Abstract
In resource-limited settings, there is increased demand for human immunodeficiency virus type 1 drug resistance testing. Because preservation of plasma specimens is often not feasible in resource-limited settings, use of dried blood spots (DBSs) is being adopted. We used 2 panels of DBSs for genotyping assay validation and proficiency testing in selected laboratories in the World Health Organization laboratory network in 14 countries. An amplification sensitivity of 1000 copies/mL was achieved by 2 laboratories. Reproducibility and accuracy of nucleotide sequence determination and resistance-associated mutation identification from DBSs was similar to that previously determined for plasma. International shipping at ambient temperature had no significant effect on amplification success. These studies indicate that DBS-based genotyping is equally reproducible and reliable, although slightly less sensitive, compared with plasma.
Development and transmission of drug-resistant human immunodeficiency virus (HIV) in resource-limited settings is a potential negative consequence of the international effort to provide antiretroviral treatment to millions of persons living with HIV infection [1]. HIV drug resistance (HIVDR) is associated with increased risk of therapeutic failures; transmission of drug-resistant virus; and decreased therapeutic options, treatment program effectiveness, and survival. The World Health Organization (WHO) global strategy for prevention and assessment of HIVDR [2] is a coordinated effort to assess HIVDR worldwide. The laboratory testing component of this strategy promotes the generation of accurate and reproducible genotyping data from countries implementing HIVDR surveillance [3].
HIVDR genotyping is usually performed using cryopreserved plasma as the specimen type. However, preservation and transportation of frozen plasma specimens is often not feasible in resource-limited settings. Dried blood spots (DBS) have been proposed as an alternative [4, 5], although there are uncertainties regarding genotyping assay method validation and standardization and there is a lack of international external quality-assurance programs using DBSs. To assess reproducibility, accuracy, and sensitivity of in-house genotyping assays, 2 DBS specimen panels (a validation panel and a proficiency panel) were distributed for testing to several WHO/HIVResNet laboratories.
METHODS
DBS Validation Panel
To enable testing of multiple replicates of the same specimen in several laboratories, a large number of DBS cards were prepared under ideal conditions at 2 sites. For sensitivity testing, serial dilutions of cell-free HIV (HXB2, subtype B) were prepared using whole blood from an HIV-negative donor to achieve final target viral loads (VLs) of 100–1 000 000 RNA copies/mL. To achieve a similar range in HIV proviral DNA concentrations, U1 cells, derived from the U937 promonocyte cell line chronically infected with subtype B HIV [6], were used to prepare serial dilutions in whole blood. Blood was spotted (50 μL per spot, 5 spots per card) on Whatman 903 filter paper and left to dry overnight in a biosafety cabinet. DBS cards were packaged with desiccant in zip-top plastic bags and stored at −80°C until shipping to 10 different genotyping laboratories. Each laboratory tested 2 (100000 copies/mL and 1000000 copies/mL) to 4 (all others) replicates of the dilution series (Table 1). The majority of laboratories used 2 spots per extraction. The sensitivity of each assay was defined as the lowest VL at which at least half of the replicates yielded sufficient polymerase chain reaction (PCR) product for full sequencing of protease (PR) and reverse transcriptase (RT).
Table 1.
Validation Panel Sensitivity Specimens and Amplification Results
| Amplification Results (No. Positive/No. of Replicates) |
|||||||
| Viral Load |
RNA |
DNA |
|||||
| Copies/mL | Copies/DBS | Min | Max | No. of laboratories ≥ 50% | Min | Max | No. of laboratories ≥ 50% |
| 1 000 000 | 50 000 | 1/2 | 2/2 | 10 | 1/2 | 2/2 | 10 |
| 100 000 | 5000 | 1/2 | 2/2 | 10 | 1/2 | 2/2 | 10 |
| 10 000 | 500 | 0/4 | 4/4 | 7 | 0/4 | 4/4 | 9 |
| 1000 | 50 | 0/4 | 3/4 | 2 | 0/4 | 4/4 | 4 |
| 100 | 5 | 0/4 | 1/4 | 0 | 0/4 | 2/4 | 1 |
| 0 | 0 | 0/4 | 0/4 | 0 | 0/4 | 0/4 | 0 |
Abbreviation: DBS, dried blood spot.
For assay reproducibility and accuracy testing, blood specimens were collected using venipuncture with ethylenediaminetetraacetic acid as an anticoagulant from 6 HIV-infected donors. Informed consent was obtained from all donors in accordance with local institutional ethics guidelines. Plasma specimens were prepared for VL testing, and DBS cards (20–50 per patient) were prepared from the whole blood specimens and stored as described above. VL was determined using the COBAS Amplicor Monitor assay (Roche), and genotyping was performed using an in-house method. Specimens with VL >1000 copies/mL from multiple subtypes and with a variety of drug resistance–associated mutations (DRMs) were selected for the reproducibility testing. Each laboratory tested 5 replicates of each specimen from nucleic acid extraction (2 spots each) through assay completion.
Two DBS cards from each panel (sensitivity and reproducibility) were shipped from the Netherlands, desiccated, and frozen on dry ice, to participating testing laboratories located in the following countries: Australia, Canada, France (2 laboratories), Puerto Rico, Spain, United Kingdom, United States, and South Africa. After receipt, DBS were stored at −80°C until testing. In 1 case (laboratory 5), the shipment arrived thawed as a result of delays in customs clearance.
DBS Proficiency Panel
A proficiency panel was developed in collaboration with National Institutes of Health through the National Institute of Allergy and Infectious Diseases Virology Quality Assurance program and was modeled after similar panels made using plasma [7]. The panel was composed of 5 clinical specimens, with 2 of the specimens being represented twice, at different VLs, resulting in a 7-specimen proficiency panel with VLs of 1700–20 000 copies/mL.
Specimens were prepared by dilution of existing plasma from previous Virology Quality Assurance program genotyping panels into HIV-negative whole blood and spotting on Whatman 903 filter paper, as described above, except that 4 spots per card were made. Viral loads were verified using plasma prepared from the diluted virus, in triplicate, using the COBAS Amplicor Monitor assay (Roche).
DBS cards, packaged in separate glassine bags and with desiccant and humidity indicators, were shipped from Chicago, Illinois, to 17 laboratories located in 14 countries: Australia (2), Cameroon, Canada, France (2), India, Kenya, Martinique, the Netherlands, Puerto Rico, Senegal, South Africa (2), Spain, United Kingdom, and United States. One set of DBS cards was shipped on dry ice, and a second set was shipped at ambient temperature. After arrival in the testing laboratory, all DBS cards were stored at −80°C until processing. One laboratory had technical issues and could not generate quality sequence data; only the amplification results are included for this laboratory.
Genotyping
Specimens were tested primarily using a variety of in-house methods. For the proficiency panel, 2 laboratories (1 and 2) used the TruGene (Siemens) assay. All in-house methods rely on RNA extraction using silica-based chemistry, reverse transcription-PCR amplification, and bulk PCR product automated sequencing of PR (at least codons 10–99) and RT (at least codons 41–236); manual editing of sequence data; and generation of a contiguous PR-RT sequence.
Sequence Analysis
All sequence analysis was limited to codons 10–99 in PR and 38–240 in RT. Consensus sequences for each specimen were generated on the basis of >80% concordance from alignments of sequence results from all participating laboratories. If 80% concordance was not achieved at a given position, an “N” was inserted in the consensus sequence and this position was ignored during the scoring. DRM codons containing the ambiguous base were also ignored.
Sequence reproducibility was measured by comparing results from each replicate with all other replicates for that specimen from each laboratory. The mean percentage nucleotide identity from all pairwise comparisons (10 comparisons if all 5 replicates were successful) was then calculated. Mixed vs unmixed bases were counted as discrepancies. Because of differences in the precise boundaries of regions sequenced in the different laboratories, the length of sequences was variable among them. If <5 replicates were available for a given specimen, the mean identity score was calculated using fewer pairwise comparisons. No penalty was assigned for missing sequence.
Sequence accuracy was assessed by calculating an overall sequence identity score, defined as the percentage of matching nucleotides of the total number of unambiguous nucleotides in the consensus, and concordance at DRM codons as the percentage of concordant nucleotides in DRM codons, as defined by International Antiviral Society-USA [8], of the total number of nucleotides in DRM codons not containing an ambiguity in the consensus [7, 9]. Mixed vs unmixed bases were counted as discrepancies for the nucleotide sequence alignment score. For the DRM site score, if a mutant codon was present in the consensus and included in the reported sequence, regardless of being mixed or not, it was not counted as a discrepancy. Missing sequence in the analyzed region was counted as an error.
RESULTS
Validation Panel: Sensitivity
The lowest VL required for reproducible PCR amplification of PR and RT from DBSs was assessed using serial dilutions of a homogeneous and well-characterized subtype B HIV grown in cell culture (Table 1). Nine of the 10 laboratories were able to amplify all replicates of specimens with VL ≥100 000 copies/mL; no false-positive amplifications were observed. At a VL of 10 000 RNA copies/mL, which corresponds to 500 copies per DBS, 7 of 10 laboratories amplified at least 2 of the 4 replicates (6 laboratories amplified all 4; 1 amplified only 2 of 4); at a VL of 1000 RNA copies/mL, only 2 of 10 laboratories amplified at least 2 of the 4 replicates (2 or 3 of 4). Amplification results from the corresponding DNA panel were similar, with 2 additional laboratories being able to amplify at least half the replicates at a VL of 1000 copies/mL (Table 1).
Validation Panel: Reproducibility and Accuracy
Reproducibility of nucleotide sequences generated from DBSs was assessed through replicate testing (target of 5 replicates per specimen) of clinical specimens freshly prepared on DBS in a hospital laboratory setting. Six patient specimens were identified, representing 4 subtypes and a variety of resistance patterns (Table 2). Two of the specimens (VP4 and VP5) contained >2% mixed bases, which has been demonstrated to influence sequence reproducibility as a result of variation in sequence data quality and subjectivity in the base-calling process [9–11]. With few exceptions, all laboratories were able to amplify all 5 replicates of specimens VP1–VP5, but only laboratory 2 could amplify VP6, with a VL of 1812 copies/mL.
Table 2.
Validation Panel Reproducibility Specimens and Amplification Results
| Amplification Results (No. of Positive/No. of Replicates) |
||||||||
| Spec ID | Subtype | Viral Load, copies/mL | PR RAMs | RT RAMs | Mixed Bases, %a | Min | Max | No. of laboratories ≥80% |
| VP1 | CRF02 | 143 475 | L10V, L33V, L89M | None | 1.0 | 3/5 | 5/5 | 9/10 |
| VP2 | G | 105 264 | V82I, L89M | K103N | 0.5 | 4/5 | 5/5 | 10/10 |
| VP3 | B | 5588 | L10I | Y181C | 1.6 | 4/5 | 5/5 | 10/10 |
| VP4 | CRF02 | 158 858 | L89M | None | 3.7 | 5/5 | 5/5 | 9/9b |
| VP5 | C | 10 927 | L89M | None | 2.8 | 4/5 | 5/5 | 9/9 |
| VP6 | B | 1812 | None | M41L, D67N, L74I, V90I, K103N, Y181C, M184I/V, L210W, T215Y | 0.05 | 0/5 | 5/5 | 1/9 |
Abbreviations: PR, protease; RAMs, resistance-associated mutations (as defined by the IAS-USA [8]); RT, reverse transcriptase.
Percentage of base calls other than A, C, G, or T in the consensus sequence from all laboratories.
One laboratory did not generate results for VP4-VP6 because of a processing error.
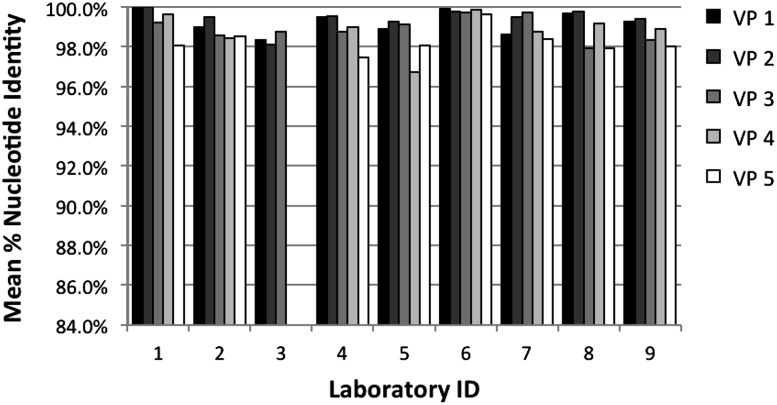
In laboratory 3, a handling error prevented completion of sequencing for specimens VP4 and VP5. In another laboratory, technical problems prevented the generation of quality sequences; only amplification results are included from this laboratory. Overall sequence data from at least 3 replicates were available for specimens VP1 to VP5 for 9 of the 10 laboratories. Sequences from each replicate were compared with those from all other replicates generated by the same laboratory. Thus, if 5 replicates were completed, there were 10 possible pairwise comparisons. The mean percentage identity scores for all such comparisons for each specimen and each laboratory are shown in Figure 1. Mean sequence identity ranged from 96.7% to 100%; 39 of the 43 results were ≥98%, and 22 were ≥99%. Mean scores across all specimens were highest for laboratory 1 (99.4%) and 6 (99.8%), and mean scores across laboratories were lowest for specimen VP4 (98.8%) and VP5 (98.3%), which were also the 2 specimens with the most mixed bases.
Figure 1.
Dried blood spot validation panel sequence reproducibility. Mean percentages of nucleotide identity from intralaboratory pairwise comparisons are shown for each specimen in bars of different shading. Laboratory 3 did not generate results for VP4 or VP5.
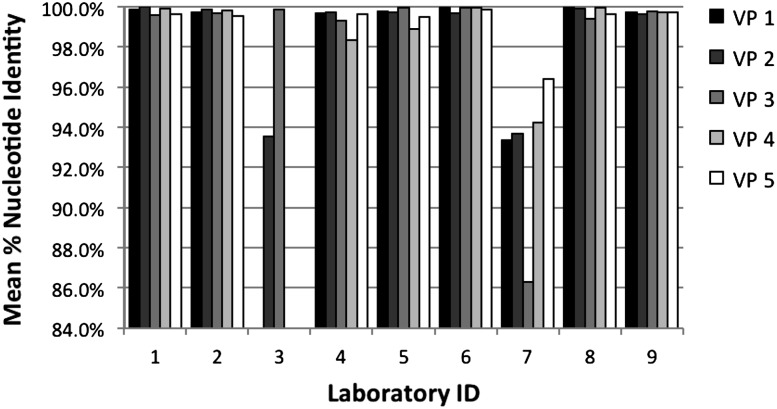
To assess sequence accuracy, individual results were compared with a consensus generated from sequences from all laboratories. The mean percentage identity for all replicates of each specimen is shown in Figure 2. Scores for laboratory 3 and 7 are low because of missing sequences. For the remaining 8 laboratories, scores ranged from 98.4% to 100%, and 33 of 35 results were ≥99%. Mean scores across all specimens were highest for laboratory 1 and 8 (99.8%) and 6 (99.9%). In addition to evaluation of the entire sequence, accuracy at the codons where DRMs have been recognized was analyzed separately; results were similar to those for the entire sequence (mean, 99.7%; range, 98.4%–100%; data not shown). Laboratories 2 and 6 achieved a score ≥99.5% for both the whole sequence and the DRM sites for all 5 specimens.
Figure 2.
Dried blood spot validation panel sequence accuracy. Mean percentages of nucleotide identity from comparison with the cross-laboratory consensus sequences are shown for each specimen in bars of different shading. Laboratory 3 did not generate results for VP4 or VP5. Sequences from laboratory 7 contained gaps in the required minimum region (see Methods).
Proficiency Panel Amplification
A blinded panel of 7 specimens was prepared and shipped to 17 laboratories around the world, both frozen on dry ice and at ambient temperature (see Methods). Specimen characteristics and the number of laboratories that successfully amplified both PR and RT are shown in Table 3. When the VL was >5000 copies/mL (ie, excluding specimens PP4 and PP7), 2–5 of the 17 laboratories were not able to amplify PR and RT when DBSs were shipped frozen, compared with 3–7 when shipped at ambient temperature. Both PR and RT were amplified by more laboratories when DBSs were shipped frozen, compared with when they were shipped at ambient temperature for 2 specimens (PP1 and PP4), whereas the opposite was true for 4 specimens (PP3, 5, 6, and 7). In paired comparisons for each individual specimen, amplification was successful from the DBSs shipped frozen, whereas it was not for the corresponding DBSs shipped at ambient temperature in 6 laboratories; the opposite was true for 6 laboratories.
Table 3.
Proficiency Panel Specimens and Amplification Results
| Amplification Results (No. Positive/No. Attempted) |
|||||||
| Spec ID | Subtype | Viral Load, copies/mL | PR RAMs | RT RAMs | Mixed Bases, %a | Frozen | Ambient |
| PP1 | F | 5963 | M36I, L89M | None | 0.0 | 12/17 | 10/17 |
| PP2 | B | 5410 | M36L | K103N, M184 M/V | 1.8 | 14/16 | 14/17 |
| PP3 | C | 19 500 | M36I | M41L, M184V, T215Y | 0.1 | 13/17 | 14/17 |
| PP4 | B | 1697 | L10I, L23I, L33F, M46L, I54V, L63P, A71I, V77I, V82A L90M | M41L, D67N, L74V, L100I, K103N, L210W, T215Y | 1.1 | 11/17 | 10/17 |
| PP5 | B | 7367 | L63P | K103N, Y188C, P225H | 1.8 | 13/17 | 14/17 |
| PP6 | B | 10 857 | L10I, L23I, L33F, M46L, I54V, L63P, A71I, V77I, V82A, L90M | M41L, D67N, L74V, L100I, K103N, L210W, T215Y | 1.1 | 13/17 | 14/17 |
| PP7 | C | 2667 | M36I | M41L, M184V, T215Y | 0.1 | 8/17 | 9/17 |
Abbreviations: PR, protease; RAMs, resistance-associated mutations (as defined by the IAS-USA [8]); RT, reverse transcriptase.
Percentage of base calls other than A, C, G, or T in the consensus sequence from all laboratories.
Proficiency Panel Accuracy
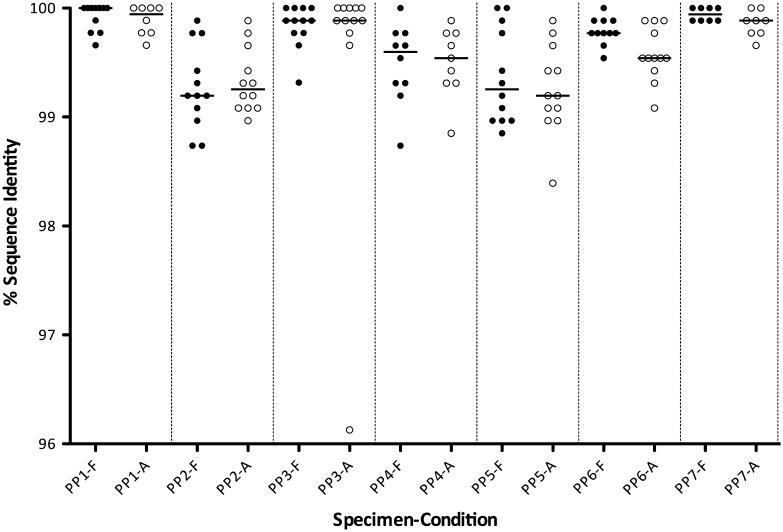
Sequence data from the proficiency panel were evaluated by comparison with a cross-laboratory consensus sequence from the corresponding plasma specimens as described in Methods. Nucleotide sequence identity scores for each specimen from the DBSs shipped frozen or at ambient temperature for each laboratory amplifying PR and RT for at least 7 of the possible 14 sequences are shown in Figure 3. Overall, scores were lowest for specimens PP2 and PP5, which were also the specimens with the most mixtures. Mean identity scores (excluding the specimens not amplified) ranged from 99.4% to 99.8% for DBS shipped frozen and from 98.8% to 99.8% for DBS shipped at ambient temperature. Paired comparisons between scores for DBS shipped frozen vs at ambient temperature were not statistically significant for any specimen except PP6 (Wilcoxon matched-pairs signed-rank test, P = .02).
Figure 3.
Dried blood spot (DBS) proficiency panel accuracy results. Each point represents the nucleotide identity score, compared with the consensus sequences for DBS that were shipped frozen on dry ice (F; filled circles) or at ambient temperature (A; open circles). Horizontal bars for each group represent the median values.
Accuracy based on DRM site scores ranged from 98.1% to 100% for DBSs shipped frozen and from 98.5% to 100% for DBSs shipped at ambient temperature. Paired comparisons between DRM site scores for DBSs shipped frozen vs at ambient temperature were not statistically significant for any specimen.
DISCUSSION
This study describes, to our knowledge, the first large-scale, multilaboratory, international comparison of HIVDR genotyping assay performance using DBS. Results provide confidence that DBS-based genotyping is similarly reproducible and reliable, although less sensitive, compared with plasma-based genotyping [10]. Reduced sensitivity is most likely attributable to reduced template copy number input for DBS, compared with plasma.
One of the goals of the DBS validation panel was to generate experimental data on which to base a recommended DBS-based genotyping protocol that could be used by all laboratories performing testing for WHO HIVDR surveys. Key variables of each assay were compared and analyzed in view of the sensitivity and reproducibility results presented here. Variables included were use of nested PCR, number (1 or 2) of PCR amplicons, amplicon length, RNA extraction chemistry or kit type, automated or manual RNA extraction, RNA extraction input (plasma volume equivalent, based on blood volume per DBS and assuming hematocrit of 50%), RT reaction input (fraction of RNA extracted), PCR input (fraction of RT reaction products), second-round PCR input (if nested, fraction of first-round PCR), RT and PCR enzyme, magnesium ion and deoxyribonucleotide triphosphate concentration in RT and PCR, number of PCR cycles, PCR product detection method, type of PCR machine (make and model), and RT and PCR primer length and sequence. However, no single variable could be clearly associated with better assay performance, most likely because of the multifactorial influence of >1 variable. Nonetheless, on the basis of the common use of several variables by all laboratories with good performance, some themes can be identified. These include use of a nested PCR amplification strategy, silica-based nucleic acid purification, a preference for manual vs automated extraction procedure, use of a 1-tube RT-PCR strategy (ie, all of the RT reaction is used for first-round PCR), and 35–40 cycles of PCR at each round. The specific procedures used by the 2 laboratories with the best performance using the validation panel that also gave good results in the proficiency panel will be made available through the WHO laboratory network for laboratories that are developing the capacity to use DBSs for genotyping; these procedures will complement the consensus protocol already available [12].
Assessment of assay performance based on the proficiency panel results was largely influenced by sensitivity (ability to amplify PR-RT for sequencing), because sequence accuracy per se was very similar to previous results obtained using plasma specimens and was heavily influenced by the degree of heterogeneity of the virus specimen used [10]. Thus, efforts to optimize assay success when using DBSs as the specimen type in laboratories already proficient at performing genotyping using plasma should be focused on this aspect of the procedure. For laboratories with no experience handling DBSs that would like to develop this capacity, WHO recommends starting with one of the protocols shown to work well through the studies described here, which are available on request.
An important caveat to conclusions based on testing of DBSs prepared by diluting cell-free virus in blood from HIV-negative donors is that sensitivity limits may be underestimated because of the lack of proviral DNA in the cellular compartment [13]. However, because the RNA component is less stable during extended storage times under suboptimal temperature or humidity conditions [4, 14], results using DBS panels that are prepared, stored, and shipped under ideal conditions may overestimate sensitivity that may be expected during implementation of HIVDR surveys in many rural settings when DBSs are prepared from HIV-infected patient anticoagulated blood or directly without venipuncture. In the present study, the possible contribution of proviral DNA to the sequence results in clinical whole blood specimens was not studied.
In the present study, international shipment at ambient temperature did not affect amplification success rates in a consistently detrimental manner. The differences observed when comparing amplification rates for specimens shipped on dry ice vs at ambient temperature were small and inconsistent, and they sometimes favored ambient shipment. However, the interpretation of the amplification success data presented here is tempered by the small number of specimens tested with VL in a range that would be expected to be on the verge of being associated with amplification failure when handled under extreme conditions and by differences among the shipping conditions (distance, time, and actual temperature) to each laboratory. Larger field studies designed to rigorously test the effect of international shipment of clinical specimens at ambient temperature are under way.
Assay validation and proficiency testing are necessary but not sufficient for WHO accreditation of a laboratory to perform genotyping using DBSs. To obtain WHO accreditation for DBS testing, a laboratory must fulfill the following criteria: existing accreditation by WHO for the performance of HIVDR genotyping of plasma specimens at least 6 months of experience in DBS-based genotyping; at least 100 DBS specimens successfully amplified and sequenced, with a success rate of at least 80% when VL is >5000 copies/mL and DBSs have been properly handled (ie, according to WHO recommendations for preparation, storage, and shipping); successful testing of a WHO-recognized proficiency panel consisting of DBS specimens; successful validation of a DBS-based in-house assay for genotyping using the standardized criteria described elsewhere [12]; and submission to WHO of a report summarizing the aforementioned elements for review.
WHO surveys of acquired HIVDR require genotyping to be performed on specimens with VL (in plasma) of ≥1000 copies/mL; it is a goal of the laboratory network to implement methods that will enable amplification of DBS specimens with VLs in this range. The laboratories with the best performance described here provide optimism that this is achievable.
Notes
Acknowledgments.
We thank Joe Fitzgibbon for his technical input and advice, and Salvatore Scianna, Brian Harty, Suzanne Granger, and Derek Weibel for assistance with proficiency panel preparation and data management.
Disclaimer.
The conclusions and opinions expressed in this article are those of the authors and do not reflect those of the World Health Organization.
Financial support.
This work was supported by The Bill & Melinda Gates Foundation (to the WHO) and the National Institutes of Health, National Institute of Allergy and Infectious Disease, Division of AIDS (NO1-AI-5004 to the Virology Quality Assurance program. Grant no. NIAID K23 AI074423-05 to MRJ).
Supplement sponsorship.
This article was published as part of a supplement entitled “The World Health Organization HIV Drug Resistance Prevention and Assessment Strategy: Global, Regional, and Country Progress,” sponsored by The Bill & Melinda Gates Foundation (38180).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Appendix. WHO DBS Genotyping Working Group:
James Brooks and Paul Sandstrom, Public Health Agency of Canada, Ottawa; Nicholas Wagar and Chunfu Yang, CDC, Atlanta, Georgia; Fatima Mouacha and Martine Peeters, IRD and UMI, Montpellier, France; Patricia Pinson and Herve Fleury, Laboratoire de Virologie, CHU, Bordeaux, France; Anna Hearps and Suzanne Crowe, Burnet Institute for Medical Research and Public Health, Melbourne, Australia; Yasuhiro Yamamura, AIDS Research Program–Immunology Reference Laboratory, Ponce, Puerto Rico; Pat Cane, Health Protection Agency, London, United Kingdom; Johanna Ledwaba and Lynn Morris, National Institute for Communicable Diseases, Johannesburg, South Africa; Philip Cunningham, NSW State Reference Laboratory for HIV and Molecular Diagnostic Medicine, Sydney, Australia; Avelin Aghokeng, IMPM-IRD/CREMER, Yaoundé, Cameroon; Srikanth Tripathy and R. S. Paranjape, National AIDS Research Institute, Pune, India; Clement Zeh, KEMRI/CDC HIV Research Laboratory, Kisumu, Kenya; Georges Dos Santos, Centre Hospitalier et Universitaire de Fort-de-France, Martinique; Wendy Stevens, CLS Genotyping Laboratory, Johannesburg, South Africa; Coumba Toure Kane, Bacteriology-Virology UTH A Le Dantec, Dakar, Senegal.
References
- 1.World Health Organization. Joint United Nations Program on HIV/AIDS, UNICEF. Towards universal access: scaling up priority HIV/AIDS interventions in the health sector. Geneva, Switzerland: World Health Organization; 2010. Available at: http://www.who.int/hiv/pub/2010progressreport/en/index.html. Accessed 5 July 2011. [Google Scholar]
- 2.Bennett DE, Bertagnolio S, Sutherland D, Gilks CF. The World Health Organization’s global strategy for prevention and assessment of HIV drug resistance. Antivir Ther. 2008;13(Suppl 2):1–13. [PubMed] [Google Scholar]
- 3.Bertagnolio S, Derdelinckx I, Parker M, et al. World Health Organization/HIVResNet drug resistance laboratory strategy. Antivir Ther. 2008;13(Suppl 2):49–57. [PubMed] [Google Scholar]
- 4.Bertagnolio S, Parkin NT, Jordan M, Brooks J, Garcia-Lerma JG. Dried blood spots for HIV-1 drug resistance and viral load testing: a review of current knowledge and WHO efforts for global HIV drug resistance surveillance. AIDS Rev. 2010;12:195–208. [PubMed] [Google Scholar]
- 5.Hamers RL, Smit PW, Stevens W, Schuurman R, Rinke de Wit TF. Dried fluid spots for HIV type-1 viral load and resistance genotyping: a systematic review. Antivir Ther. 2009;14:619–29. [PubMed] [Google Scholar]
- 6.Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science. 1987;238:800–2. doi: 10.1126/science.3313729. [DOI] [PubMed] [Google Scholar]
- 7.Huang DD, Bremer JW, Brambilla DJ, et al. Model for assessment of proficiency of human immunodeficiency virus type 1 sequencing-based genotypic antiretroviral assays. J Clin Microbiol. 2005;43:3963–70. doi: 10.1128/JCM.43.8.3963-3970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson VA, Brun-Vezinet F, Clotet B, et al. Update of the drug resistance mutations in HIV-1: December 2010. Top HIV Med. 2010;18:156–63. [PubMed] [Google Scholar]
- 9.Huang DD, Eshleman SH, Brambilla DJ, Palumbo PE, Bremer JW. Evaluation of the editing process in human immunodeficiency virus type 1 genotyping. J Clin Microbiol. 2003;41:3265–72. doi: 10.1128/JCM.41.7.3265-3272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkin N, Bremer J, Bertagnolio S. Genotyping external quality assurance in the World Health Organization HIV drug resistance laboratory network during 2007–2010. Clin Infect Dis. 2012;54(Suppl 4):S266–72. doi: 10.1093/cid/cir992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galli RA, Sattha B, Wynhoven B, O’Shaughnessy MV, Harrigan PR. Sources and magnitude of intralaboratory variability in a sequence-based genotypic assay for human immunodeficiency virus type 1 drug resistance. J Clin Microbiol. 2003;41:2900–7. doi: 10.1128/JCM.41.7.2900-2907.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization. WHO manual for HIV drug resistance testing using dried blood spot specimens. 2010. Available at: http://www.who.int/entity/hiv/topics/drugresistance/dbs_protocol.pdf. Accessed 15 December 2010. [Google Scholar]
- 13.McNulty A, Jennings C, Bennett D, et al. Evaluation of dried blood spots for human immunodeficiency virus type 1 drug resistance testing. J Clin Microbiol. 2007;45:517–21. doi: 10.1128/JCM.02016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monleau M, Montavon C, Laurent C, et al. Evaluation of different RNA extraction methods and storage conditions of dried plasma or blood spots for human immunodeficiency virus type 1 RNA quantification and PCR amplification for drug resistance testing. J Clin Microbiol. 2009;47:1107–18. doi: 10.1128/JCM.02255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]