Abstract
Little is known regarding the molecular phenotype of kidneys with AKI because biopsies are performed infrequently. However, all kidney transplants experience acute injury, making early kidney transplants an excellent model of acute injury, provided the absence of rejection, because donor kidneys should not have CKD, post-transplant biopsies occur relatively frequently, and follow-up is excellent typically. Here, we used histopathology and microarrays to compare indication biopsies from 26 transplants with acute injury with 11 pristine protocol biopsies of stable transplants. Kidneys with acute injury showed increased expression of 394 transcripts associated with the repair response to injury, including many epithelium-like injury molecules tissue, remodeling molecules, and inflammation molecules. Many other genes also predicted the phenotype, including the acute injury biomarkers HAVCR1 and IL18. Pathway analysis of the injury-repair transcripts revealed similarities to cancer, development, and cell movement. The injury-repair transcript score in kidneys with acute injury correlated with reduced graft function, future renal recovery, brain death, and need for dialysis, but not with future graft loss. In contrast, histologic features of acute tubular injury did not correlate with function or with the molecular changes. Thus, the transcripts associated with repair of injury suggest a massive coordinated response of the kidney parenchyma to acute injury, providing both an objective measure for assessing the severity of injury in kidney biopsies and validation for many biomarkers of AKI.
Although AKI is an important problem,1–3 native kidneys with AKI are rarely biopsied, limiting studies of AKI biomarkers to urine and body fluids.4 However, kidney transplants offer a unique opportunity to study the injury-repair response to AKI because all transplants experience AKI, CKD is excluded, and numerous indication biopsies are performed to exclude rejection, guided by the international Banff histopathology consensus system. Moreover, transplants have detailed function assessments and are followed indefinitely. AKI induced by transplantation can be modeled in rodent kidney isografts,5–7 as recently reviewed.8 The mouse studies showed that some transcript changes in transplants reflect AKI, but other transcripts reflect the acute phase response to the surgical procedure.5 The picture is further complicated in aging kidneys.9,10
This prospective study of kidney transplants with AKI was undertaken to define the transcripts induced by human AKI and their clinical correlations. We previously analyzed the correlates of estimated GFR (eGFR) in transplant biopsies, but those were mainly late biopsies with atrophy-fibrosis, not AKI.11 We hypothesized that indication biopsies from early kidney transplants, selected to exclude kidneys with rejection and diseases, will reveal the molecular features of AKI. Our strategy was to neutralize the ubiquitous minor changes inevitably induced by the transplant process by comparing AKI kidneys to histologically pristine protocol biopsies of stable transplants. This study aimed to elucidate the molecular changes that reflect clinically significant functional disturbance and their relationship with histopathology changes and prognosis.
Results
Selection of Biopsies from Kidney Transplants with Pure AKI
We identified AKI kidneys from a prospective study of 234 kidney transplant biopsies for clinical indications from 173 consenting patients. We excluded kidneys with rejection and kidney disease by histologic criteria, and also excluded those with nondiagnostic suspicious histologic lesions. This identified a “pure AKI” cohort of 28 biopsies from 26 patients, with mean eGFR at biopsy of 26 ml/min (range, 8–52) (Table 1). Of 28 biopsies, 21 displayed features that pathologists believe reflect acute tubular injury.12,13 Fifteen were from deceased donor kidneys, of which eight manifested delayed graft function (DGF) (i.e., initial dialysis dependency). The median follow-up after biopsy was 3.9 years. The extent of histologic tubule injury changes can be assessed for its correlation with molecular changes and function because these kidneys were not chosen on the basis of histologic criteria for tubular injury; rather, they were chosed only by time post-transplant and exclusion of diseases.
Table 1.
Demographics of patients selected for early AKI from biopsies for clinical indications and stable early transplants selected from protocol biopsies
| Demographics | Pure AKI Cohort | Pristine Protocol Biopsy Cohort | Validation Set | P Value (Pure AKI versus Pristine Protocol Biopsies) | P Value (Validation Set versus Pristine Protocol Biopsies) |
|---|---|---|---|---|---|
| Patient characteristics at diagnosis | |||||
| number of patients | 26 | 11 | 27 | ||
| mean recipient age (yr) | 52 (16–75) | 49 (29–70) | 49 (21–68) | >0.05 | >0.05 |
| primary disease | |||||
| GN | 9 (32) | 2 (18.2) | 3 (11) | >0.05 | >0.05 |
| diabetic nephropathy | 4 (14) | 1 (9.1) | 10 (37) | >0.05 | >0.05 |
| others | 9 (32) | 4 (36.4) | 14 (52) | >0.05 | >0.05 |
| unknown etiology | 6 (21) | 4 (36.4) | 13 (48) | >0.05 | >0.05 |
| mean donor age (yr) | 50 (22–69) | 53 (20–82) | 43 (16–59) | >0.05 | >0.05 |
| mean donor age (living donor) | 46 (37–56) | 44 (35–52) | 44 (16–59) | >0.05 | >0.05 |
| mean donor age (deceased donor) | 53 (22–69) | 55 (20–82) | 42 (26–56) | >0.05 | >0.05 |
| donor type deceased/living (% deceased donor transplants) | 15/ 11 (57.6) | 9/2 (81.8) | 8/19 (30) | >0.05 | 0.004 |
| delayed graft function rate | 8/26(31) | 2/11 (18) | 5/27 (19) | >0.05 | >0.05 |
| median time from transplant to biopsy (d) | 16 (6–42) | 42 | 24 (7–42) | <0.001 | <0.001 |
| median duration follow-up postbiopsy (d) | 1414 (98–2155) | 2000 (732–2932) | 727 (1–2146) | 0.006 | <0.001 |
| allograft failure | 2/26 (7.7)a | 1/11 (9.1)b | 2/27 (7.4)c | >0.05 | >0.05 |
| Clinical characteristics at time of biopsy | |||||
| number of biopsies | 28 | 11 | 28 | ||
| indication for biopsy as indicated by clinician | |||||
| concerns about renal function | 20 (71) | (protocol) | 25 (89) | ||
| other or unknown | 8 (29) | (protocol) | 3 (11) | ||
| histologic diagnosis | |||||
| acute tubular injury (≥5% acute tubular injury score) | 21 (75) | 0 (0) | 18 (64) | <0.001 | <0.001 |
| no major abnormalities | 7 (25) | 11 (100) | 10 (36) | <0.001 | <0.001 |
| mean eGFRd at the time of biopsy | 26 (8–52) | 49 (23–89) | 31 (10–46) | <0.001 | <0.001 |
| immunosuppressive drug regimen | |||||
| MMF, tacrolimus, steroids | 11 (39.3) | 0 (0) | 5 (18) | 0.017 | >0.05 |
| MMF, tacrolimus | 8 (28.6) | 0 (0) | 7 (25) | >0.05 | >0.05 |
| MMF, cyclosporine, steroids | 3 (10.7) | 8 (72.7) | 7 (25) | <0.001 | 0.010 |
| other | 6 (21.4) | 3 (27.3) | 9 (32) | >0.05 | >0.05 |
Data are shown as n (%) or median (range) unless otherwise indicated. MMF, mycophenolate mofetil.
The cause of failure: death with functioning graft, and ABMR in a nonadherent patient 30 months post-transplant.
The cause of failure: death with functioning graft.
The cause of failure: death with functioning graft, and membranoproliferative GN at 60 months post-transplant.
eGFR obtained using the four-variable MDRD formula (in milliliters per minute per 1.73 m2).
During follow-up, only one kidney from the AKI cohort progressed to end stage kidney failure at 30 months, attributed to antibody-mediated rejection (ABMR) caused by nonadherence, and one patient died with a functioning graft.
A validation set was generated to test the molecular features (Table 1). The results below refer exclusively to the original AKI cohort, and the validation set is analyzed at the end of the Results section.
Selection of Controls
Our strategy was to distinguish clinically significant AKI from the minor injury that accompanies every transplant. Thus, controls consisted of protocol kidney transplant biopsies selected from a previously published cohort14 with clinically appropriate stable function at 6 weeks post-transplant, with no indications for biopsy. After excluding biopsies with any histologic abnormalities, including acute tubular injury changes (Table 1), we were left with a set of 11 pristine protocol biopsies similar in age to the AKI kidneys, including both living and deceased donor kidneys.
Histopathology, Function, and Survival of Kidneys with AKI
Histology scores for inflammation (i-Banff, i-total) and atrophy fibrosis (ci, ct) were compared for biopsies from the AKI cohort versus those with T cell–mediated rejection (TCMR), borderline rejection, other diseases and pristine protocol biopsies (Table 2). Because of the selection criteria, the pure AKI biopsies and pristine protocol biopsies had little inflammation or atrophy fibrosis, and the pristine protocol biopsies had no acute tubular injury features.
Table 2.
Histology and kidney function in 28 AKI kidney biopsies compared with 206 other transplant biopsies for clinical indications
| Feature | Pure AKI (n=28) | TCMR (n=35) | Borderline (n=23) | Remaining Biopsies (n=148)a | Pristine Protocol Biopsies (n=11) |
|---|---|---|---|---|---|
| Histologic lesions (continuous scores, % of cortex) | |||||
| inflammation (i-Banff) | 4 | 26b | 8.9 | 7.9 | 3.9 |
| inflammation (i-total) | 7 | 45b | 28b | 23b | 3.9 |
| scarring (ci) | 7 | 24c | 27b | 22b | 0b |
| atrophy (ct) | 6 | 23b | 26b | 22b | 0.5c |
| Grafts failed during follow-up (excluding death with function)d | 1 | 3 | 1 | 34 | 0 |
| Median follow-up time after first biopsy (yr) | 3.9 | 4.0 | 4.2 | 3.8 | 6.3 |
| Renal function (eGFR, ml/min) | |||||
| eGFR at biopsy | 26 | 33 | 34 | 35c | 49c |
| eGFR at 6 mo | 51e | 42e | 40 | 38c | 55 |
| ΔeGFR by 6 mo | 25 | 9f | 5b | 2b | 6b |
AKI biopsies were compared with the remaining biopsy groups. Kruskal–Wallis test with Dunn’s correction for multiple comparisons.
Remaining biopsies included diagnoses of ABMR, mixed ABMR/TCMR, GN, polyoma virus, and other nonrejecting biopsies.
P<0.001.
P<0.01.
Failed grafts are represented by one biopsy per patient.
Indicates significant difference between eGFR at biopsy and 6-month eGFR, by the paired t test.
P<0.05.
Three of 35 grafts with TCMR failed, but all had later biopsies showing ABMR or mixed rejection. The 34 other kidney failures after indication biopsies reflected diseases that emerged late after transplantation such as ABMR or recurrent GN.15,16
The mean eGFR at the time of biopsy was low in the AKI cohort (mean 26) and in TCMR and borderline rejection biopsies, compared with other indication biopsies (Table 2). However, by 6 months after biopsy, these kidneys showed the most improvement (ΔeGFR). As expected, the pristine protocol biopsy cohort had higher mean eGFR at the time of biopsy than the AKI cohort, and showed little improvement over time.
Kidneys with pure AKI in the early post-transplant period manifest impaired function at biopsy but good recovery and prognosis, and are suitable for defining the injury-repair response to clinically significant AKI.
Defining the Injury-Repair Response Associated Transcripts in the Pure AKI Biopsies
We compared microarray results from AKI biopsies versus pristine protocol biopsies to define the features of clinically significant injury. This comparison identified 946 probe sets, including 394 nonredundant genes whose expression was increased at least ≥1.5-fold higher in AKI than in pristine protocol biopsies (Supplemental Table 1), at the false discovery rate (FDR) of 0.05. We named these injury-repair response–associated transcripts (IRRATs).
For detailed study, we selected from the 394 IRRATs the 30 with the highest fold increase compared with the pristine protocol biopsies (Table 3). The expression of the individual top 30 IRRATs in AKI biopsies was 2.7- to 7.9-fold higher than in pristine protocol biopsies. Four of the top 30 IRRATs (LCN2, AKAP12, FOS, and EGR1) had previously been identified as biomarkers of AKI, on the basis of detection of the product in urine or blood,17–19 or mRNA expression in human kidney.20,21 The complete set of 394 IRRATs included four other known AKI biomarkers (CLU, CXCL1, CYR61, and THSB1) but did not include some known biomarkers such as HAVCR1 and IL18.
Table 3.
Top 30 IRRATs ordered by fold change in AKI biopsies versus pristine protocol biopsies
| Gene Symbol | Gene Name | Fold Change AKI versus Protocol Biopsies | P Value | Raw Signal in Protocol Biopsies | Correlation with eGFR at Biopsy in AKI | Correlation with ΔGFR at 6 Mo in AKI |
|---|---|---|---|---|---|---|
| ITGB6 | Integrin β6 | 7.9 | 0.001 | 72 | −0.69 | 0.54 |
| SERPINA3 | Serine (or cysteine) proteinase inhibitor, clade A | 6.1 | 0.001 | 252 | −0.71 | 0.47 |
| MTND6 | NADH dehydrogenase, subunit 6 (complex I) | 5.6 | 0.003 | 67 | −0.51 | 0.43 |
| OLFM4 | Olfactomedin 4 | 5.5 | 0.001 | 53 | −0.74 | 0.46 |
| PTX3 | Pentaxin-related gene, rapidly induced by IL-1β | 5.3 | 0.002 | 15 | −0.58 | 0.52 |
| LCN2a | lipocalin 2 (oncogene 24p3) | 4.3 | 0.004 | 89 | −0.57 | 0.50 |
| VCAN | chondroitin sulfate proteoglycan 2 (versican) | 4.1 | 0.004 | 47 | −0.35b | 0.38 |
| LTF | Lactotransferrin | 4.1 | 0.032 | 441 | −0.63 | 0.54 |
| SLPI | Secretory leukocyte protease inhibitor (antileukoproteinase) | 4.1 | 0.013 | 334 | −0.70 | 0.52 |
| ADAMTS1 | A disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 1 | 4.0 | 0.000 | 110 | −0.64 | 0.52 |
| MEGF11 | Multiple EGF-like domains | 3.8 | 0.001 | 12 | −0.68 | 0.59 |
| CDH6 | cadherin 6, type 2, K-cadherin (fetal kidney) | 3.7 | 0.000 | 105 | −0.72 | 0.56 |
| PI15 | Peptidase inhibitor 15 | 3.6 | 0.008 | 17 | −0.40 | 0.23b |
| FCGR3A | Fc fragment of IgG, low affinity IIIa, receptor for (CD16) | 3.4 | 0.002 | 41 | −0.30b | 0.30b |
| NNMT | Nicotinamide N-methyltransferase | 3.3 | 0.005 | 155 | −0.56 | 0.52 |
| S100A8 | S100 calcium binding protein A8 (calgranulin A) | 3.2 | 0.004 | 175 | −0.11b | 0.07b |
| SOD2 | SOD2, mitochondrial | 3.2 | 0.001 | 264 | −0.61 | 0.57 |
| ITGB3 | Integrin β3 (platelet glycoprotein IIIa, antigen CD61) | 3.0 | 0.006 | 45 | −0.74 | 0.71 |
| NFKBIZ | NF of κ light polypeptide gene enhancer in B cells inhibitor, ζ | 3.0 | 0.004 | 141 | −0.67 | 0.59 |
| AKAP12a | A kinase (PRKA) anchor protein (gravin) 12 | 3.0 | 0.007 | 57 | −0.60 | 0.32b |
| VMP1 | Likely ortholog of rat vacuole membrane protein 1 | 2.9 | 0.003 | 375 | −0.71 | 0.59 |
| ADAM9 | A disintegrin and metalloproteinase domain 9 (meltrin γ) | 2.9 | 0.009 | 103 | −0.57 | 0.41 |
| PTPRC | Protein tyrosine phosphatase, receptor type, C | 2.9 | 0.002 | 56 | −0.53 | 0.37b |
| FOSa | V-fos FBJ murine osteosarcoma viral oncogene homolog | 2.9 | 0.000 | 29 | −0.48 | 0.49 |
| OSMR | Oncostatin M receptor | 2.8 | 0.001 | 65 | −0.80 | 0.73 |
| C9orf71 | Chromosome 9 open reading frame 71 | 2.8 | 0.005 | 283 | −0.41 | 0.38 |
| EGR1a | Early growth response 1 | 2.8 | 0.009 | 184 | −0.48 | 0.56 |
| CTSS | Cathepsin S | 2.7 | 0.021 | 190 | −0.50 | 0.43 |
| RARRES1 | Retinoic acid receptor responder (tazarotene induced) 1 | 2.7 | 0.025 | 76 | −0.55 | 0.54 |
| VI2A | Ecotropic viral integration site 2A | 2.7 | 0.008 | 59 | −0.32b | 0.25b |
Nonredundant and annotated genes are shown. In case of multiple transcripts per gene, the one with highest fold change was selected. Spearman correlations of gene expression with eGFR at the time of biopsy and with change in eGFR (ΔeGFR) at 6 months were considered significant at P<0.05.
Prototypical markers of kidney injury.
P value not significant (P>0.05).
Although the IRRATs were defined without reference to GFR measurements, they correlated strongly with eGFR. Thus, expression of 26 of the top 30 individual IRRATs correlated with reduced eGFR at biopsy, and 24 of 30 also correlated with eGFR recovery 6 months after biopsy (Table 3). After excluding the top 30 IRRATs, there were 138 additional IRRATs whose expression correlated with eGFR, with correlation coefficients ranging from −0.82 to −0.50 (Table 4).
Table 4.
Examples of other IRRATs anticorrelating with eGFR at the time of biopsy in AKI biopsies
| Gene Symbol | Gene Title | Correlation with eGFR |
|---|---|---|
| TRIM38 | Tripartite motif-containing 38 | −0.82 |
| CLDN1 | Claudin 1 | −0.78 |
| CDC42SE2 | CDC42 small effector 2 | −0.75 |
| ICAM1 | Intercellular adhesion molecule 1 | −0.73 |
| IFITM2 | IFN induced transmembrane protein 2 (1-8D) | −0.73 |
| C2CD4A | C2 calcium-dependent domain containing 4A | −0.73 |
| KPNA2 | Karyopherin α2 (RAG cohort 1, importin α1) | −0.72 |
| CASP1 | Caspase 1, apoptosis-related cysteine peptidase (IL-1, β, convertase) | −0.72 |
| RASD1 | RAS, dexamethasone-induced 1 | −0.71 |
| UGCG | UDP-glucose ceramide glucosyltransferase | −0.71 |
| TGM2 | Transglutaminase 2 (C polypeptide, protein-glutamine-γ-glutamyltransferase) | −0.71 |
| CLIC1 | Chloride intracellular channel 1 | −0.70 |
| TFPI | Tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | −0.70 |
| CASP8 | Caspase 8, apoptosis-related cysteine peptidase | −0.69 |
| SERPINA1 | Serpin peptidase inhibitor, clade A (α-1 antiproteinase, antitrypsin), member 1 | −0.69 |
| KLF6 | Kruppel-like factor 6 | −0.69 |
| AMACR///C1QTNF3 | α-methylacyl-CoA racemase///C1q and TNF-related protein 3 | −0.68 |
| TNIK | TRAF2 and NCK interacting kinase | −0.68 |
| HN1 | Hematologic and neurologic expressed 1 | −0.67 |
| MT1M | Metallothionein 1M | −0.67 |
| PTP4A1 | Protein tyrosine phosphatase type IVA, member 1 | −0.67 |
| NFKBIZ | NF of κ light polypeptide gene enhancer in B cells inhibitor, ζ | −0.67 |
| SEC23IP | SEC23 interacting protein | −0.67 |
| GBP2 | Guanylate binding protein 2, IFN-inducible | −0.67 |
| MCL1 | Myeloid cell leukemia sequence 1 (BCL2-related) | −0.66 |
| MET | Met proto-oncogene (hepatocyte growth factor receptor) | −0.66 |
| ABCC4 | ATP-binding cassette, subfamily C (CFTR/MRP), member 4 | −0.66 |
| SFN | Stratifin | −0.66 |
| SOCS3 | Suppressor of cytokine signaling 3 | −0.66 |
| CPD | Carboxypeptidase D | −0.66 |
Genes previously identified as top 30 IRRATs by the fold change in AKI biopsies versus pristine PBx were excluded because their correlations with eGFR are shown in Table 3.
Mouse orthologs of the 394 IRRATs had extensive overlap with transcripts previously shown to be increased in mouse AKI, either isografts (n=124, P<0.001) or native kidneys with ischemic ATN (n=254, P<0.0001).5 Of 18 mouse orthologs of the top 30 human IRRATs, 17 were increased in mouse AKI models5 (Table 5). Six of those belonged to the previously published injury-repair induced sets IRIT(I) and IRIT(L).5 Thus, IRRAT changes in AKI are largely conserved between human and mouse.
Table 5.
All 18 identified mouse orthologs of the top 30 IRRATs show elevated expression in mouse AKI models (isografts and/or ATN)
| Gene Symbol | Isografts and ATN | Isografts Only | ATN Only | IRIT (I) | IRIT (L) |
|---|---|---|---|---|---|
| Adamts1 | Yes | Yes | |||
| Akap12 | Yes | Yes | |||
| Ctss | Yes | Yes | |||
| Egr1 | Yes | ||||
| Evi2a | Yes | ||||
| Itgb3 | Yes | ||||
| Fos | Yes | ||||
| Fcgr4 | Yes | ||||
| Olfm4 | Yes | ||||
| Ltf | Yes | Yes | |||
| Lcn2 | Yes | ||||
| Nnmt | Yes | ||||
| Osmr | Yes | Yes | |||
| Ptprc | Yes | ||||
| S100a8 | Yes | ||||
| Serpina3n | Yes | Yes | |||
| Sod2 | Yes | ||||
| Vcan | Yes | ||||
| Havcr1a | Yes | Yes | |||
| Il18a | No | No | |||
AKI biomarkers that are absent in the human IRRAT set.
Relationship of the IRRAT Score to Histology and Function in Injured Transplants
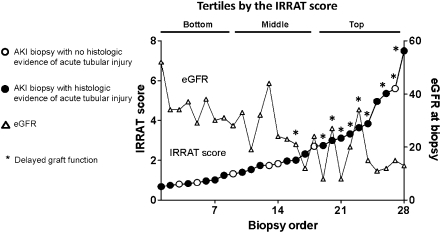
The 28 AKI biopsies were arranged by increasing IRRAT score (geometric mean of the fold increase in the top 30 IRRATs versus control nephrectomies) to compare the IRRAT scores with DGF, eGFR, and histopathology features of acute tubular injury (Figure 1). We discretized the IRRAT scores into tertiles (see top of Figure 1). Nine of 10 kidneys with DGF had IRRAT scores in the top tertile (P<0.05). The top tertile also contained nine biopsies from deceased donors (P<0.05). Kidneys from deceased donors had higher IRRAT scores than kidneys from living donors (3.0 versus 1.6, P<0.05).
Figure 1.
Relationship between IRRAT scores, histopathology, delayed graft function, and eGFR at the time of biopsy for clinical indication in kidneys with AKI. Biopsies (open circles) are ordered by the IRRAT score, and the graph illustrates the relationship of IRRAT scores to the histologic assessment of tubular injury (filled circles), delayed graft function (*), and eGFR (triangles).
Whereas high IRRAT scores were associated with low eGFR, acute tubular injury features were not; they were present in 6 of 9 in the lowest tertile, 6 of 9 in the middle tertile, and 9 of 10 in the upper tertile (not significant). Histologic features of acute tubular injury were also not associated with DGF or donor type. However, inflammation in AKI biopsies, albeit mild by definition, was nevertheless highest in the top tertile: the mean inflammation scores (i-total) were 2% for the lowest tertile, 5% for the middle tertile, and 13% for the top tertile (P<0.05).
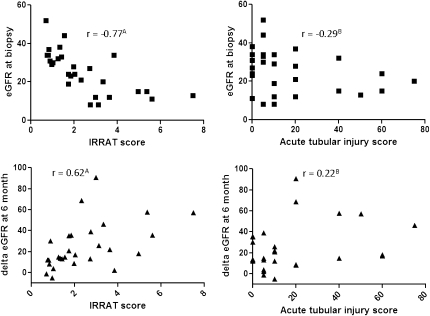
The IRRAT score in the AKI biopsies anticorrelated with the eGFR at biopsy (−0.77, P<0.001) and correlated with the increase in eGFR by 6 months (0.62, P<0.001). The acute tubular injury score by histology did not correlate with eGFR or the increase in GFR (Figure 2).
Figure 2.
Correlation of IRRAT scores (left panel) and AKI scores (right panel) with eGFR at the time of biopsy (upper panels) or change in eGFR (ΔeGFR) at 6 months after biopsy (lower panels) in biopsies for clinical indication with AKI. Values are Spearman correlation coefficients. AP<0.001; BP>0.05.
In AKI kidneys, the IRRAT score is associated with DGF, brain death, impairment in eGFR, future recovery of function, and interstitial inflammation. The histologic features that pathologists believe represent acute tubular injury were not related to DGF, deceased donation, eGFR, recovery of function, or IRRAT scores.
Cellular Expression and Functional Annotation of IRRATs
Functional annotation of the top 30 IRRATs using Ingenuity Pathway Analysis (Table 6) indicated that the pathways overrepresented included cancer; cellular movement, including migration of tumor cells; tissue development, including cell adhesion; and acute phase response. The complete list of 394 nonredundant IRRATs yielded similar results. Most IRRATs are expressed in parenchymal cells of kidney or in a broad expression in many cells, making kidney parenchymal expression likely. Relatively few were expressed in inflammatory cells.
Table 6.
Functional annotation of IRRATs in human transplants with AKI by IPA
| Annotation Tool | Function or Pathway | Number of Genes Present in All 394 IRAATs (% of All IRRATs) | Number of Genes Present in the Top 30 IRRATs (% of Top 30 IRRATs) |
|---|---|---|---|
| IPA diseases and disorders | Cancer (n=6087) | 215 (55) | 25 (83) |
| IPA molecular and cellular function | Cellular movement (n=2755) | 131 (33) | 19 (63) |
| IPA development and function | Tissue development (n=959) | 100 (25) | 11 (36) |
| IPA canonical pathway | Acute phase response signaling (n=172) | 19 (5) | 4 (13) |
All IRRATs ≥1.5-fold in AKI versus protocol biopsies (394, nonredundant) and 30 top IRRATs were used for annotation. Enrichment in Ingenuity Pathway Analysis (IPA) terms was tested by the Fisher's exact test. All enrichments were highly significant, P<0.0001, and remained significant after the multiple test correction (P values ranged from 0.023 to <0.0001).
Many Other Transcripts Correlate with IRRATs and Predict AKI
Using our statistical filtering rules, some reported biomarkers of AKI were not found in the top 30 or the complete 394 IRRAT lists, including HAVCR1 (KIM1), IL18, UMOD, CTS3, or FABP1. HAVCR1 did not meet the fold cutoff criteria, and IL18 did not pass the statistical filter. Nevertheless, HAVCR1 and IL18 expression correlated with the IRRAT score in the AKI biopsies (r=0.9 and r=0.7, P<0.0001, respectively). UMOD, CTS3, and FABP1 had no positive correlation with the IRRAT score.
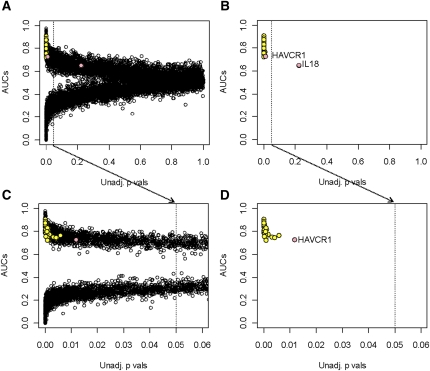
We re-examined all probe sets passing the interquartile range filter (n=12,552) to identify transcripts missed by the initial filtering rules, comparing the binary phenotype of AKI versus pristine protocol biopsies. Our goal was to identify all transcripts that differ between these phenotypes by their P value and by their ability to predict these phenotypes. In Figure 3, the x-axis gives the uncorrected P values for the comparison of phenotypes, whereas the y-axis gives the area under the curve for prediction (AUC). The transcripts positively associated with the phenotype have an AUC >0.5. If negatively associated, the AUC is <0.5. An AUC at or near 0.5 signifies no discriminatory power.
Figure 3.
Prediction of AKI by individual transcripts. (A) Comparison of AKI biopsies to pristine protocol biopsies. All transcripts that went into the analysis are shown. (B) Only the top 30 IRRATs, HAVCR1, and IL18 are shown. (C) Comparison of AKI biopsies to pristine protocol biopsies. Only transcripts significant at the false discovery rate of 0.05 are shown. (D) Vertical line indicates the uncorrected P value of 0.05. The top 30 IRRATs are represented by yellow dots, and HAVCR1 and IL18 by pink dots.
Apart from the top 30 IRRATs (shown as yellow dots), other redundant transcripts (n=1424) had high predictive values for the AKI (AUC ≥0.7). HAVCR1 and IL18 (pink dots) now showed correlations similar to many top 30 IRRATs. After adjustment for the FDR, 930 transcripts remained, including all IRRATs. Another 1785 transcripts (after the FDR adjustment) were highly negatively associated with the AKI phenotype (AUC ≤0.3).
As an additional control, we examined these relationships after random shuffling of the phenotype labels of AKI and protocol biopsies. No transcript was significant at FDR=0.05 when the labels were randomly shuffled.
Relationship between IRRATs, Histology, and Function in the Validation Set
For validation of the molecular studies, we identified a second, independent set of kidneys with AKI, selected only as being the next consecutive kidney biopsies meeting the same criteria in this ongoing study (Table 1). The validation set of 27 kidneys were similar to the original set, differing only in longer median time to biopsy (24 versus 16 days), fewer deceased donors (30% versus 58%), and fewer grafts with DGF (19% versus 31%). Because of these differences, the eGFR was higher in the validation set (31 versus 26 ml/min), reflecting the fact that the extent of AKI was less than in the original AKI set. In the validation set, only one graft was lost, due to membranoproliferative GN, 60 months after the transplant, and one patient died with a functioning graft.
The correlation of the top 30 IRRAT score in the validation set with the eGFR at biopsy was −0.62 (P<0.001) and with the increase in eGFR by 6 months was 0.45 (P<0.05). The recovery of eGFR was highest in kidneys with high IRRAT scores (Table 7). The acute tubular injury features did not correlate with IRRAT scores. Inflammation was again highest in the top tertile by the IRRAT score. Thus, the relationships among kidney function, histology, and IRRAT score expression were confirmed in the validation set biopsies.
Table 7.
Relationship of the IRRAT score to the histology and function of kidneys with AKI in the validation set
| Bottom Tertile (n=9) | Middle Tertile (n=9) | Top Tertile (n=10) | |
|---|---|---|---|
| eGFR at biopsya | 39.8b | 31.3 | 23.2 |
| eGFR at 6 mo | 47.2 | 50.5c | 47.7c |
| ΔeGFR by 6 mo | 7.4b | 20.1 | 24.9 |
| Features of ATI (yes/no) | 4/5 | 7/2 | 7/3 |
| i-total (%) | 6.3b | 2.6d | 18.8 |
| Delayed/immediate graft functione | 0/9 | 2/5 | 3/6 |
| Deceased/living donor | 2/7 | 3/5 | 3/7 |
Kruskal–Wallis test with Dunn’s correction for multiple comparisons. The top tertile was compared with the remaining tertiles. Graft function and the donor type are represented by one biopsy per patient.
eGFR and ΔeGFR are in milliliters per minute per 1.73 m2.
P<0.01.
Indicates significant difference between eGFR at biopsy and 6-month eGFR, by the paired t test.
P<0.001.
Missing data for two grafts.
The IRRAT score was higher in the original set than in the validation set (2.5 versus 1.6, P=0.02), because the kidneys from the validation set were less injured (i.e., had better eGFR and less DGF). Analysis of gene expression revealed that 19 of the top 30 IRRATs and 212 of the 394 IRRATs were significantly increased (FDR=0.05) in the validation set (Supplemental Table 1).
Discussion
Although all kidney transplants experience AKI, some are more injured and require an indication biopsy. When rejection and diseases are excluded, such kidneys constitute a pure AKI cohort with varying degrees of functional impairment. We compared this cohort to pristine protocol biopsies of similar ages, thus neutralizing the mild injury present even in well functioning transplants and identifying the IRRATs as the signal evoked in human kidneys by recent major parenchymal injury. In the AKI cohort, the IRRATs correlated with a history of DGF and with deceased donors, depression of eGFR at time of biopsy, future eGFR recovery, and interstitial inflammation. Histologic features considered typical of acute tubular injury did not correlate with these measurements or with the IRRATs. An independent validation set confirmed all of the major relationships, as well as IRRAT expression. The IRRATs extensively overlapped the transcripts previously identified in injured mouse kidneys, as well as many biomarkers previously described in body fluids. Thus, the results present a comprehensive picture of molecular events in clinically significant human transplant AKI that will be useful in supporting diagnostic applications.
The elucidation of the transcripts expressed in human transplant kidneys with AKI offers new support to and understanding of developing biomarkers, refining their interpretation, and establishing how expression of AKI biomarkers in body fluids correlates with expression in kidney tissue. Some biomarkers are more sensitive to minor injury or more persistent after injury resolves, whereas others may be more quantitative at distinguishing current functional impairment.22,23 For example, increased expression of HAVCR1 is reported in primary diseases24 and in the zero-hour transplant biopsies.25 However, in one study, HAVCR1 lacked the ability to distinguish major from minor injury states in kidney transplants.26 Moreover, the expression of biomarkers such as HAVCR1 and IL18 in body fluids may also be a sign of systemic trauma.10,27–29 The fact that two AKI, biomarkers HAVCR1 (KIM1) or IL18 were not identified as IRRATs by our initial statistical filtering strategy (fold change and FDR) yet were nevertheless associated with AKI phenotype, when re-analyzed, illustrates a caveat about reaching negative conclusions in microarray data. Arbitrary selection criteria may miss important associations when many features are highly correlated, and a comprehensive examination of the phenotype associations of all transcripts should be included.
Although IRRAT expression accurately reflected the functional disturbance in transplants with AKI, the histologic features believed to reflect acute tubular injury did not. Various focal features of acute tubular necrosis in humans have previously been reported in native kidney AKI,30 the ability of these features to predict current GFR impairment or future functional recovery has been poor, indicating that focal abnormalities are not quantitatively related to overall parenchymal function. The emergence of the IRRATs as a reliable measurement of AKI in human biopsies can be used in place of histopathology, but can also be used to guide the identification of new histologic features. For example, we recently showed using confocal microscopy that kidney transplants with AKI undetected by conventional features have a profound diffuse loss of solute carriers in the brush border.7 A systematic search for new histologic features that predict IRRAT expression and GFR is now warranted. Moreover, one histologic feature that correlates with IRRAT expression in kidney transplants with pure AKI, mild interstitial inflammation, should probably be given more weight. Interstitial inflammation is an inherent component of the injury-repair response, like the inflammation in a healing wound, and has been described in reperfusion31,32 and DGF.33
The IRRATs show considerable similarity to the transcripts induced in late kidneys with diseases that predict progression to renal failure in such kidneys and that can be used to define a risk score (e.g., ITGB6, VCAN, NNMT).34 Thus, 11 of the top 30 IRRATs were previously reported by us as frequently used by the molecular risk score classifier for predicting a graft loss in late kidney transplant biopsies.34 Although the significance of this observation is the subject of a separate study, it seems likely that the IRRATs are an important signal in a variety of renal diseases other than AKI, but their significance will reflect whether that disease is treatable or self-limited or untreatable and relentlessly progressive. In this respect, the close relationship between the top 30 IRRATs and cancer provides insights, illustrating the ubiquitous nature of the injury-repair/wound-healing response, and recalling the concept of cancers as “wounds that do not heal.”35,36 Thus, the IRRATs represent major general mechanisms such as development37,38 and cancer,37 particularly invasive39 and metastatic cancers,40 as well as fibrosis.41,42
Pure AKI in transplants did not lead to late graft failures in our analyses, either in the first AKI cohort or the validation set, consistent with recent studies indicating that most late kidney transplant failures reflect the onset of diseases such as late ABMR or recurrent disease.16,43 All of the kidneys in the AKI set and the validation set recovered, and the only two kidney failures were due to rejection triggered by nonadherence and recurrent GN. Thus, AKI evokes the injury-repair response, like wound healing, and does not lead to progressive deterioration in humans. At first glance this may seem to contradict some analyses of DGF in transplant registry data. However, DGF in registry data reflects many different disease states, including not only AKI but also technical failures and undetected ABMR.44,45 Moreover, kidneys with DGF carry the weight of many covariates, such as donor and recipient age, that increase the probability of both DGF and future failure.46 Thus, the impaired long-term survival of kidneys with DGF in registry data are likely not an effect of pure AKI, which is always undesirable but nevertheless inherently reversible.
Concise Methods
Human Biopsies for Clinical Indications, Histopathology, and Diagnoses
The collection of 234 human kidney biopsies for clinical indications and eight normal nephrectomies was previously described.47 Biopsies were obtained under ultrasound guidance by spring-loaded needles (ASAP Automatic Biopsy; Microvasive, Watertown, MA). This study was approved by the University of Alberta Health Research Ethics Board (Issue #5299), the University of Illinois Chicago Office for the Protection of Research Subjects (protocol #2006-0544), and the University of Minnesota (protocol HSR#0606 M87646).
Biopsies were assessed by a pathologist (M.M.), blinded to the results of molecular studies. The total interstitial inflammation (i total score, recorded as a continuous variable48) was scored in both nonscarred and fibrotic cortical parenchyma. In addition, we assessed the interstitial fibrosis and tubular atrophy in the parenchyma. We previously described the Banff histopathologic diagnoses.47 We thus analyzed two independent sets of 28 nonrejecting biopsies each with early injury (see Results), 35 biopsies with Banff TCMR, 23 biopsies with borderline rejection, and 148 remaining biopsies representing 46 with ABMR (including 17 C4d-positive and 29 suspicious for ABMR, aka C4d-negative ABMR), 10 with mixed (ABMR/TCMR) rejection, 24 with GN, and 68 late nonrejecting biopsies. All diagnoses reflect updated donor-specific antibody status.
Protocol biopsies were taken around 6 weeks post-transplant and were described previously.14 From those, we selected a subset of biopsies that represented kidneys with a stable future function (at least 2 years of follow-up), no evidence for AKI and rejection by histology, and no clinical indication for biopsy (clinical or subclinical, before or after biopsy). Thus, this subset of the pristine protocol biopsies, 11 in total, represents histologically normal kidneys whose only injury pertains to the transplantation process. The mean eGFR in these biopsies was 51.2 compared with a mean of 48.8 in remaining protocol biopsies.
The histologic features of kidney injury assessed as the percentage of involved cortical tubules were as follows: loss of brush border, epithelium-like vacuolization, cytoplasmic lucency, and/or cell necrosis. Biopsies with ≥5% tubules showing such histologic features were labeled as having acute tubular injury by histology.
Renal Function
Renal function was defined by estimated creatinine clearance, using a four-variable Modified Diet in Renal Disease equation at the time of biopsy and 6 months later. We also calculated the change in eGFR at 6 months after the biopsy, and studied the graft losses in the AKI cohort.
RNA Extraction and Microarrays
As previously reported,49 one 18-gauge biopsy core was collected for gene expression analysis. RNA extraction, quality control, and HG_U133_Plus_2.0 GeneChip (Affymetrix Santa Clara, CA) processing were described previously.49 Detailed protocols are available in the Affymetrix Technical Manual (www.affymetrix.com).
Microarray Data Analyses
Microarray data files from 28 AKI biopsies, 11 protocol biopsies,14 and 8 nephrectomies were preprocessed using robust multichip averaging in Bioconductor and interquartile range filtering was used to eliminate probe sets with low variation across the dataset.34 The details of microarray expression data are posted on the Gene Expression Omnibus website (GSE30718).
Discovery of transcripts differentially expressed between binary phenotypes (kidneys with AKI and pristine protocol biopsies) of both the test set and the validation set was based on the Welch t test with Benjamini–Hochberg false discovery rate correction (GeneSpring 7.3; Agilent, Santa Clara CA). A corrected P value ≤0.05 was considered significant.
Summarized expression of transcripts in the IRRAT set (set score) was calculated as the geometric mean of the fold change values versus nephrectomies across all transcripts in a set.
Functional annotations of transcript sets were done using the Ingenuity Pathway Analysis Global Molecular Network. Enrichment in functional categories was tested by Fisher’s exact test. A P value <0.05 was considered significant.
Statistical Analyses
Statistical significance of differences between the groups of biopsies was tested by Kruskal–Wallis test with Dunn’s correction for multiple comparisons. The Spearman two-tailed correlation was used to analyze relationship between features and gene set scores. The chi-squared test was used for contingency tables. P values <0.05 were considered significant. AUC analysis tested the ability of transcripts to predict AKI.
Disclosures
P.F.H. holds shares in Transcriptome Sciences Inc, a company with an interest in molecular diagnostics. The other authors have no competing financial interests.
Acknowledgments
The authors thank Anna Hutton and Vido Ramassar for providing technical support and Zija Jacaj for collecting the clinical data. The authors also thank Dr. Bruce Kaplan and Dr. Arthur Matas for biopsy material.
The Alberta Transplant Applied Genomic Centre has been supported by Genome Canada, University of Alberta, University of Alberta Hospital Foundation, Alberta Advanced Education and Technology, Roche Molecular Systems, Hoffmann-La Roche Canada Ltd, Alberta Ministry of Advanced Education and Technology, Roche Organ Transplant Research Foundation (grants to B.S. and M.M.), Kidney Foundation of Canada, Stromedix Inc, and Astellas Canada. P.F.H. also holds a Canada Research Chair in Transplant Immunology and the Muttart Chair in Clinical Immunology.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2011090887/-/DCSupplemental.
References
- 1.Venkatachalam MA, Griffin KA, Lan RP, Geng H, Saikumar P, Bidani AK: Acute kidney injury: A springboard for progression in chronic kidney disease. Am J Physiol Renal Physiol 298: F1078–F1094, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharfuddin AA, Molitoris BA: Pathophysiology of ischemic acute kidney injury. Nat Rev Nephrol 7: 189–200, 2011 [DOI] [PubMed] [Google Scholar]
- 3.Devarajan P: Update on mechanisms of ischemic acute kidney injury. J Am Soc Nephrol 17: 1503–1520, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Siew ED, Ware LB, Ikizler TA: Biological markers of acute kidney injury. J Am Soc Nephrol 22: 810–820, 2011 [DOI] [PubMed] [Google Scholar]
- 5.Famulski KS, Broderick G, Einecke G, Hay K, Cruz J, Sis B, Mengel M, Halloran PF: Transcriptome analysis reveals heterogeneity in the injury response of kidney transplants. Am J Transplant 7: 2483–2495, 2007 [DOI] [PubMed] [Google Scholar]
- 6.Edemir B, Kurian SM, Eisenacher M, Lang D, Müller-Tidow C, Gabriëls G, Salomon DR, Schlatter E: Activation of counter-regulatory mechanisms in a rat renal acute rejection model. BMC Genomics 9: 71, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Einecke G, Kayser D, Vanslambrouck JM, Sis B, Reeve J, Mengel M, Famulski KS, Bailey CG, Rasko JEJ, Halloran PF: Loss of solute carriers in T cell-mediated rejection in mouse and human kidneys: An active epithelial injury-repair response. Am J Transplant 10: 2241–2251, 2010 [DOI] [PubMed] [Google Scholar]
- 8.Halloran PF, de Freitas DG, Einecke G, Famulski KS, Hidalgo LG, Mengel M, Reeve J, Sellares J, Sis B: The molecular phenotype of kidney transplants. Am J Transplant 10: 2215–2222, 2010 [DOI] [PubMed] [Google Scholar]
- 9.Westhoff JH, Schildhorn C, Jacobi C, Hömme M, Hartner A, Braun H, Kryzer C, Wang CF, von Zglinicki T, Kränzlin B, Gretz N, Melk A: Telomere shortening reduces regenerative capacity after acute kidney injury. J Am Soc Nephrol 21: 327–336, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson S, Eldadah B, Halter JB, Hazzard WR, Himmelfarb J, Horne FM, Kimmel PL, Molitoris BA, Murthy M, O’Hare AM, Schmader KE, High KP: Acute kidney injury in older adults. J Am Soc Nephrol 22: 28–38, 2011 [DOI] [PubMed] [Google Scholar]
- 11.Bunnag S, Einecke G, Reeve J, Jhangri GS, Mueller TF, Sis B, Hidalgo LG, Mengel M, Kayser D, Kaplan B, Halloran PF: Molecular correlates of renal function in kidney transplant biopsies. J Am Soc Nephrol 20: 1149–1160, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gwinner W, Hinzmann K, Erdbruegger U, Scheffner I, Broecker V, Vaske B, Kreipe H, Haller H, Schwarz A, Mengel M: Acute tubular injury in protocol biopsies of renal grafts: Prevalence, associated factors and effect on long-term function. Am J Transplant 8: 1684–1693, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Racusen L, Kashgarian M: Ischemic and toxic acute tubular injury and other ischemic renal injury. In: Heptinstall's Pathology of the Kidney, edited by Jennette JC, Olson JL, Schwartz MM, Silva FG, 6th Ed., Philadelphia, Lippincott Williams and Wilkins, 2007, pp 1139–1198 [Google Scholar]
- 14.Mengel M, Chang J, Kayser D, Gwinner W, Schwarz A, Einecke G, Broecker V, Famulski K, de Freitas DG, Guembes-Hidalgo L, Sis B, Haller H, Halloran PF: The molecular phenotype of 6-week protocol biopsies from human renal allografts: Reflections of prior injury but not future course. Am J Transplant 11: 708–718, 2011 [DOI] [PubMed] [Google Scholar]
- 15.Einecke G, Sis B, Reeve J, Mengel M, Campbell PM, Hidalgo LG, Kaplan B, Halloran PF: Antibody-mediated microcirculation injury is the major cause of late kidney transplant failure. Am J Transplant 9: 2520–2531, 2009 [DOI] [PubMed] [Google Scholar]
- 16.Sellarés J, de Freitas DG, Mengel M, Sis B, Hidalgo LG, Matas AJ, Kaplan B, Halloran PF: Inflammation lesions in kidney transplant biopsies: Association with survival is due to the underlying diseases. Am J Transplant 11: 489–499, 2011 [DOI] [PubMed] [Google Scholar]
- 17.Coca SG, Yalavarthy R, Concato J, Parikh CR: Biomarkers for the diagnosis and risk stratification of acute kidney injury: A systematic review. Kidney Int 73: 1008–1016, 2008 [DOI] [PubMed] [Google Scholar]
- 18.Vaidya VS, Ozer JS, Dieterle F, Collings FB, Ramirez V, Troth S, Muniappa N, Thudium D, Gerhold D, Holder DJ, Bobadilla NA, Marrer E, Perentes E, Cordier A, Vonderscher J, Maurer G, Goering PL, Sistare FD, Bonventre JV: Kidney injury molecule-1 outperforms traditional biomarkers of kidney injury in preclinical biomarker qualification studies. Nat Biotechnol 28: 478–485, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dieterle F, Sistare F, Goodsaid F, Papaluca M, Ozer JS, Webb CP, Baer W, Senagore A, Schipper MJ, Vonderscher J, Sultana S, Gerhold DL, Phillips JA, Maurer G, Carl K, Laurie D, Harpur E, Sonee M, Ennulat D, Holder D, Andrews-Cleavenger D, Gu YZ, Thompson KL, Goering PL, Vidal JM, Abadie E, Maciulaitis R, Jacobson-Kram D, Defelice AF, Hausner EA, Blank M, Thompson A, Harlow P, Throckmorton D, Xiao S, Xu N, Taylor W, Vamvakas S, Flamion B, Lima BS, Kasper P, Pasanen M, Prasad K, Troth S, Bounous D, Robinson-Gravatt D, Betton G, Davis MA, Akunda J, McDuffie JE, Suter L, Obert L, Guffroy M, Pinches M, Jayadev S, Blomme EA, Beushausen SA, Barlow VG, Collins N, Waring J, Honor D, Snook S, Lee J, Rossi P, Walker E, Mattes W: Renal biomarker qualification submission: A dialog between the FDA-EMEA and Predictive Safety Testing Consortium. Nat Biotechnol 28: 455–462, 2010 [DOI] [PubMed] [Google Scholar]
- 20.Perco P, Pleban C, Kainz A, Lukas A, Mayer B, Oberbauer R: Gene expression and biomarkers in renal transplant ischemia reperfusion injury. Transpl Int 20: 2–11, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Mühlberger I, Perco P, Fechete R, Mayer B, Oberbauer R: Biomarkers in renal transplantation ischemia reperfusion injury. Transplantation 88[Suppl]: S14–S19, 2009 [DOI] [PubMed] [Google Scholar]
- 22.Zhang PL, Rothblum LI, Han WK, Blasick TM, Potdar S, Bonventre JV: Kidney injury molecule-1 expression in transplant biopsies is a sensitive measure of cell injury. Kidney Int 73: 608–614, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall IE, Yarlagadda SG, Coca SG, Wang Z, Doshi M, Devarajan P, Han WK, Marcus RJ, Parikh CR: IL-18 and urinary NGAL predict dialysis and graft recovery after kidney transplantation. J Am Soc Nephrol 21: 189–197, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaidya VS, Niewczas MA, Ficociello LH, Johnson AC, Collings FB, Warram JH, Krolewski AS, Bonventre JV: Regression of microalbuminuria in type 1 diabetes is associated with lower levels of urinary tubular injury biomarkers, kidney injury molecule-1, and N-acetyl-β-D-glucosaminidase. Kidney Int 79: 464–470, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korbély R, Wilflingseder J, Perco P, Kainz A, Langer RM, Mayer B, Oberbauer R: Molecular biomarker candidates of acute kidney injury in zero-hour renal transplant needle biopsies. Transpl Int 24: 143–149, 2011 [DOI] [PubMed] [Google Scholar]
- 26.Schröppel B, Krüger B, Walsh L, Yeung M, Harris S, Garrison K, Himmelfarb J, Lerner SM, Bromberg JS, Zhang PL, Bonventre JV, Wang Z, Farris AB, Colvin RB, Murphy BT, Vella JP: Tubular expression of KIM-1 does not predict delayed function after transplantation. J Am Soc Nephrol 21: 536–542, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parikh CR, Devarajan P, Zappitelli M, Sint K, Thiessen-Philbrook H, Li S, Kim RW, Koyner JL, Coca SG, Edelstein CL, Shlipak MG, Garg AX, Krawczeski CD, TRIBE-AKI Consortium : Postoperative biomarkers predict acute kidney injury and poor outcomes after pediatric cardiac surgery. J Am Soc Nephrol 22: 1737–1747, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parikh CR, Devarajan P, Zappitelli M, Sint K, Thiessen-Philbrook H, Li S, Kim RW, Koyner JL, Coca SG, Edelstein CL, Shlipak MG, Garg AX, Krawczeski CD, Garg AX, TRIBE-AKI Consortium : Postoperative biomarkers predict acute kidney injury and poor outcomes after adult cardiac surgery. J Am Soc Nephrol 22: 1748–1757, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarjou A, Agarwal A: Sepsis and acute kidney injury. J Am Soc Nephrol 22: 999–1006, 2011 [DOI] [PubMed] [Google Scholar]
- 30.Solez K, Morel-Maroger L, Sraer JD: The morphology of “acute tubular necrosis” in man: Analysis of 57 renal biopsies and a comparison with the glycerol model. Medicine (Baltimore) 58: 362–376, 1979 [PubMed] [Google Scholar]
- 31.Avihingsanon Y, Ma N, Pavlakis M, Chon WJ, Uknis ME, Monaco AP, Ferran C, Stillman I, Schachter AD, Mottley C, Zheng XX, Strom TB: On the intraoperative molecular status of renal allografts after vascular reperfusion and clinical outcomes. J Am Soc Nephrol 16: 1542–1548, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bodonyi-Kovacs G, Putheti P, Marino M, Avihingsanon Y, Uknis ME, Monaco AP, Strom TB, Pavlakis M: Gene expression profiling of the donor kidney at the time of transplantation predicts clinical outcomes 2 years after transplantation. Hum Immunol 71: 451–455, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mas VR, Archer KJ, Yanek K, Dumur CI, Capparuccini MI, Mangino MJ, King A, Gibney EM, Fisher R, Posner M, Maluf D: Gene expression patterns in deceased donor kidneys developing delayed graft function after kidney transplantation. Transplantation 85: 626–635, 2008 [DOI] [PubMed] [Google Scholar]
- 34.Einecke G, Reeve J, Sis B, Mengel M, Hidalgo L, Famulski KS, Matas A, Kasiske B, Kaplan B, Halloran PF: A molecular classifier for predicting future graft loss in late kidney transplant biopsies. J Clin Invest 120: 1862–1872, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riss J, Khanna C, Koo S, Chandramouli GVR, Yang HH, Hu Y, Kleiner DE, Rosenwald A, Schaefer CF, Ben-Sasson SA, Yang LM, Powell J, Kane DW, Star RA, Aprelikova O, Bauer K, Vasselli JR, Maranchie JK, Kohn KW, Buetow KH, Linehan WM, Weinstein JN, Lee MP, Klausner RD, Barrett JC: Cancers as wounds that do not heal: Differences and similarities between renal regeneration/repair and renal cell carcinoma. Cancer Res 66: 7216–7224, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Schäfer M, Werner S: Cancer as an overhealing wound: An old hypothesis revisited. Nat Rev Mol Cell Biol 9: 628–638, 2008 [DOI] [PubMed] [Google Scholar]
- 37.Shimazui T, Oosterwijk-Wakka J, Akaza H, Bringuier PP, Ruijter E, Debruyne FM, Schalken JA, Oosterwijk E: Alterations in expression of cadherin-6 and E-cadherin during kidney development and in renal cell carcinoma. Eur Urol 38: 331–338, 2000 [DOI] [PubMed] [Google Scholar]
- 38.Shindo T, Kurihara H, Kuno K, Yokoyama H, Wada T, Kurihara Y, Imai T, Wang Y, Ogata M, Nishimatsu H, Moriyama N, Oh-hashi Y, Morita H, Ishikawa T, Nagai R, Yazaki Y, Matsushima K: ADAMTS-1: A metalloproteinase-disintegrin essential for normal growth, fertility, and organ morphology and function. J Clin Invest 105: 1345–1352, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng L, Pu J, Jiang G, Weng M, He J, Mei H, Hou X, Tong Q: Abnormal expression of early growth response 1 in gastric cancer: Association with tumor invasion, metastasis and heparanase transcription. Pathol Int 60: 268–277, 2010 [DOI] [PubMed] [Google Scholar]
- 40.Marshall FF: The level of cadherin-6 mRNA in peripheral blood is associated with the site of metastasis and with the subsequent occurrence of metastases in renal cell carcinoma. J Urol 173: 1919, 2005 [PubMed] [Google Scholar]
- 41.Hahm K, Lukashev ME, Luo Y, Yang WJ, Dolinski BM, Weinreb PH, Simon KJ, Chun Wang L, Leone DR, Lobb RR, McCrann DJ, Allaire NE, Horan GS, Fogo A, Kalluri R, Shield CF, 3rd, Sheppard D, Gardner HA, Violette SM: Alphav beta6 integrin regulates renal fibrosis and inflammation in Alport mouse. Am J Pathol 170: 110–125, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trevillian P, Paul H, Millar E, Hibberd A, Agrez MV: alpha(v)beta(6) Integrin expression in diseased and transplanted kidneys. Kidney Int 66: 1423–1433, 2004 [DOI] [PubMed] [Google Scholar]
- 43.Choy BY, Chan TM, Lai KN: Recurrent glomerulonephritis after kidney transplantation. Am J Transplant 6: 2535–2542, 2006 [DOI] [PubMed] [Google Scholar]
- 44.Halloran PF, Hunsicker LG: Delayed graft function: State of the art, November 10-11, 2000. Summit meeting, Scottsdale, Arizona, USA. Am J Transplant 1: 115–120, 2001 [PubMed] [Google Scholar]
- 45.Coca SG: Long-term outcomes of acute kidney injury. Curr Opin Nephrol Hypertens 19: 266–272, 2010 [DOI] [PubMed] [Google Scholar]
- 46.Terasaki PI, Gjertson DW, Cecka JM, Takemoto S, Cho YW: Significance of the donor age effect on kidney transplants. Clin Transplant 11: 366–372, 1997 [PubMed] [Google Scholar]
- 47.Famulski KS, Einecke G, Sis B, Mengel M, Hidalgo LG, Kaplan B, Halloran PF: Defining the canonical form of T-cell-mediated rejection in human kidney transplants. Am J Transplant 10: 810–820, 2010 [DOI] [PubMed] [Google Scholar]
- 48.Mengel M, Reeve J, Bunnag S, Einecke G, Jhangri GS, Sis B, Famulski K, Guembes-Hidalgo L, Halloran PF: Scoring total inflammation is superior to the current Banff inflammation score in predicting outcome and the degree of molecular disturbance in renal allografts. Am J Transplant 9: 1859–1867, 2009 [DOI] [PubMed] [Google Scholar]
- 49.Mueller TF, Einecke G, Reeve J, Sis B, Mengel M, Jhangri GS, Bunnag S, Cruz J, Wishart D, Meng C, Broderick G, Kaplan B, Halloran PF: Microarray analysis of rejection in human kidney transplants using pathogenesis-based transcript sets. Am J Transplant 7: 2712–2722, 2007 [DOI] [PubMed] [Google Scholar]