathy exhibits aberrant O-glycosylation: observations in three patients. Kidney Int 60: 969–973, 2001 [DOI] [PubMed] [Google Scholar]
- 34.Hiki Y, Odani H, Takahashi M, Yasuda Y, Nishimoto A, Iwase H, Shinzato T, Kobayashi Y, Maeda K: Mass spectrometry proves under-O-glycosylation of glomerular IgA1 in IgA nephropathy. Kidney Int 59: 1077–1085, 2001 [DOI] [PubMed] [Google Scholar]
- 35.Roccatello D, Picciotto G, Torchio M, Ropolo R, Ferro M, Franceschini R, Quattrocchio G, Cacace G, Coppo R, Sena LM, et al. : Removal systems of immunoglobulin A and immunoglobulin A containing complexes in IgA nephropathy and cirrhosis patients. The role of asialoglycoprotein receptors. Lab Invest 69: 714–723, 1993 [PubMed] [Google Scholar]
- 36.Novak J, Tomana M, Matousovic K, Brown R, Hall S, Novak L, Julian BA, Wyatt RJ, Mestecky J: IgA1-containing immune complexes in IgA nephropathy differentially affect proliferation of mesangial cells. Kidney Int 67: 504–513, 2005 [DOI] [PubMed] [Google Scholar]
- 37.Moura IC, Arcos-Fajardo M, Sadaka C, Leroy V, Benhamou M, Novak J, Vrtovsnik F, Haddad E, Chintalacharuvu KR, Monteiro RC: Glycosylation and size of IgA1 are essential for interaction with mesangial transferrin receptor in IgA nephropathy. J Am Soc Nephrol 15: 622–634, 2004 [DOI] [PubMed] [Google Scholar]
- 38.Moura IC, Arcos-Fajardo M, Gdoura A, Leroy V, Sadaka C, Mahlaoui N, Lepelletier Y, Vrtovsnik F, Haddad E, Benhamou M, Monteiro RC: Engagement of transferrin receptor by polymeric IgA1: Evidence for a positive feedback loop involving increased receptor expression and mesangial cell proliferation in IgA nephropathy. J Am Soc Nephrol 16: 2667–2676, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Peppard JV, Kaetzel CS, Russell MW: Phylogeny and comparative physiology of IgA. In: Mucosal Immunology, 3rd ed., edited by Mestecky J, Bienenstock J, Lamm, ME, Mayer L, McGhee JR, Strober W, Academic Press, Burlington, MA, 2005, pp 195–210 [Google Scholar]
- 40.Mestecky J, Moro I, Kerr MA, Woof JM: Mucosal immunoglobulins. In: Mucosal Immunology, 3rd ed., edited by Mestecky J, Bienenstock J, Lamm, ME, Mayer L, McGhee JR, Strober W, Academic Press, Burlington, MA, 2005, pp 153–181 [Google Scholar]
- 41.Manno C, Strippoli GF, D’Altri C, Torres D, Rossini M, Schena FP: A novel simpler histological classification for renal survival in IgA nephropathy: A retrospective study. Am J Kidney Dis 49: 763–775, 2007 [DOI] [PubMed] [Google Scholar]
- 42.Storey JD, Tibshirani R: Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ: miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34[Database issue]: D140–D144, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedman RC, Farh KK, Burge CB, Bartel DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N: Combinatorial microRNA target predictions. Nat Genet 37: 495–500, 2005 [DOI] [PubMed] [Google Scholar]
- 46.Barenboim M, Zoltick BJ, Guo Y, Weinberger DR: MicroSNiPer: A web tool for prediction of SNP effects on putative microRNA targets. Hum Mutat 31: 1223–1232, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinitz M, Klein G: EBV-transformation of surface IgA-positive human lymphocytes. J Immunol 125: 194–196, 1980 [PubMed] [Google Scholar]
- 48.Moore JS, Kulhavy R, Tomana M, Moldoveanu Z, Suzuki H, Brown R, Hall S, Kilian M, Poulsen K, Mestecky J, Julian BA, Novak J: Reactivities of N-acetylgalactosamine-specific lectins with human IgA1 proteins. Mol Immunol 44: 2598–2604, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moldoveanu Z, Wyatt RJ, Lee JY, Tomana M, Julian BA, Mestecky J, Huang WQ, Anreddy SR, Hall S, Hastings MC, Lau KK, Cook WJ, Novak J: Patients with IgA nephropathy have increased serum galactose-deficient IgA1 levels. Kidney Int 71: 1148–1154, 2007 [DOI] [PubMed] [Google Scholar]